Investigating Genetic Diversity and Population Structure in Rice Breeding from Association Mapping of 116 Accessions Using 64 Polymorphic SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. SSR Genotyping and Data Analysis
2.3. Genetic Variability
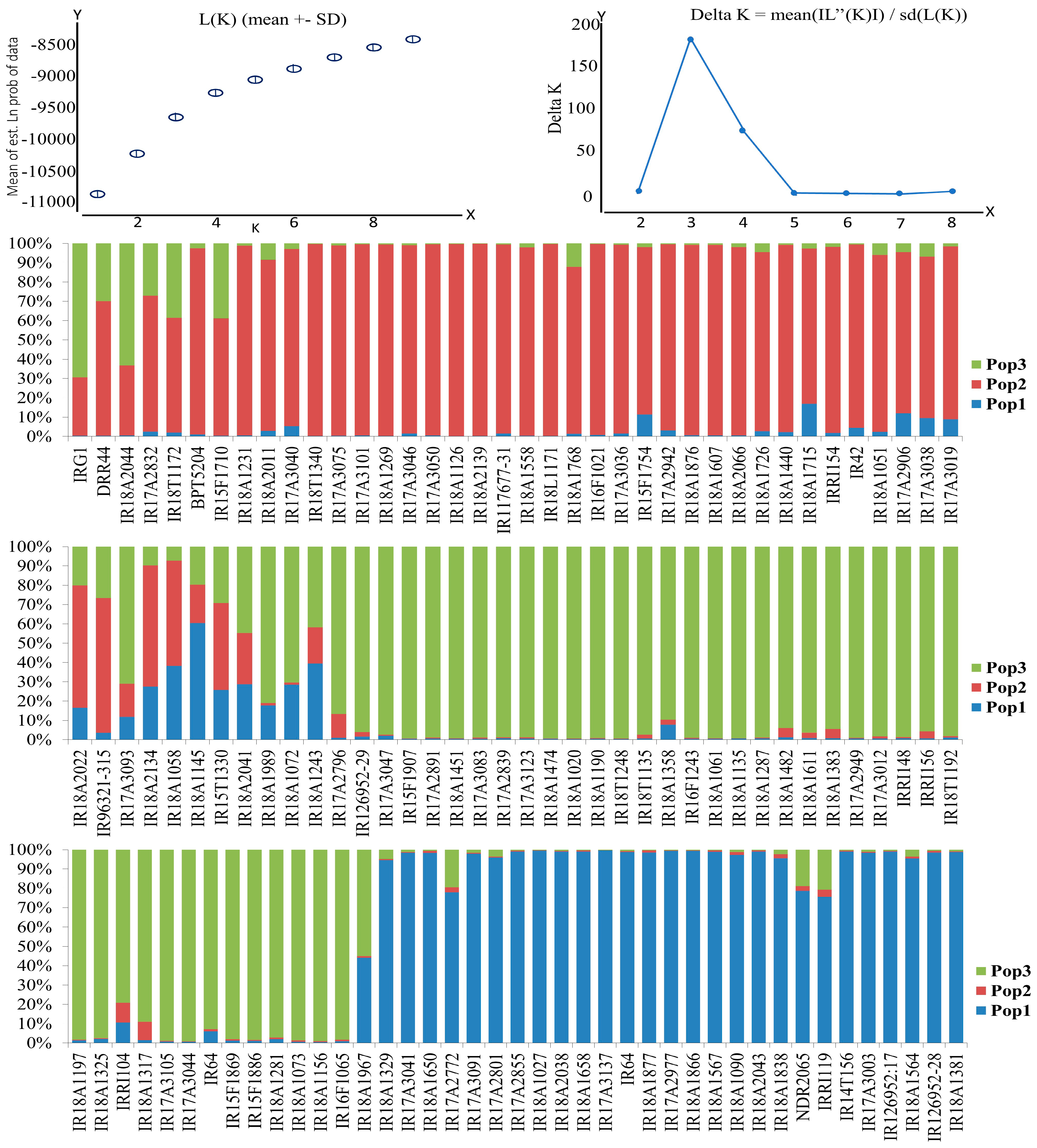
2.4. Structure Analysis
3. Results
3.1. Allelic Diversity and SSR Marker Informativeness
3.2. Chromosomal Distribution and Molecular Weight Analysis of SSR Markers
3.3. Genetic Variability
3.4. Distinct Subgroup Identification through Population Structure
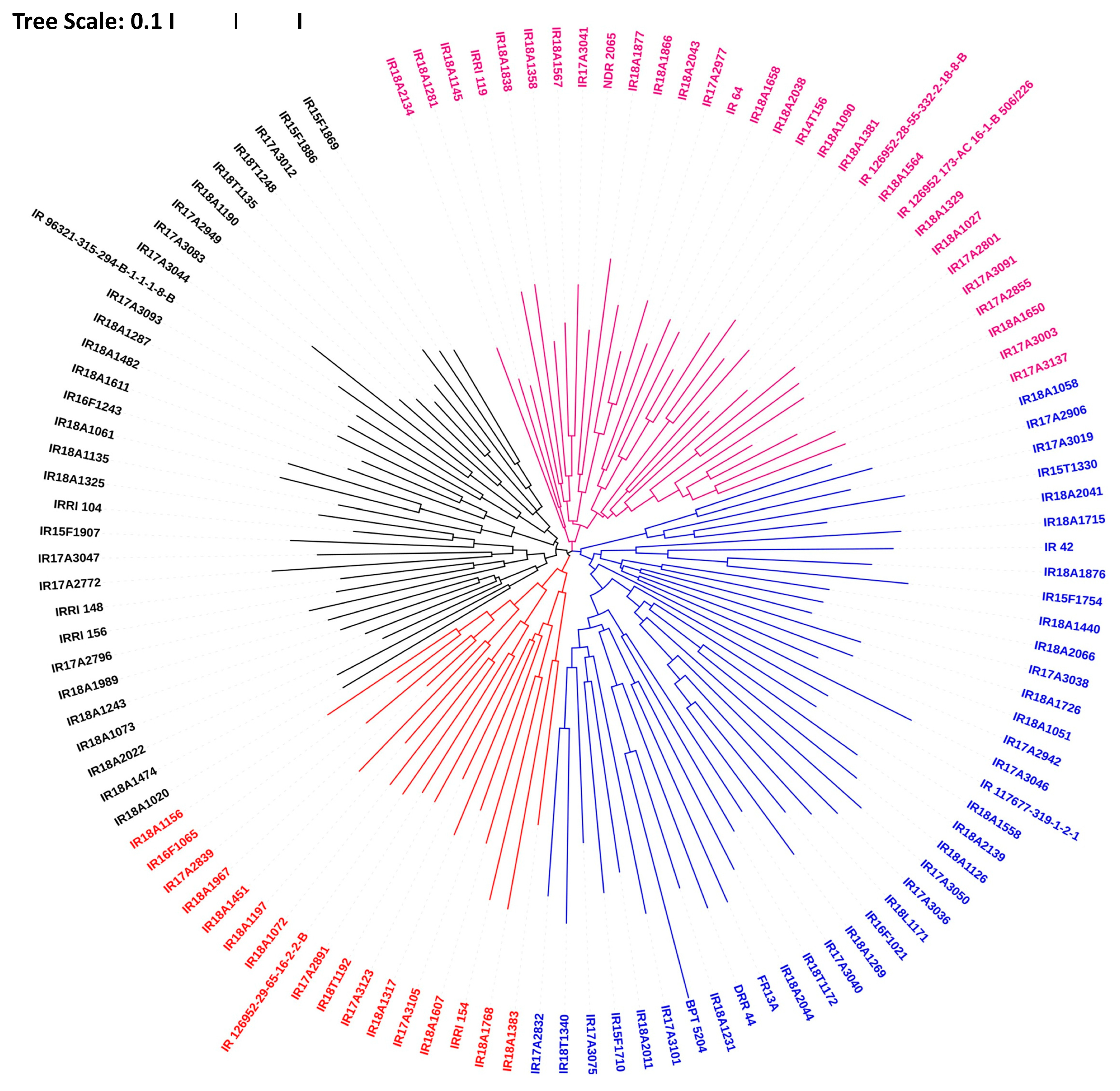
3.5. Genetic Relatedness and Diversity Assessment
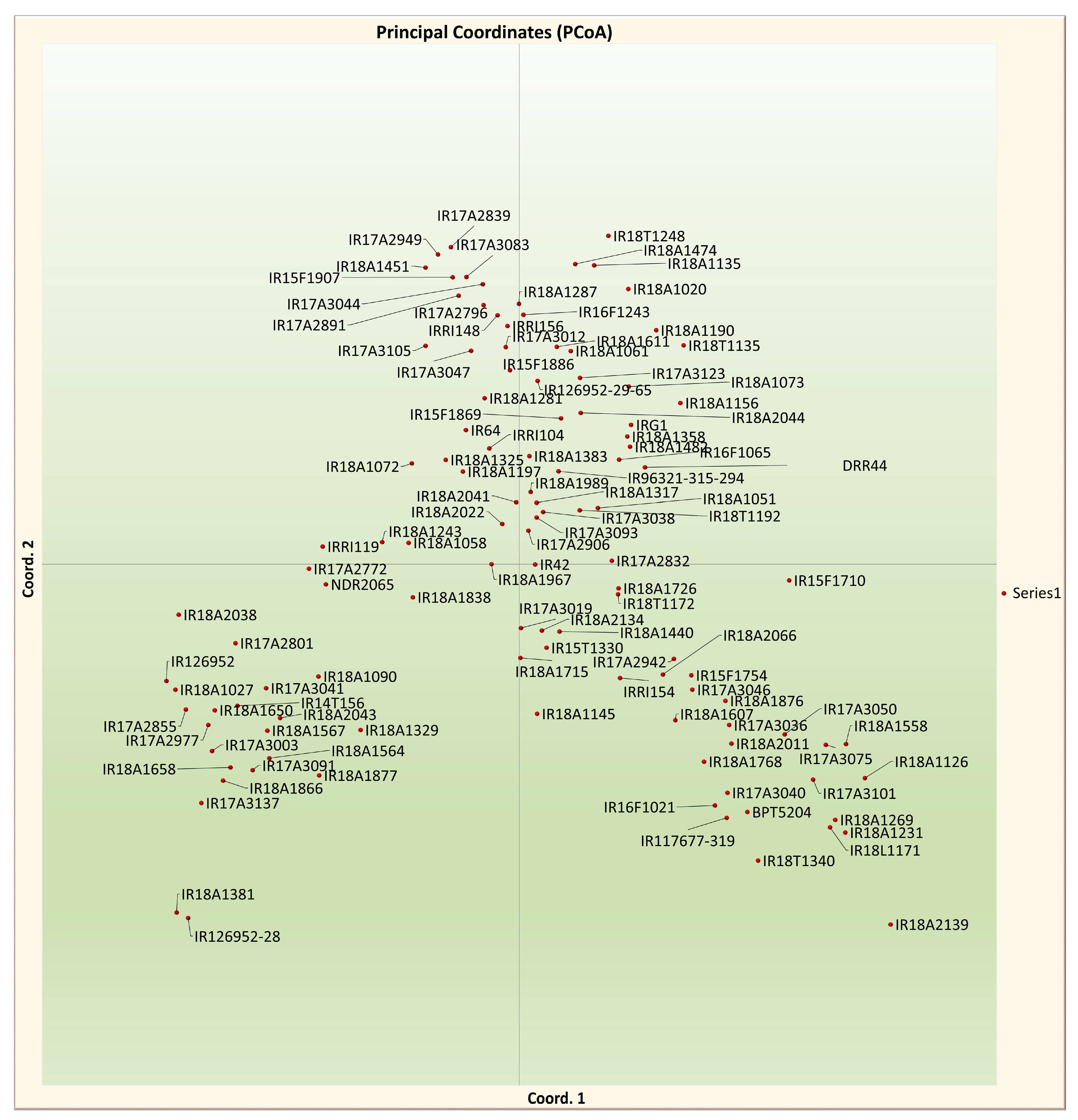
3.6. Principal Coordinate Analysis (PCoA)
3.7. Genetic Differentiation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rasheed, A.; Fahad, S.; Hassan, M.; Tahir, M.; Aamer, M.; Wu, Z. A review on aluminum toxicity and quantitative trait loci mapping in rice (Oryza sativa L). Appl. Ecol. Environ. Res. 2020, 18, 3951–3964. [Google Scholar] [CrossRef]
- Rasheed, A.; Fahad, S.; Aamer, M.; Hassan, M.; Tahir, M.; Wu, Z. Role of genetic factors in regulating cadmium uptake, transport and accumulation mechanisms and quantitative trait loci mapping in rice. A review. Appl. Ecol. Environ. Res. 2020, 18, 4005–4023. [Google Scholar] [CrossRef]
- FAO. World Food and Agriculture – Statistical Yearbook 2022; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Collard, B.C.; Gregorio, G.B.; Thomson, M.J.; Islam, R.; Vergara, G.V.; Laborte, A.G.; Nissila, E.; Kretzschmar, T.; Cobb, J.N. Transforming rice breeding: Re-designing the irrigated breeding pipeline at the International Rice Research Institute (IRRI). Crop Breed. Genet. Genom. 2019, 1, e190008. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Xu, K.; Mackill, D.J. A major locus for submergence tolerance mapped on rice chromosome 9. Mol. Breed. 1996, 2, 219–224. [Google Scholar] [CrossRef]
- Nandi, S.; Subudhi, P.; Senadhira, D.; Manigbas, N.; Sen-Mandi, S.; Huang, N. Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol. Gen. Genet. MGG 1997, 255, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Siangliw, M.; Toojinda, T.; Tragoonrung, S.; Vanavichit, A. Thai jasmine rice carrying QTLch9 (Sub QTL) is submergence tolerant. Ann. Bot. 2003, 91, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Toojinda, T.; Siangliw, M.; Tragoonrung, S.; Vanavichit, A. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann. Bot. 2003, 91, 243–253. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Collard, B.C.; Heuer, S.; Bailey-Serres, J.; Ismail, A.M.; Mackill, D.J. Applying genomics tools for breeding submergence tolerance in rice. Transl. Genom. Crop Breed. Abiotic Stress Yield Qual. 2013, 2, 9–30. [Google Scholar]
- Gonzaga, Z.J.C.; Carandang, J.; Sanchez, D.L.; Mackill, D.J.; Septiningsih, E.M. Mapping additional QTLs from FR13A to increase submergence tolerance in rice beyond SUB1. Euphytica 2016, 209, 627–636. [Google Scholar] [CrossRef]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef]
- Chakravarthi, B.K.; Naravaneni, R. SSR marker based DNA fingerprinting and diversity study in rice (Oryza sativa L). Afr. J. Biotechnol. 2006, 5, 684–688. [Google Scholar]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Mondini, L.; Noorani, A.; Pagnotta, M.A. Assessing plant genetic diversity by molecular tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- McCouch, S.R.; Chen, X.; Panaud, O.; Temnykh, S.; Xu, Y.; Cho, Y.G.; Huang, N.; Ishii, T.; Blair, M. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol. Biol. 1997, 35, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Kumar, A.; Sharma, S.; Singh, B.; Poonam; Sood, S.; Dipta, B.; Singh, R.; Siddappa, S.; Thakur, A.K.; et al. Analysis of Genetic Diversity, Population Structure and Association Mapping for Late Blight Resistance in Potato (Solanum tuberosum L.) Accessions Using SSR Markers. Agronomy 2023, 13, 294. [Google Scholar] [CrossRef]
- Jin, L.; Lu, Y.; Xiao, P.; Sun, M.; Corke, H.; Bao, J. Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor. Appl. Genet. 2010, 121, 475–487. [Google Scholar] [CrossRef]
- Ilyas, M.Z.; Park, H.; Jang, S.J.; Cho, J.; Sa, K.J.; Lee, J.K. Association Mapping for Evaluation of Population Structure, Genetic Diversity, and Physiochemical Traits in Drought-Stressed Maize Germplasm Using SSR Markers. Plants 2023, 12, 4092. [Google Scholar] [CrossRef]
- Li, R.; Huang, Y.; Yang, X.; Su, M.; Xiong, H.; Dai, Y.; Wu, W.; Pei, X.; Yuan, Q. Genetic Diversity and Relationship of Shanlan Upland Rice Were Revealed Based on 214 Upland Rice SSR Markers. Plants 2023, 12, 2876. [Google Scholar] [CrossRef]
- Vats, G.; Das, D.; Gupta, R.; Singh, A.; Maurya, A.; Rajkumar, S.; Singh, A.K.; Bharadwaj, R.; Kumar, S.; Kaushik, S.K.; et al. Validation of Genome-Wide SSR Markers Developed for Genetic Diversity and Population Structure Study in Grain Amaranth (Amaranthus hypochondriacus). Agriculture 2023, 13, 431. [Google Scholar] [CrossRef]
- Ma, M.; Lei, E.; Wang, T.; Meng, H.; Zhang, W.; Lu, B. Genetic Diversity and Association Mapping of Grain-Size Traits in Rice Landraces from the Honghe Hani Rice Terraces System in Yunnan Province. Plants 2023, 12, 1678. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.; Yan, W.; Fjellstrom, R.; Jia, M.; McClung, A. Genetic Diversity and Relationships Assessed by SSRs in the USDA World-Wide Rice Germplasm Collection. In Proceedings of the 2008 Joint Annual Meeting, American Society of Agronomy Abstracts, Houston, TX, USA, 5–9 October 2008; Dale Bumpers National Rice Research Center: Stuttgart, Germany; pp. 552–555. [Google Scholar]
- Das, B.; Sengupta, S.; Parida, S.K.; Roy, B.; Ghosh, M.; Prasad, M.; Ghose, T.K. Genetic diversity and population structure of rice landraces from Eastern and North Eastern States of India. BMC Genet. 2013, 14, 71. [Google Scholar] [CrossRef]
- Choudhury, B.; Khan, M.L.; Dayanandan, S. Genetic structure and diversity of indigenous rice (Oryza sativa) varieties in the Eastern Himalayan region of Northeast India. SpringerPlus 2013, 2, 228. [Google Scholar] [CrossRef]
- Sow, M.; Ndjiondjop, M.-N.; Sido, A.; Mariac, C.; Laing, M.; Bezançon, G. Genetic diversity, population structure and differentiation of rice species from Niger and their potential for rice genetic resources conservation and enhancement. Genet. Resour. Crop Evol. 2014, 61, 199–213. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, L.; Zhao, Q.; Chen, T.; Yao, S.; Zhu, Z.; Zhou, L.; Nadaf, A.B.; Liang, W.; Lu, K. Genetic dissection of eating and cooking qualities in different subpopulations of cultivated rice (Oryza sativa L.) through association mapping. BMC Genet. 2020, 21, 119. [Google Scholar] [CrossRef] [PubMed]
- Courtois, B.; Frouin, J.; Greco, R.; Bruschi, G.; Droc, G.; Hamelin, C.; Ruiz, M.; Clément, G.; Evrard, J.C.; Van Coppenole, S. Genetic diversity and population structure in a European collection of rice. Crop Sci. 2012, 52, 1663–1675. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software, 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 8 July 2023).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef]
- Singh, A.V.; Shelar, A.; Rai, M.; Laux, P.; Thakur, M.; Dosnkyi, I.; Santomauro, G.; Singh, A.K.; Luch, A.; Patil, R.; et al. Harmonization Risks and Rewards: Nano-QSAR for Agricultural Nanomaterials. J. Agric. Food Chem. 2024, 72, 2835–2852. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Varma, M.; Rai, M.; Pratap Singh, S.; Bansod, G.; Laux, P.; Luch, A. Advancing Predictive Risk Assessment of Chemicals via Integrating Machine Learning, Computational Modeling, and Chemical/Nano-Quantitative Structure-Activity Relationship Approaches. Adv. Intell. Syst. 2024, 6, 2300366. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Malik, N.; Kumar, D.; Babu, B.K. Analysis of genetic divergence and population structure through microsatellite markers in normal and quality protein maize genotypes from NW Himalayan region of India. Vegetos 2020, 33, 194–202. [Google Scholar] [CrossRef]
- Singh, A.K.; Dwivedi, D.K.; Kumar, D.; Singh, A.; Dixit, S.; Khan, N.A.; Kumar, A. Genetic variability, character association and path coefficient analysis in rice (Oryza sativa) genotypes of semi-arid region of India. Indian J. Agric. Sci. 2023, 93, 844–849. [Google Scholar] [CrossRef]
- Swarup, S.; Cargill, E.J.; Crosby, K.; Flagel, L.; Kniskern, J.; Glenn, K.C. Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci. 2021, 61, 839–852. [Google Scholar] [CrossRef]
- Singh, S.K.; Singh, P.; Khaire, A.R.; Korada, M.; Singh, D.K.; Majhi, P.K.; Jayasudha, S. Genetic Variability, Character Association and Path Analysis for Yield and its Related Traits in Rice (Oryza sativa L.) Genotypes. Int. J. Plant Soil Sci. 2021, 33, 437–446. [Google Scholar] [CrossRef]
- Biswajit, P.; Sritama, K.; Anindya, S.; Moushree, S.; Sabyasachi, K. Breeding for submergence tolerance in rice (Oryza sativa L.) and its management for flash flood in rainfed low land area: A review. Agric. Rev. 2017, 38, 167–179. [Google Scholar] [CrossRef]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Ram, S.G.; Thiruvengadam, V.; Vinod, K.K. Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J. Appl. Genet. 2007, 48, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; He, H.; Zou, Y.; Chen, W.; Yu, R.; Liu, X.; Yang, Y.; Gao, Y.-M.; Xu, J.-L.; Fan, L.-M.; et al. Development and application of a set of breeder-friendly SNP markers for genetic analyses and molecular breeding of rice (Oryza sativa L.). Theor. Appl. Genet. 2011, 123, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Agrama, H.; Eizenga, G. Molecular diversity and genome-wide linkage disequilibrium patterns in a worldwide collection of Oryza sativa and its wild relatives. Euphytica 2008, 160, 339–355. [Google Scholar] [CrossRef]
- Ni, J.; Colowit, P.; Mackill, D. Evaluation of Genetic Diversity in Rice Subspecies Using Microsatellite Markers. Crop Sci. 2002, 42, 601–607. [Google Scholar] [CrossRef]
- Agrama, H.A.; Eizenga, G.C.; Yan, W. Association mapping of yield and its components in rice cultivars. Mol. Breed. 2007, 19, 341–356. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Wei, X.; Qi, Y.; Wang, M.; Sun, J.; Ding, L.; Tang, S.; Qiu, Z.E.; Cao, Y.; et al. Genetic structure and diversity of Oryza sativa L. in Guizhou, China. Chin. Sci. Bull. 2007, 52, 343–351. [Google Scholar] [CrossRef]
- Zhao, K.; Wright, M.; Kimball, J.; Eizenga, G.; McClung, A.; Kovach, M.; Tyagi, W.; Ali, M.L.; Tung, C.W.; Reynolds, A.; et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE 2010, 5, e10780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, H.; Wang, M.; Sun, J.; Qi, Y.; Wang, F.; Wei, X.; Han, L.; Wang, X.; Li, Z. Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor. Appl. Genet. 2009, 119, 1105–1117. [Google Scholar] [CrossRef]
- Caicedo, A.L.; Williamson, S.H.; Hernandez, R.D.; Boyko, A.; Fledel-Alon, A.; York, T.L.; Polato, N.R.; Olsen, K.M.; Nielsen, R.; McCouch, S.R.; et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007, 3, 1745–1756. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, H.; Zhang, D.; Wang, M.; Sun, J.; Ding, L.; Wang, F.; Li, Z. Assessing indica-japonica differentiation of improved rice varieties using microsatellite markers. J. Genet. Genom. 2009, 36, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]

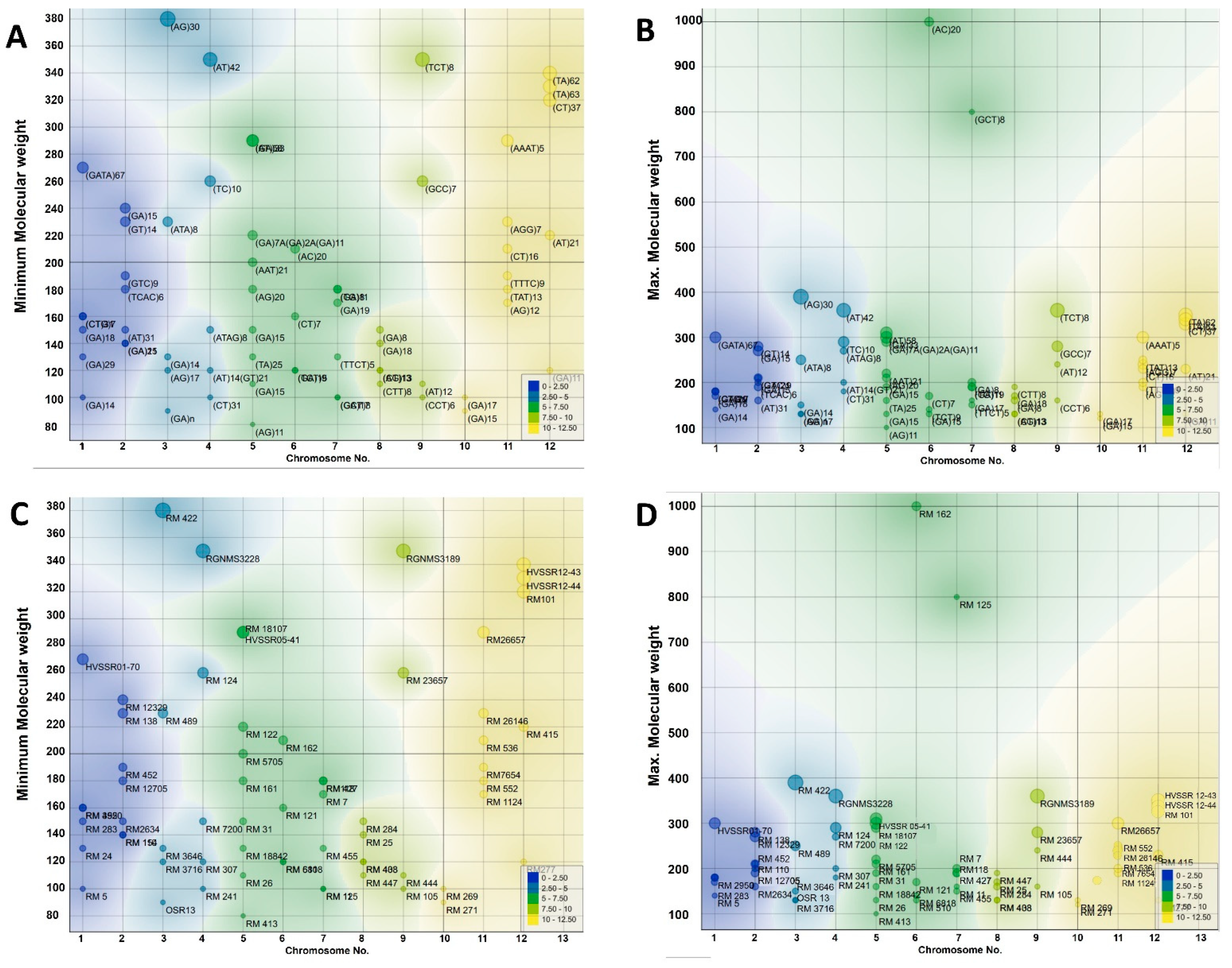
| Marker | Chromosome No. | SSR Motif | Mini. Mol. Weight | Maxi. Mol Weight | Number of Alleles | Heterozygosity | Gene Diversity | PIC Value |
|---|---|---|---|---|---|---|---|---|
| RM 495 | 1 | (CTG)7 | 160 | 180 | 3 | 0.494 | 0.497 | 0.722 |
| RM 283 | 1 | (GA)18 | 150 | 170 | 3 | 0.138 | 0.139 | 0.566 |
| RM 24 | 1 | (GA)29 | 130 | 180 | 6 | 0.811 | 0.815 | 0.969 |
| RM 5 | 1 | (GA)14 | 100 | 140 | 5 | 0.674 | 0.677 | 0.870 |
| HVSSR01-70 | 1 | (GATA)67 | 270 | 300 | 3 | 0.500 | 0.502 | 0.803 |
| RM 3520 | 1 | (CT)31 | 160 | 180 | 5 | 0.540 | 0.543 | 0.763 |
| RM 12329 | 2 | (GA)15 | 240 | 270 | 4 | 0.694 | 0.698 | 0.908 |
| RM 154 | 2 | (GA)21 | 140 | 210 | 8 | 0.819 | 0.823 | 0.925 |
| RM 110 | 2 | (GA)15 | 140 | 200 | 7 | 0.754 | 0.758 | 0.924 |
| RM 12705 | 2 | (TCAC)6 | 180 | 190 | 3 | 0.583 | 0.585 | 0.815 |
| RM 452 | 2 | (GTC)9 | 190 | 210 | 3 | 0.253 | 0.254 | 0.623 |
| RM2634 | 2 | (AT)31 | 150 | 160 | 3 | 0.584 | 0.586 | 0.830 |
| RM 138 | 2 | (GT)14 | 230 | 280 | 6 | 0.518 | 0.520 | 0.836 |
| RM 489 | 3 | (ATA)8 | 230 | 250 | 3 | 0.047 | 0.048 | 0.860 |
| RM 3716 | 3 | (AG)17 | 120 | 130 | 3 | 0.437 | 0.439 | 0.983 |
| OSR13 | 3 | (GA)n | 90 | 130 | 3 | 0.338 | 0.340 | 0.701 |
| RM 3646 | 3 | (GA)14 | 130 | 150 | 3 | 0.383 | 0.384 | 0.734 |
| RM 422 | 3 | (AG)30 | 380 | 390 | 2 | 0.393 | 0.395 | 0.729 |
| RM 307 | 4 | (AT)14(GT)21 | 120 | 200 | 3 | 0.339 | 0.341 | 0.740 |
| RM 7200 | 4 | (ATAG)8 | 150 | 270 | 8 | 0.868 | 0.872 | 0.980 |
| RGNMS3228 | 4 | (AT)42 | 350 | 360 | 3 | 0.405 | 0.407 | 0.984 |
| RM 241 | 4 | (CT)31 | 100 | 180 | 4 | 0.582 | 0.585 | 0.845 |
| RM 124 | 4 | (TC)10 | 260 | 290 | 3 | 0.323 | 0.325 | 0.959 |
| RM 122 | 5 | (GA)7A(GA)2A(GA)11 | 220 | 290 | 7 | 0.758 | 0.761 | 0.942 |
| RM 413 | 5 | (AG)11 | 80 | 100 | 3 | 0.560 | 0.563 | 0.925 |
| RM 18107 | 5 | (GA)33 | 290 | 300 | 2 | 0.436 | 0.438 | 0.755 |
| RM 5705 | 5 | (AAT)21 | 200 | 220 | 3 | 0.624 | 0.626 | 0.828 |
| HVSSR05-41 | 5 | (AT)58 | 290 | 310 | 3 | 0.400 | 0.402 | 0.799 |
| RM 161 | 5 | (AG)20 | 180 | 210 | 3 | 0.075 | 0.076 | 0.507 |
| RM 26 | 5 | (GA)15 | 110 | 130 | 0.620 | 0.623 | 0.694 | |
| RM 18842 | 5 | (TA)25 | 130 | 160 | 3 | 0.482 | 0.485 | 0.776 |
| RM 31 | 5 | (GA)15 | 150 | 190 | 6 | 0.482 | 0.485 | 0.846 |
| RM 510 | 6 | (GA)15 | 120 | 130 | 2 | 0.454 | 0.456 | 0.515 |
| RM 121 | 6 | (CT)7 | 160 | 170 | 2 | 0.224 | 0.225 | 0.640 |
| RM 6818 | 6 | (TCT)9 | 120 | 140 | 3 | 0.430 | 0.432 | 0.718 |
| RM 162 | 6 | (AC)20 | 210 | 1000 | 5 | 0.167 | 0.168 | 0.495 |
| RM 427 | 7 | (TG)11 | 180 | 190 | 2 | 0.334 | 0.336 | 0.701 |
| RM 11 | 7 | (GA)17 | 100 | 160 | 5 | 0.569 | 0.572 | 0.816 |
| RM 7 | 7 | (GA)19 | 170 | 190 | 2 | 0.238 | 0.239 | 0.684 |
| RM 455 | 7 | (TTCT)5 | 130 | 150 | 3 | 0.277 | 0.278 | 0.649 |
| RM118 | 7 | (GA)8 | 180 | 200 | 2 | 0.035 | 0.034 | 0.035 |
| RM 125 | 7 | (GCT)8 | 100 | 800 | 7 | 0.413 | 0.415 | 0.682 |
| RM 408 | 8 | (CT)13 | 120 | 130 | 2 | 0.017 | 0.017 | 0.517 |
| RM 25 | 8 | (GA)18 | 140 | 170 | 4 | 0.486 | 0.488 | 0.719 |
| RM 284 | 8 | (GA)8 | 150 | 160 | 2 | 0.176 | 0.177 | 0.600 |
| RM 433 | 8 | (AG)13 | 120 | 130 | 2 | 0.031 | 0.031 | 0.663 |
| RM 447 | 8 | (CTT)8 | 110 | 190 | 4 | 0.278 | 0.279 | 0.710 |
| RM 23657 | 9 | (GCC)7 | 260 | 280 | 3 | 0.216 | 0.217 | 0.690 |
| RGNMS3189 | 9 | (TCT)8 | 350 | 360 | 2 | 0.423 | 0.425 | 0.771 |
| RM 444 | 9 | (AT)12 | 110 | 240 | 6 | 0.691 | 0.694 | 0.909 |
| RM 105 | 9 | (CCT)6 | 100 | 160 | 4 | 0.406 | 0.408 | 0.703 |
| RM 271 | 10 | (GA)15 | 90 | 120 | 3 | 0.137 | 0.137 | 0.564 |
| RM 269 | 10 | (GA)17 | 100 | 130 | 4 | 0.620 | 0.623 | 0.834 |
| RM 26146 | 11 | (AGG)7 | 230 | 240 | 3 | 0.251 | 0.252 | 0.581 |
| RM 1124 | 11 | (AG)12 | 170 | 190 | 3 | 0.130 | 0.131 | 0.656 |
| RM 552 | 11 | (TAT)13 | 180 | 250 | 6 | 0.291 | 0.292 | 0.692 |
| RM 536 | 11 | (CT)16 | 210 | 230 | 3 | 0.434 | 0.436 | 0.709 |
| RM26657 | 11 | (AAAT)5 | 290 | 300 | 3 | 0.603 | 0.606 | 0.846 |
| RM7654 | 11 | (TTTC)9 | 190 | 200 | 3 | 0.550 | 0.553 | 0.800 |
| RM 415 | 12 | (AT)21 | 220 | 230 | 2 | 0.175 | 0.176 | 0.678 |
| RM101 | 12 | (CT)37 | 320 | 330 | 3 | 0.564 | 0.567 | 0.817 |
| RM277 | 12 | (GA)11 | 120 | 130 | 2 | 0.480 | 0.483 | 0.648 |
| HVSSR12-43 | 12 | (TA)62 | 340 | 350 | 2 | 0.655 | 0.658 | 0.734 |
| HVSSR12-44 | 12 | (TA)63 | 330 | 340 | 2 | 0.227 | 0.228 | 0.871 |
| Characters | Mean | Range | Var (g) | Var (p) | GCV (%) | PCV (%) | ECV (%) |
|---|---|---|---|---|---|---|---|
| DFF | 107.65 | 86.60–125.27 | 93.72 | 95.03 | 8.99 | 9.06 | 1.32 |
| SV | 32.33 | 17.41–56.75 | 70.32 | 75.97 | 25.96 | 26.98 | 5.65 |
| PH (cm) | 118.51 | 71.80–171.80 | 225.48 | 226.44 | 12.67 | 12.70 | 0.97 |
| PL | 113.96 | 52.21–179.08 | 554.43 | 615.66 | 20.65 | 21.76 | 61.23 |
| SPP | 93.84 | 0.00–165.21 | 54.39 | 71.64 | 8.66 | 9.93 | 17.25 |
| BYP | 28.50 | 14.61–48.50 | 28.91 | 31.94 | 18.86 | 19.82 | 3.03 |
| HI | 22.32 | 16.43–35.03 | 6.94 | 9.29 | 11.81 | 13.66 | 2.35 |
| Source | df | SS | MS | Est. Var. | Percent |
|---|---|---|---|---|---|
| Among the Population | 3 | 351.793 | 117.264 | 1.575 | 10% |
| Among Individuals | 112 | 3137.005 | 28.009 | 13.099 | 79% |
| Within Individuals | 116 | 210.000 | 1.810 | 1.810 | 11% |
| Total | 231 | 3698.797 | 16.484 | 100% | |
| F-Statistics | Value | p (r and >= data) | |||
| Fst | 0.096 | 0.001 | |||
| Fis | 0.879 | 0.001 | |||
| Fit | 0.890 | 0.001 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.K.; Kumar, D.; Gemmati, D.; Ellur, R.K.; Singh, A.; Tisato, V.; Dwivedi, D.K.; Singh, S.K.; Kumar, K.; Khan, N.A.; et al. Investigating Genetic Diversity and Population Structure in Rice Breeding from Association Mapping of 116 Accessions Using 64 Polymorphic SSR Markers. Crops 2024, 4, 180-194. https://doi.org/10.3390/crops4020014
Singh AK, Kumar D, Gemmati D, Ellur RK, Singh A, Tisato V, Dwivedi DK, Singh SK, Kumar K, Khan NA, et al. Investigating Genetic Diversity and Population Structure in Rice Breeding from Association Mapping of 116 Accessions Using 64 Polymorphic SSR Markers. Crops. 2024; 4(2):180-194. https://doi.org/10.3390/crops4020014
Chicago/Turabian StyleSingh, Alok Kumar, Devendra Kumar, Donato Gemmati, Ranjith Kumar Ellur, Ashutosh Singh, Veronica Tisato, Devendra Kumar Dwivedi, Sanjay Kumar Singh, Kishor Kumar, Nawaz Ahmad Khan, and et al. 2024. "Investigating Genetic Diversity and Population Structure in Rice Breeding from Association Mapping of 116 Accessions Using 64 Polymorphic SSR Markers" Crops 4, no. 2: 180-194. https://doi.org/10.3390/crops4020014
APA StyleSingh, A. K., Kumar, D., Gemmati, D., Ellur, R. K., Singh, A., Tisato, V., Dwivedi, D. K., Singh, S. K., Kumar, K., Khan, N. A., & Singh, A. V. (2024). Investigating Genetic Diversity and Population Structure in Rice Breeding from Association Mapping of 116 Accessions Using 64 Polymorphic SSR Markers. Crops, 4(2), 180-194. https://doi.org/10.3390/crops4020014









