Revolutionizing Detection of Minimal Residual Disease in Breast Cancer Using Patient-Derived Gene Signature
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Processing and RNA Extraction
2.2. Overexpressed Biomarker Identification and Validation
2.3. Functional Enrichment GO and KEGG Analysis
2.4. Correlation of Gene Expression
2.5. The OncoMRD BREAST Scoring Algorithm
2.6. The Proof-of-Concept Clinical Study
3. Results
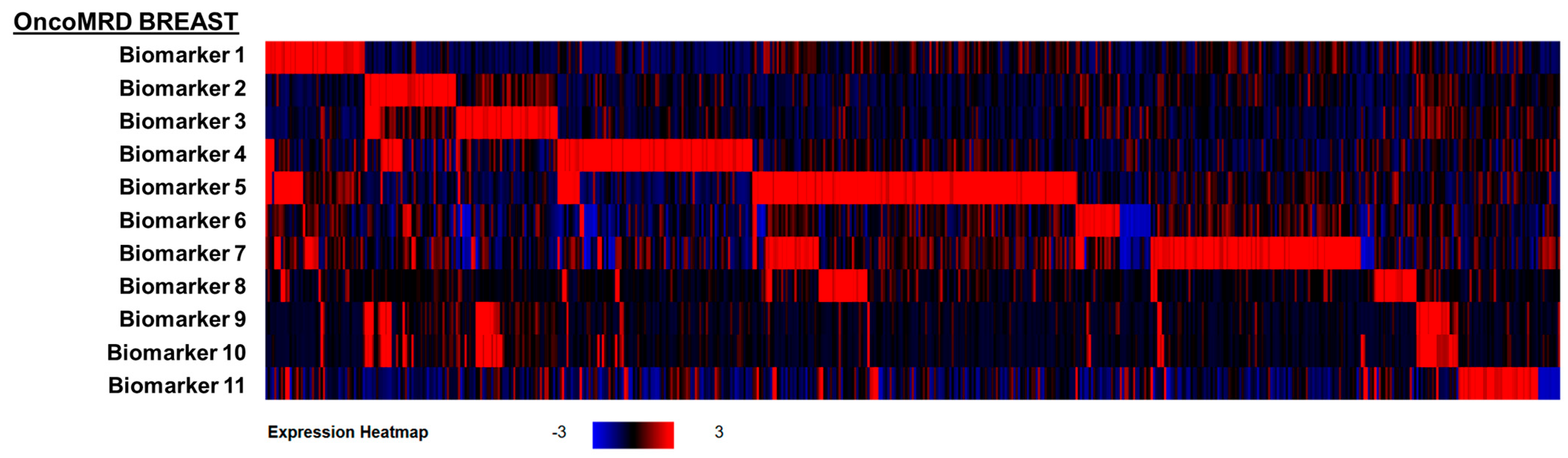
3.1. Identification and Validation of Hyperactive OncoMRD BREAST Biomarkers
3.2. Association of OncoMRD BREAST Gene Signature with Tumor Activity
3.3. Strong Correlation Between OncoMRD BREAST Gene Signature and Genomic Alterations
3.4. Gene Expression Correlation Between OncoMRD BREAST Biomarkers and Key Breast Cancer-Associated Genes
3.5. Correlation Between Tumor Clinicopathological Parameters and Expression of OncoMRD BREAST Biomarkers
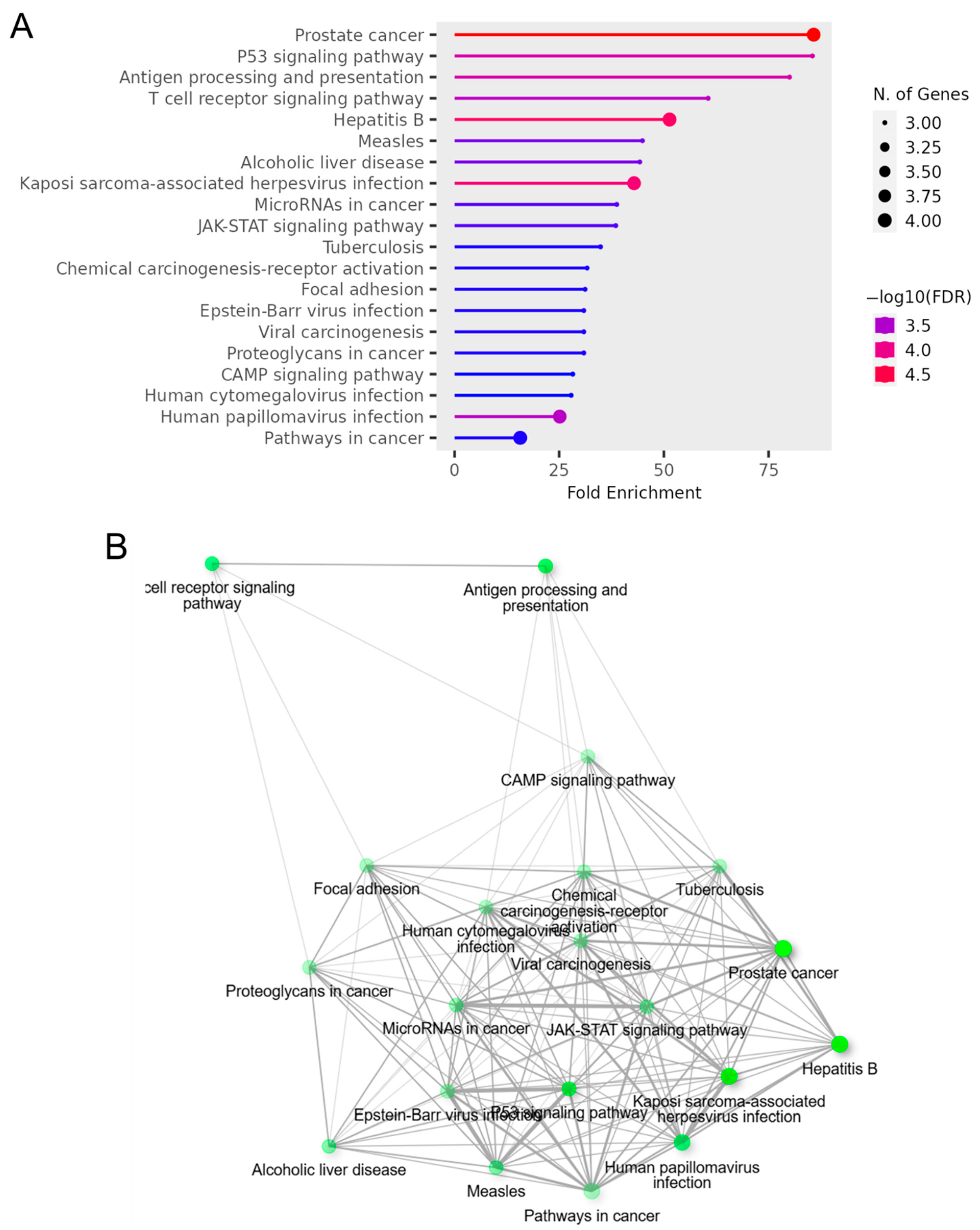
3.6. Gene Ontology Term Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analyses of OncoMRD BREAST Gene Signature
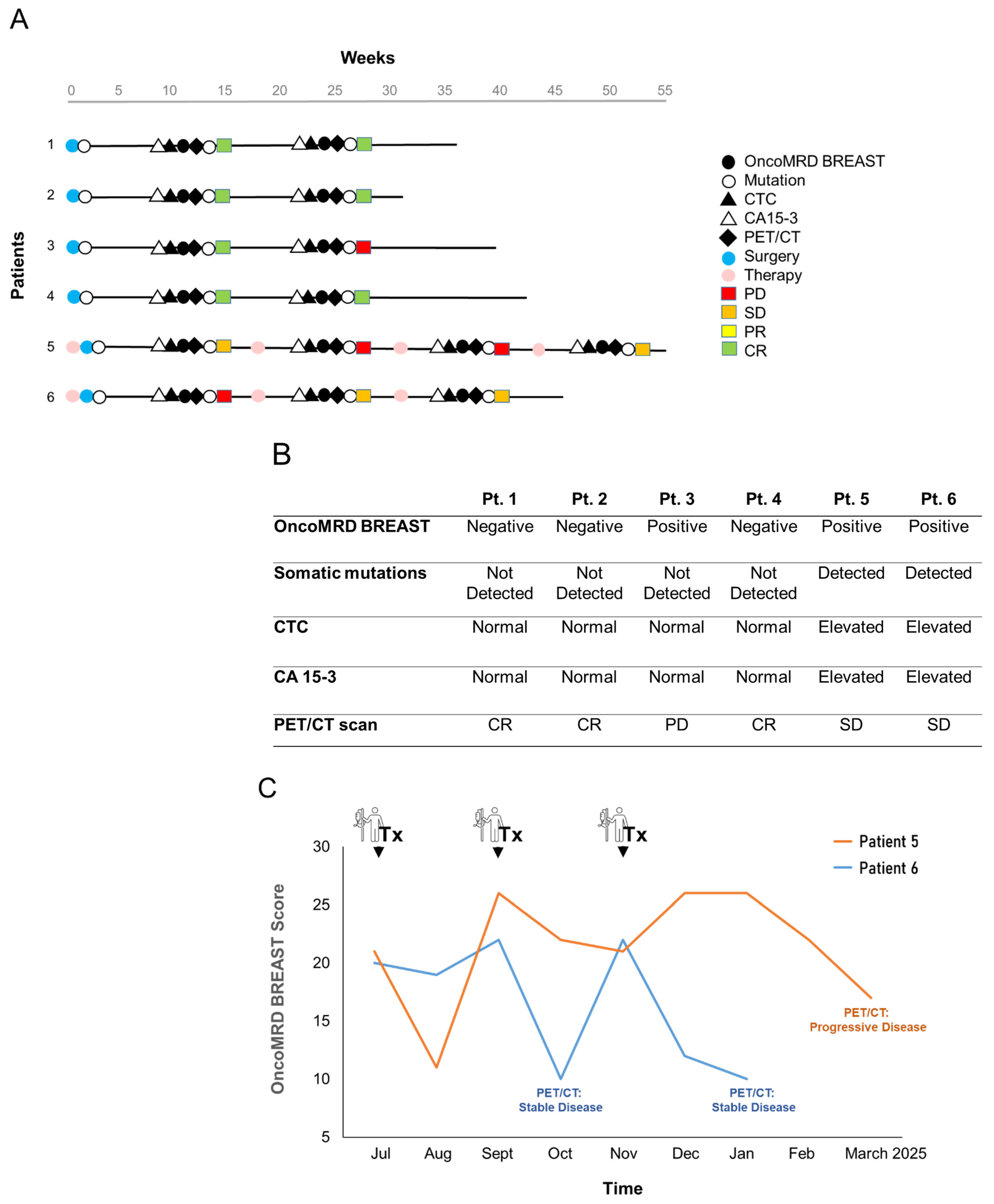
3.7. OncoMRD BREAST Minimal Residual Disease Detection and Treatment Monitoring of Breast Cancer Patients
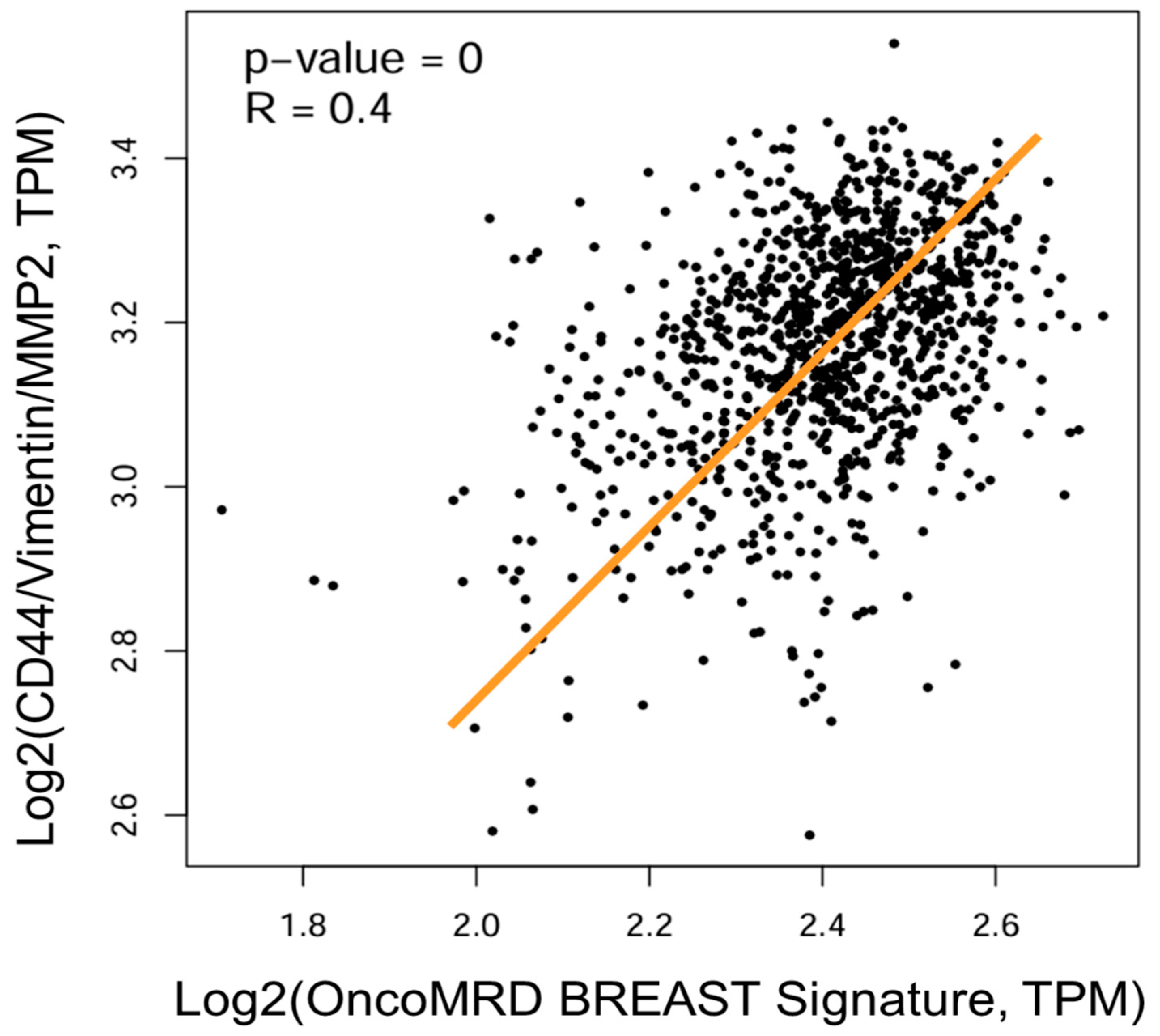
3.8. Strong Correlation of OncoMRD BREAST Gene Signature with a Key Gene Signature Associated with Residual Breast Tumors After Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Colleoni, M.; Sun, Z.; Price, K.N.; Karlsson, P.; Forbes, J.F.; Thürlimann, B.; Gianni, L.; Castiglione, M.; Gelber, R.D.; Coates, A.S.; et al. Annual Hazard Rates of Recurrence for Breast Cancer During 24 Years of Follow-Up: Results From the International Breast Cancer Study Group Trials I to V. J. Clin. Oncol. 2016, 34, 927–935. [Google Scholar] [CrossRef]
- Stergiopoulou, D.; Markou, A.; Strati, A.; Zavridou, M.; Tzanikou, E.; Mastoraki, S.; Kallergi, G.; Georgoulias, V.; Lianidou, E. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci. Rep. 2023, 13, 1258. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.N.; Esen, B.Ö.; Mellemkjær, L.; Christiansen, P.; Ejlertsen, B.; Lash, T.L.; Nørgaard, M.; Cronin-Fenton, D. The Incidence of Breast Cancer Recurrence 10–32 Years After Primary Diagnosis. J. Natl. Cancer Inst. 2022, 114, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Tachtsidis, A.; McInnes, L.M.; Jacobsen, N.; Thompson, E.W.; Saunders, C.M. Minimal residual disease in breast cancer: An overview of circulating and disseminated tumour cells. Clin. Exp. Metastasis 2016, 33, 521–550. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, Q. Detecting liquid remnants of solid tumors treated with curative intent: Circulating tumor DNA as a biomarker of minimal residual disease (Review). Oncol. Rep. 2023, 49, 106. [Google Scholar] [CrossRef]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Yeh, J.-S.; Lin, S.-T. Minimal Residual Disease in Solid Tumors: Shifting the Focus from Cell-Free DNA to Cell-Free RNA. J. Oncogenom. Oncotarget 2022, 1, 1–9. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Yuguang, C. Therapeutic Response Monitoring: Minimal Residual Disease Testing in Cancer Management. Amer. J. Physiol. Biochem. Pharmacol. 2024, 14, 1–4. [Google Scholar] [CrossRef]
- Werner, S.; Heidrich, I.; Pantel, K. Clinical management and biology of tumor dormancy in breast cancer. Semin. Cancer Biol. 2022, 78, 49–62. [Google Scholar] [CrossRef]
- Zheng, J.; Qin, C.; Wang, Q.; Tian, D.; Chen, Z. Circulating tumour DNA-Based molecular residual disease detection in resectable cancers: A systematic review and meta-analysis. EBioMedicine 2024, 103, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Chin, R.-I.; Chen, K.; Usmani, A.; Chua, C.; Harris, P.K.; Binkley, M.S.; Azad, T.D.; Dudley, J.C.; Chaudhuri, A.A. Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol. Diagn. Ther. 2019, 23, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Rahal, Z.; Scheet, P.; Kadara, H. Somatic Mutations in Normal Tissues: Calm before the Storm. Cancer Discov. 2024, 14, 605–609. [Google Scholar] [CrossRef]
- Yeh, C.H.; Ford, A.; Brown, C. Somatic Mutations in Cancer-Free Individuals: A Liquid Biopsy Connection. Open Access J. Oncol. Med. 2018, 1, 1–4. [Google Scholar] [CrossRef]
- Yates, L.; Seoane, J.; Le Tourneau, C.; Siu, L.; Marais, R.; Michiels, S.; Soria, J.; Campbell, P.; Normanno, N.; Scarpa, A.; et al. The European Society for Medical Oncology (ESMO) Precision Medicine Glossary. Ann. Oncol. 2017, 29, 30–35. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Seifert, S.; Gundlach, S.; Junge, O.; Szymczak, S.; Martelli, L. Integrating biological knowledge and gene expression data using pathway-guided random forests: A benchmarking study. Bioinformatics 2020, 36, 4301–4308. [Google Scholar] [CrossRef]
- Yeh, C. Circulating cell-free transcriptomics in cancer. J. Lung Pulm Respir. Res. 2023, 10, 23–25. [Google Scholar] [CrossRef]
- Yeh, C.; Lin, S.-T.; Lai, H.-C. A Transformative Technology Linking Patient’s mRNA Expression Profile to Anticancer Drug Efficacy. Onco 2024, 4, 143–162. [Google Scholar] [CrossRef]
- Lin, S.; Lai, H.; Yeh, C. Single-tube two-pronged approach using both cell-free DNA and RNA for multimodal biomarker tests at the time of biopsy. Precis. Med. Sci. 2023, 12, 233–241. [Google Scholar] [CrossRef]
- Lin, S.T.; Yip, C.H. Harnessing the Power of CpG Methylation Biomarkers for Early Cancer Detection. J. Mol. Genet. Med. 2022, 16, 537. [Google Scholar]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Schramm, A.; Friedl, T.W.P.; Schochter, F.; Scholz, C.; de Gregorio, N.; Huober, J.; Rack, B.; Trapp, E.; Alunni-Fabbroni, M.; Müller, V.; et al. Therapeutic intervention based on circulating tumor cell phenotype in metastatic breast cancer: Concept of the DETECT study program. Arch. Gynecol. Obstet. 2015, 293, 271–281. [Google Scholar] [CrossRef]
- Kostecka, A.; Nowikiewicz, T.; Olszewski, P.; Koczkowska, M.; Horbacz, M.; Heinzl, M.; Andreou, M.; Salazar, R.; Mair, T.; Madanecki, P.; et al. High prevalence of somatic PIK3CA and TP53 pathogenic variants in the normal mammary gland tissue of sporadic breast cancer patients revealed by duplex sequencing. npj Breast Cancer 2022, 8, 76. [Google Scholar] [CrossRef]
- Britschgi, A.; Bill, A.; Brinkhaus, H.; Rothwell, C.; Clay, I.; Duss, S.; Rebhan, M.; Raman, P.; Guy, C.T.; Wetzel, K.; et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 1026–1034. [Google Scholar] [CrossRef]
- Dancau, A.; Wuth, L.; Waschow, M.; Holst, F.; Krohn, A.; Choschzick, M.; Terracciano, L.; Politis, S.; Kurtz, S.; Lebeau, A.; et al. PPFIA1 and CCND1 are frequently coamplified in breast cancer. Genes Chromosom Cancer 2009, 49, 1–8. [Google Scholar] [CrossRef]
- Xu, L.; Li, P.; Hao, X.; Lu, Y.; Liu, M.; Song, W.; Shan, L.; Yu, J.; Ding, H.; Chen, S.; et al. SHANK2 is a frequently amplified oncogene with evolutionarily conserved roles in regulating Hippo signaling. Protein Cell 2020, 12, 174–193. [Google Scholar] [CrossRef]
- Zhou, R.; Zhu, X.; Peng, Y.; Zhong, L.; Peng, L.; Yang, B.; Meng, Y.; Chen, X.; Lu, Y. Clinical Impact of 11q13.3 Amplification on Immune Cell Infiltration and Prognosis in Breast Cancer. Int. J. Gen. Med. 2022, 15, 4037–4052. [Google Scholar] [CrossRef]
- Guo, Z.-H.; Yao, L.-T.; Guo, A.-Y. Clinical and biological impact of LINC02544 expression in breast cancer after neoadjuvant chemotherapy. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10573–10585. [Google Scholar] [CrossRef]
- Santolla, M.F.; Maggiolini, M. The FGF/FGFR System in Breast Cancer: Oncogenic Features and Therapeutic Perspectives. Cancers 2020, 12, 3029. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gonzalez, L.; Cendra, A.S.; Cendra, C.S.; Cervantes, E.D.R.; Espinosa, J.C.; Pekarek, T.; Fraile-Martinez, O.; García-Montero, C.; Rodriguez-Slocker, A.M.; Jiménez-Álvarez, L.; et al. Exploring Biomarkers in Breast Cancer: Hallmarks of Diagnosis, Treatment, and Follow-Up in Clinical Practice. Medicina 2024, 60, 168. [Google Scholar] [CrossRef] [PubMed]
- Ruth, J.R.; Pant, D.K.; Pan, T.C.; Seidel, H.E.; Baksh, S.C.; Keister, B.A.; Singh, R.; Sterner, C.J.; Bakewell, S.J.; Moody, S.E.; et al. Cellular dormancy in minimal residual disease following targeted therapy. Breast Cancer Res. 2021, 23, 63. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, 466. [Google Scholar] [CrossRef]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef]
- Sadeh, R.; Sharkia, I.; Fialkoff, G.; Rahat, A.; Gutin, J.; Chappleboim, A.; Nitzan, M.; Fox-Fisher, I.; Neiman, D.; Meler, G.; et al. ChIP-seq of plasma cell-free nucleosomes identifies gene expression programs of the cells of origin. Nat. Biotechnol. 2021, 39, 586–598. [Google Scholar] [CrossRef]
- Perou, C.M.; Jeffrey, S.S.; van de Rijn, M.; Rees, C.A.; Eisen, M.B.; Ross, D.T.; Pergamenschikov, A.; Williams, C.F.; Zhu, S.X.; Lee, J.C.F.; et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA 1999, 96, 9212–9217. [Google Scholar] [CrossRef]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L.; et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006, 7, 96. [Google Scholar] [CrossRef]



| OncoMRD BREAST Biomarkers | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| BRCA1 | R = 0.23 p = 1.7E−14 | R = 0.05 p = 0.098 | R = −0.14 p = 2.1E−6 | R = 0.25 p = 0 | R = 0.31 p = 0 | R = 0.36 p = 0 | R = 0.3 p = 0 |
| BRCA2 | R = −0.0045 p = 0.88 | R = 0.20 p = 2.4E−11 | R = 0.0093 p = 0.76 | R = 0.21 p = 9.9E−13 | R = 0.13 p = 2.4E−5 | R = 0.46 p = 0 | R = 0.25 p = 0 |
| ATM | R = 0.22 p = 8.0E−13 | R = 0.4 p = 0 | R = 0.26 p = 0 | R = −0.16 p = 5.0E−8 | R = −0.03 p = 0.32 | R = 0.65 p = 0 | R = 0.55 p = 0 |
| ESR1 | R = 0.50 p = 0 | R = −0.22 p = 1.4E−13 | R = −0.26 p = 7.6E−19 | R = −0.086 p = 4.7E−3 | R = 0.32 p = 0 | R = 0.20 p = 8.4E−11 | R = 0.36 p = 0 |
| PIK3CA | R = 0.15 p = 1.3E−6 | R = 0.30 p = 0 | R = 0.11 p = 1.9E−4 | R = −0.018 p = 0.56 | R = 0.055 p = 0.07 | R = 0.56 p = 0 | R = 0.42 p = 0 |
| ERBB2 (HER2) | R = 0.13 p = 1.0E−5 | R = −0.052 p = 0.087 | R = −0.041 p = 0.17 | R = −0.087 p = 0.22 | R = −0.039 p = 0.20 | R = 0.089 p = 3.5E−3 | R = −0.03 p = 0.33 |
| CDK4 | R = −0.059 p = 0.052 | R = 0.047 p = 0.12 | R = −0.054 p = 0.078 | R = 0.16 p = 6.7E−8 | R = −0.014 p = 0.63 | R = 0.11 p = 3.4E−4 | R = 0.045 p = 0.14 |
| OncoMRD BREAST Biomarkers | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation Count Spearman p-value | 1.78E−41 | 0.994 | 6.35E−06 | 6.68E−11 | 2.93E−13 | 0.176 | 3.88E−07 | 0.389 | 1.02E−05 | 4.21E−09 |
| TMB Spearman p-value | 2.70E−32 | 0.977 | 3.88E−07 | 2.03E−07 | 9.97E−11 | 0.641 | 3.11E−05 | 0.686 | 1.99E−05 | 4.60E−07 |
| Winter Hypoxia Score Spearman p-value | 4.07E−81 | 0.136 | 0.039 | 6.94E−34 | 4.78E−12 | 1.85E−28 | 4.48E−35 | 6.32E−05 | 2.27E−05 | 0.336 |
| MSIsensor Score Spearman p-value | 1.04E−05 | 3.54E−13 | 5.67E−18 | 2.23E−09 | 0.824 | 1.21E−05 | 0.324 | 0.044 | 9.46E−15 | 2.07E−11 |
| Tumor Break Load Spearman p-value | 6.42E−38 | 1.01E−03 | 0.437 | 1.11E−16 | 0.012 | 0.360 | 3.55E−10 | 0.589 | 0.524 | 0.0915 |
| Progress Free Survival Spearman p-value | 2.15E−03 | 2.66E−04 | 0.661 | 0.195 | 0.764 | 6.24E−05 | 0.0685 | 0.507 | 1.09E−03 | 4.82E−03 |
| Overall Survival Spearman p-value | 4.67E−03 | 2.71E−04 | 0.440 | 0.343 | 0.985 | 3.57E−04 | 0.153 | 0.463 | 1.04E−03 | 4.36E−03 |
| Biomarkers | Sample Requirement | Sample Processing | Need for Baseline Sample | Sensitivity | Turnaround and Complexity | Clinical Validation | Clonal Consideration | Cost Per Sample |
|---|---|---|---|---|---|---|---|---|
| ctDNA | Cell-free portion of blood (Plasma) | Cell-free DNA extraction | Yes/No | <0.001% mutant allele frequency | Labor intensive; requires robust bioinformatics support; could take 2–4 weeks | Yes | Covers major and minor clones | High |
| CTCs | Require >5 million cells (7.5 mL blood) | Needs assessment within 24–48 h (requires a fresh sample) | No | ≥1 in 100,000 cells | Labor intensive with long hands-on time; may take 1–2 days | Yes | Considers all clones with a similar phenotype | Low |
| cfmRNAs | Cell-free portion of blood (Plasma) | Cell-free RNA extraction | No | 0.01–0.001% | Requires development of cancer type-specific biomarkers; could take 3–5 days | No | Detects tumor gene activities | Low |
| miRNAs | Cell-free portion of blood (Plasma) | Cell-free RNA extraction | No | 0.01–0.001% | Requires development of cancer type-specific biomarkers; could take 3–5 days | No | Detects expression signature | Low |
| Extracellular vesicles (EV) | Cell-free portion of blood (Plasma) | Specialized EV isolation; isolating and analyzing technologies not yet fully developed | No | Solely depending on purity | Requires development of cancer type-specific biomarkers; could take 1–2 weeks | No | Detects expression signature | High |
| Circulating cancer antigens | Cell-free portion of blood (Serum) | No | Yes | Moderate sensitivity depending on the type and stage of cancer | Low complexity; fast turnaround | Yes | Detects a specific antigen | Low |
| 18F-FDG (fluorodeoxyglucose) uptake in PET/CT imaging | Patient preparation required | No | Yes | Resolution of 4–5 mm | High complexity; requires professional operation and interpretation; patient radiation exposure concerns | Yes | Detects tumor metabolic activity | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, C.; Lai, H.-C.; Grabbe, N.; Willett, X.; Lin, S.-T. Revolutionizing Detection of Minimal Residual Disease in Breast Cancer Using Patient-Derived Gene Signature. Onco 2025, 5, 35. https://doi.org/10.3390/onco5030035
Yeh C, Lai H-C, Grabbe N, Willett X, Lin S-T. Revolutionizing Detection of Minimal Residual Disease in Breast Cancer Using Patient-Derived Gene Signature. Onco. 2025; 5(3):35. https://doi.org/10.3390/onco5030035
Chicago/Turabian StyleYeh, Chen, Hung-Chih Lai, Nathan Grabbe, Xavier Willett, and Shu-Ti Lin. 2025. "Revolutionizing Detection of Minimal Residual Disease in Breast Cancer Using Patient-Derived Gene Signature" Onco 5, no. 3: 35. https://doi.org/10.3390/onco5030035
APA StyleYeh, C., Lai, H.-C., Grabbe, N., Willett, X., & Lin, S.-T. (2025). Revolutionizing Detection of Minimal Residual Disease in Breast Cancer Using Patient-Derived Gene Signature. Onco, 5(3), 35. https://doi.org/10.3390/onco5030035






