Association of JAK2 Haplotype GGCC_46/1 with the Response to Onco-Drug in MPNs Patients Positive for JAK2V617F Mutation
Abstract
Simple Summary
Abstract
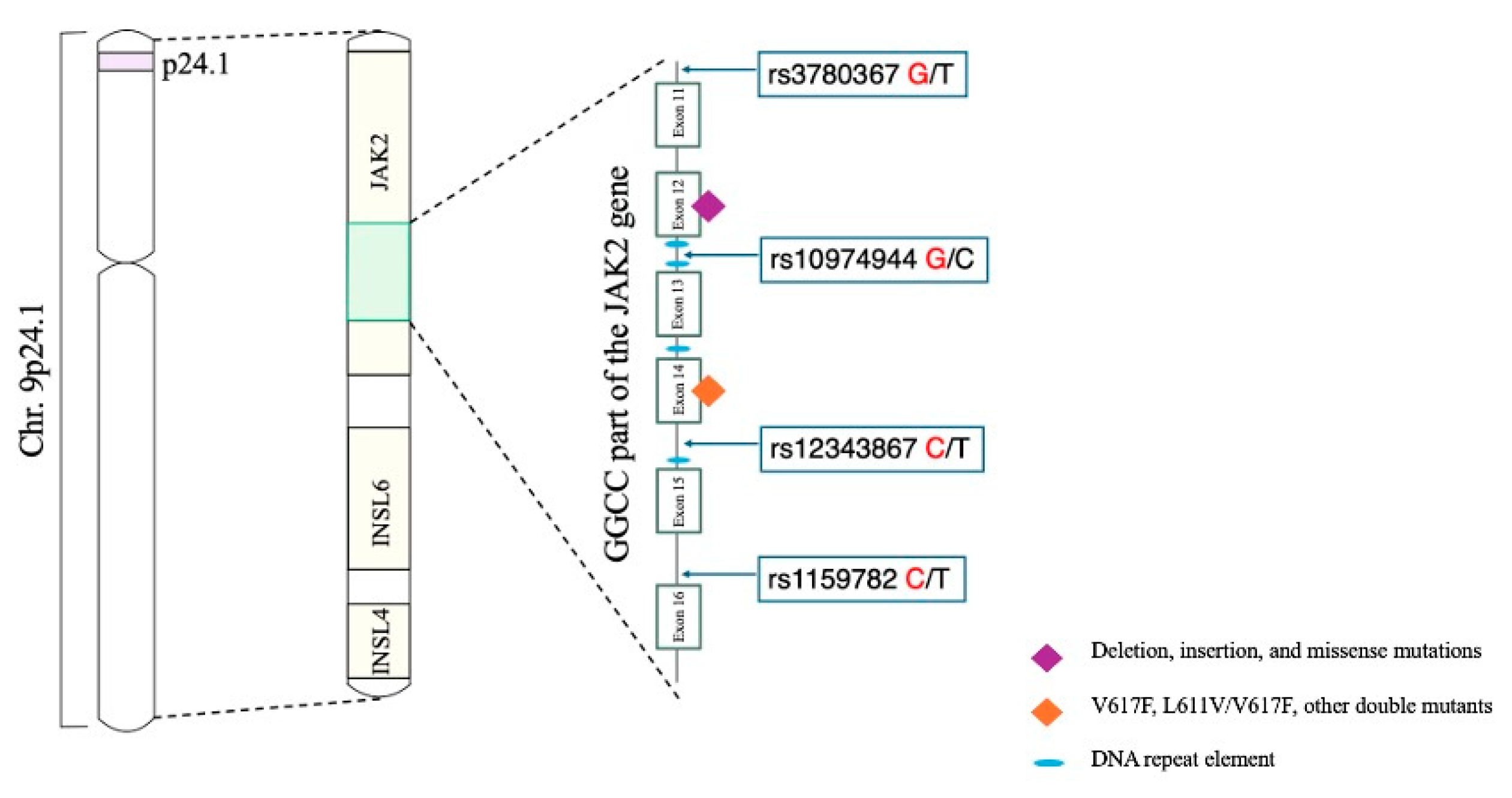
1. Introduction
2. Materials and Methods
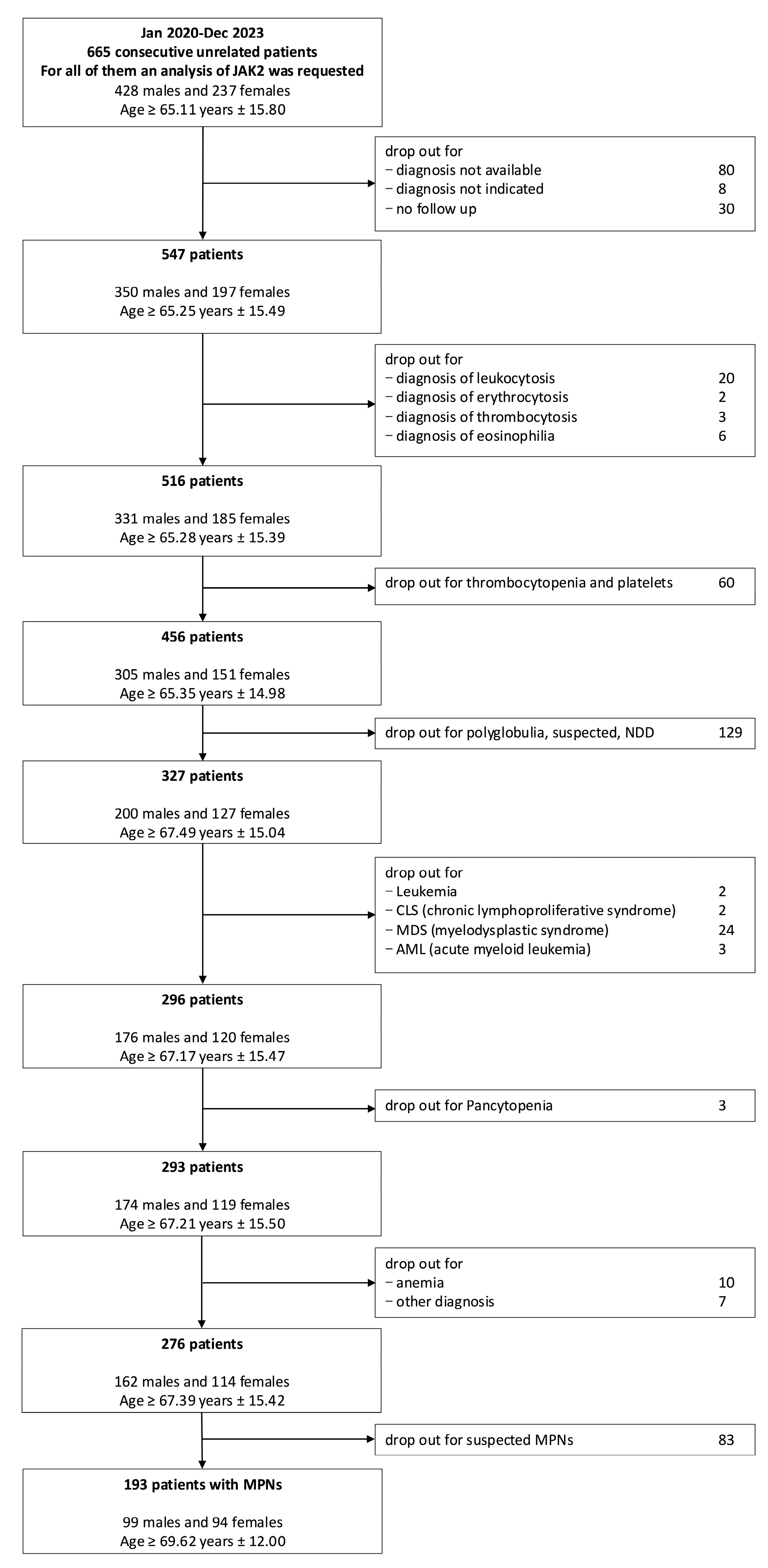
2.1. Selection of Patients
2.2. DNA Extraction from Fresh Blood Samples with NucleoSpin DX Blood*
2.3. JAK2 MutaQuant Analysis
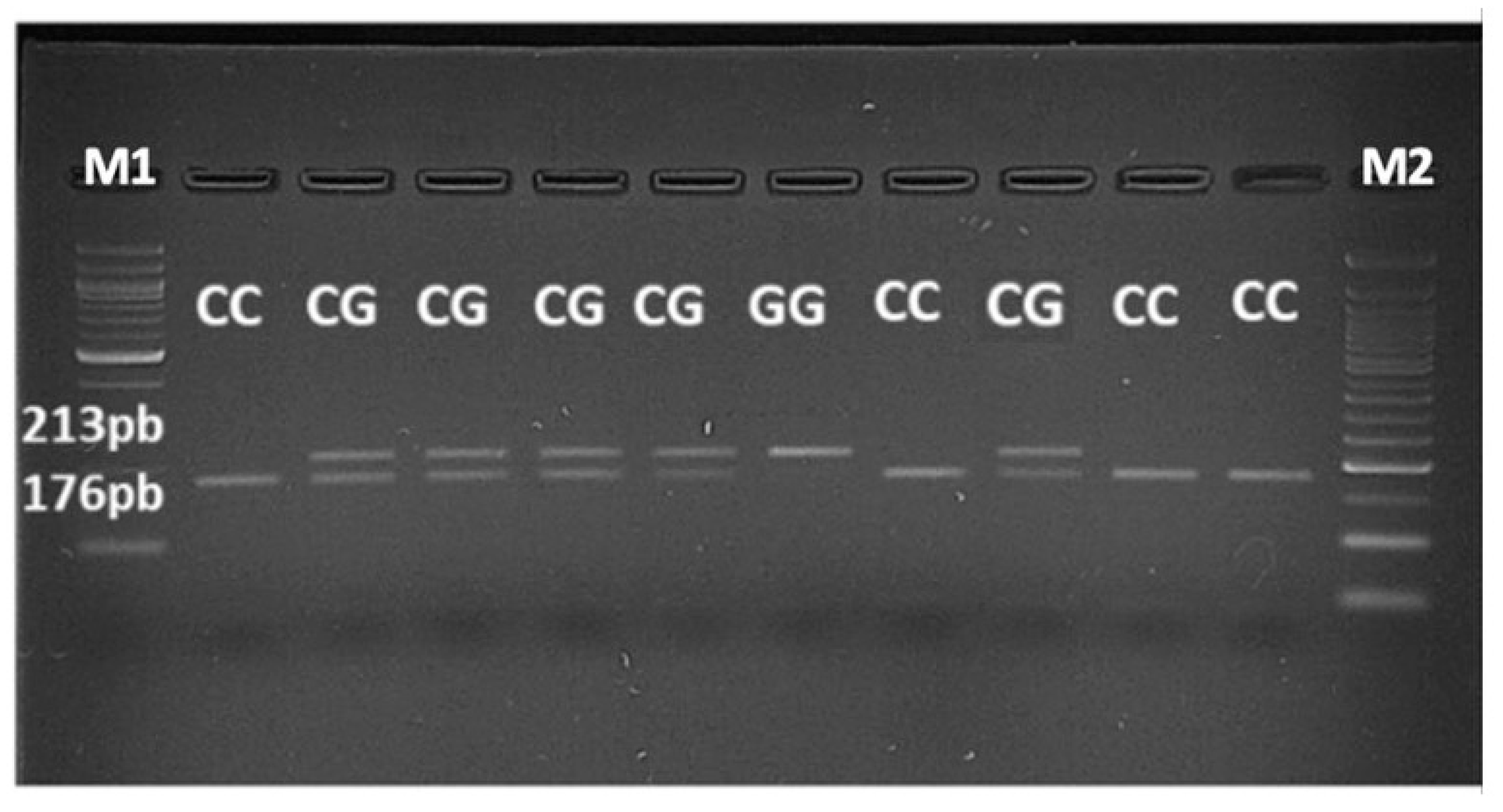
2.4. PCR-RFLP Assay
2.5. Statistical Analysis
3. Results
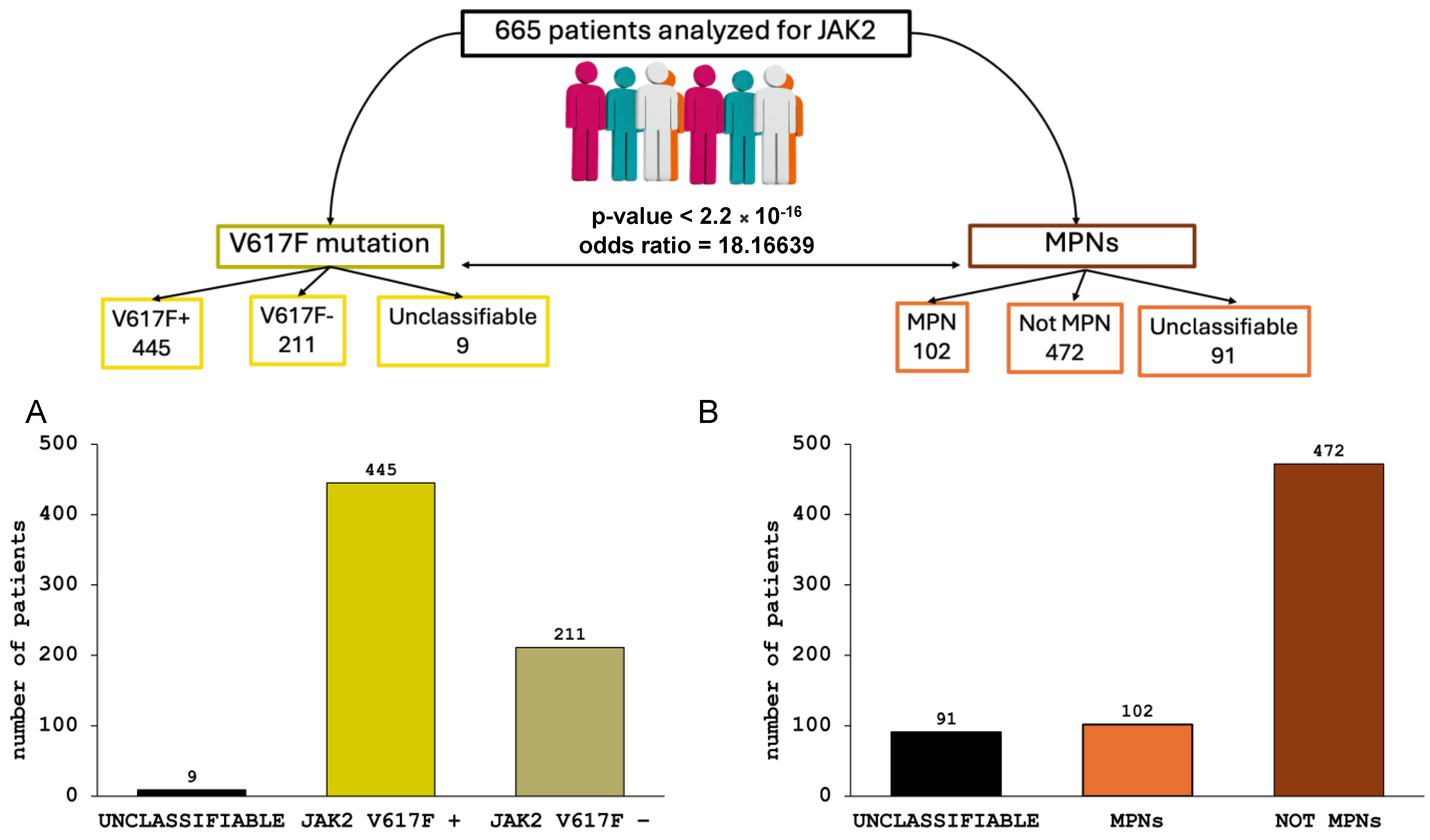
3.1. Prevalence and Correlations of JAK2–V617F Mutation in MPN Diseases
3.2. JAK2 Haplotype 46/1 Distribution in MPN Patients Tested Positive for JAK2 V617F Mutation
3.3. JAK2 Haplotype 46/1 Association with Therapy Response in MPNs Patients Positive for JAK2V617F Mutation
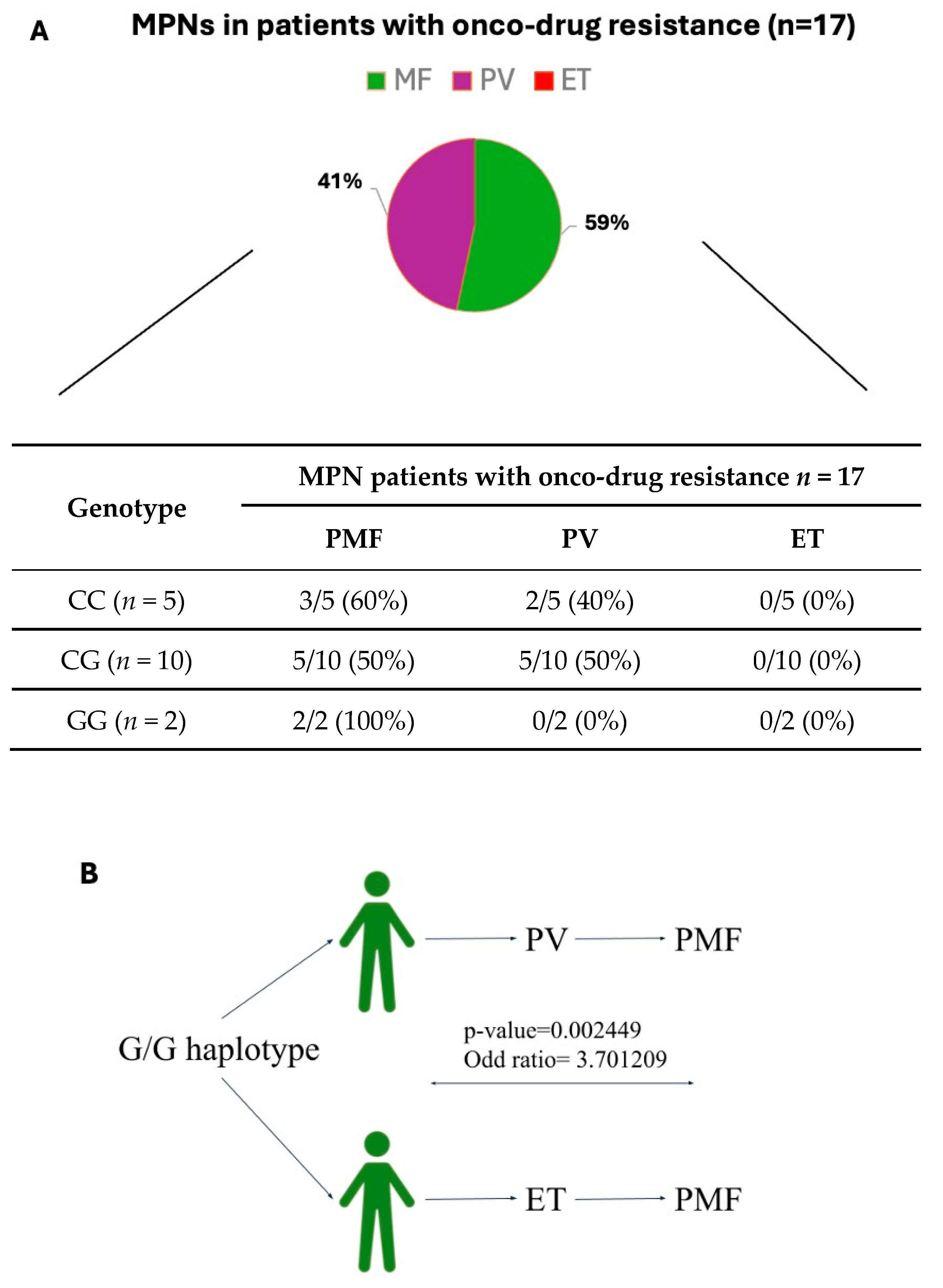
3.4. JAK2 Haplotype 46/1 Association with Onco-Drug Resistance Symptoms in JAK2V617F-Positive MPN Patients
3.5. JAK2 Haplotype 46/1 Contribution in MPN Patients with Drug Resistance
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paes, J.; Silva, G.A.V.; Tarragô, A.M.; Mourão, L.P.d.S. The Contribution of JAK2 46/1 Haplotype in the Predisposition to Myeloproliferative Neoplasms. Int. J. Mol. Sci. 2022, 23, 12582. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Pardanani, A.; Tefferi, A.; Gilliland, D.G. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat. Rev. Cancer 2007, 7, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, K.; Saharinen, P.; Pesu, M.; Holt, V.E.; Silvennoinen, O.; O’Shea, J.J. The Janus kinases (Jaks). Genome Biol. 2004, 5, 253. [Google Scholar] [CrossRef] [PubMed]
- Staerk, J.; Constantinescu, S.N. The JAK-STAT pathway and hematopoietic stem cells from the JAK2 V617F perspective. JAK-STAT 2012, 1, 184–190. [Google Scholar] [CrossRef]
- Torres, D.G.; Paes, J.; da Costa, A.G.; Malheiro, A.; Silva, G.V.; Mourão, L.P.d.S.; Tarragô, A.M. JAK2 Variant Signaling: Genetic, Hematologic and Immune Implication in Chronic Myeloproliferative Neoplasms. Biomolecules 2022, 12, 291. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dunbar, A.J.; Bowman, R.L.; Park, Y.C.; O’Connor, K.; Izzo, F.; Myers, R.M.; Karzai, A.; Zaroogian, Z.; Kim, W.J.; Fernández-Maestre, I.; et al. Jak2V617F Reversible Activation Shows Its Essential Requirement in Myeloproliferative Neoplasms. Cancer Discov. 2024, 14, 737–751. [Google Scholar] [CrossRef]
- Josil, J.; Thuillier, E.; Chambrun, L.; Plo, I. Décryptage de l’histoire naturelle des néoplasmes myéloprolifératifs grâce à une approche d’arbres phylogénétiques. M S-Med. Sci. 2024, 40, 209–211. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Mudireddy, M.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Pardanani, A. The germlineJAK2GGCC (46/1) haplotype and survival among 414 molecularly-annotated patients with primary myelofibrosis. Am. J. Hematol. 2018, 94, 299–305. [Google Scholar] [CrossRef]
- Jones, A.V.; Chase, A.; Silver, R.T.; Oscier, D.; Zoi, K.; Wang, Y.L.; Cario, H.; Pahl, H.L.; Collins, A.; Reiter, A.; et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 2009, 41, 446–449. [Google Scholar] [CrossRef]
- Olcaydu, D.; Harutyunyan, A.; Jäger, R.; Berg, T.; Gisslinger, B.; Pabinger, I.; Gisslinger, H.; Kralovics, R. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 2009, 41, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Kilpivaara, O.; Mukherjee, S.; Schram, A.M.; Wadleigh, M.; Mullally, A.; Ebert, B.L.; Bass, A.; Marubayashi, S.; Heguy, A.; Garcia-Manero, G.; et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat. Genet. 2009, 41, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Guglielmelli, P. The JAK2 46/1 (GGCC) MPN-predisposing haplotype: A risky haplotype, after all. Am. J. Hematol. 2018, 94, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Hermouet, S.; Vilaine, M. The JAK2 46/1 haplotype: A marker of inappropriate myelomonocytic response to cytokine stimulation, leading to increased risk of inflammation, myeloid neoplasm, and impaired defense against infection? Haematologica 2011, 96, 1575–1579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. The JAK2 GGCC (46/1) Haplotype in Myeloproliferative Neoplasms: Causal or Random? Int. J. Mol. Sci. 2018, 19, 1152. [Google Scholar] [CrossRef]
- Pagliarini-e-Silva, S.; Santos, B.C.; Pereira, E.M.d.F.; Ferreira, M.E.; Baraldi, E.C.; Sell, A.M.; Visentainer, J.E.L. Evaluation of the association between the JAK2 46/1 haplotype and chronic myeloproliferative neoplasms in a Brazilian population. Clinics 2013, 68, 5–9. [Google Scholar] [CrossRef]
- Macedo, L.C.; Santos, B.C.; Pagliarini-E-Silva, S.; Pagnano, K.B.B.; Rodrigues, C.; Quintero, F.C.; Ferreira, M.E.; Baraldi, E.C.; Ambrosio-Albuquerque, E.P.; Sell, A.M.; et al. JAK2 46/1 haplotype is associated with JAK2 V617F—Positive myeloproliferative neoplasms in Brazilian patients. Int. J. Lab. Hematol. 2015, 37, 654–660. [Google Scholar] [CrossRef]
- Ohyashiki, J.H.; Yoneta, M.; Hisatomi, H.; Iwabuchi, T.; Umezu, T.; Ohyashiki, K. The C allele of JAK2 rs4495487 is an additional candidate locus that contributes to myeloproliferative neoplasm predisposition in the Japanese population. BMC Med Genet. 2012, 13, 6. [Google Scholar] [CrossRef]
- Li, S.-L.; Zhang, P.-J.; Sun, G.-X.; Lu, Z.-J. The JAK2 46/1 haplotype (GGCC) in myeloproliferative neoplasms and splanchnic vein thrombosis: A pooled analysis of 26 observational studies. Ann. Hematol. 2014, 93, 1845–1852. [Google Scholar] [CrossRef]
- Koh, S.P.; Yip, S.P.; Lee, K.K.; Chan, C.C.; Lau, S.M.; Kho, C.S.; Lau, C.K.; Lin, S.Y.; Lau, Y.M.; Wong, L.G.; et al. Genetic association between germline JAK2 polymorphisms and myeloproliferative neoplasms in Hong Kong Chinese population: A case-control study. BMC Genet. 2014, 15, 147. [Google Scholar] [CrossRef][Green Version]
- Paes, J.F.; Torres, D.G.; Aquino, D.C.; Alves, E.V.B.; Mesquita, E.A.; Sousa, M.A.; Fraiji, N.A.; Passos, L.N.M.; Abreu, R.S.; Silva, G.A.V.; et al. Exploring hematological alterations and genetics linked to SNV rs10974944 in myeloproliferative neoplasms among Amazon patients. Sci. Rep. 2024, 14, 9389. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, X.; Hu, T.; Wu, Z.; Kang, Z.; Liu, W.; Guan, M. The JAK2 46/1 haplotype is a risk factor for myeloproliferative neoplasms in Chinese patients. Int. J. Hematol. 2012, 96, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Andrikovics, H.; Nahajevszky, S.; Koszarska, M.; Meggyesi, N.; Bors, A.; Halm, G.; Lueff, S.; Lovas, N.; Matrai, Z.; Csomor, J.; et al. JAK2 46/1 haplotype analysis in myeloproliferative neoplasms and acute myeloid leukemia. Leukemia 2010, 24, 1809–1813. [Google Scholar] [CrossRef] [PubMed]
- Pardanani, A.; Lasho, T.L.; Finke, C.M.; Gangat, N.; Wolanskyj, A.P.; Hanson, C.A.; Tefferi, A. The JAK2 46/1 haplotype confers susceptibility to essential thrombocythemia regardless of JAK2V617F mutational status—Clinical correlates in a study of 226 consecutive patients. Leukemia 2009, 24, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Biamonte, F.; Spolverini, A.; Pieri, L.; Isgrò, A.; Antonioli, E.; Pancrazzi, A.; Bosi, A.; Barosi, G.; Vannucchi, A.M. Frequency and clinical correlates of JAK2 46/1 (GGCC) haplotype in primary myelofibrosis. Leukemia 2010, 24, 1533–1537. [Google Scholar] [CrossRef]
- Verstovsek, S.; Mesa, R.A.; Gotlib, J.; Levy, R.S.; Gupta, V.; DiPersio, J.F.; Catalano, J.V.; Deininger, M.; Miller, C.; Silver, R.T.; et al. A Double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med. 2012, 366, 799–807. [Google Scholar] [CrossRef]
- Sobas, M.; Ianotto, J.-C.; Kiladjian, J.-J.; Harrison, C. Myeloproliferative neoplasms: Young patients, current data and future considerations. Ann. Hematol. 2024, 103, 3287–3291. [Google Scholar] [CrossRef]
- Passamonti, F.; Maffioli, M. The role of JAK2 inhibitors in MPNs 7 years after approval. Blood 2018, 131, 2426–2435. [Google Scholar] [CrossRef]
- Barbui, T.; Barosi, G.; Birgegard, G.; Cervantes, F.; Finazzi, G.; Griesshammer, M.; Harrison, C.; Hasselbalch, H.C.; Hehlmann, R.; Hoffman, R.; et al. Philadelphia-negative classical myeloproliferative neoplasms: Critical concepts and management recommendations from European LeukemiaNet. J. Clin. Oncol. 2011, 29, 761–770. [Google Scholar] [CrossRef]
- Trifa, A.P.; Cucuianu, A.; Popp, R.A. Development of a reliable PCR-RFLP assay for investigation of the JAK2 rs10974944 SNP, which might predispose to the acquisition of somatic mutation JAK2V617F. Acta Haematol. 2009, 123, 84–87. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Levine, R. The Spliceosome as an indicted conspirator in myeloid malignancies. Cancer Cell 2011, 20, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Duletic, A.N.; Dekanic, A.; Hadzisejdic, I.; Kusen, I.; Matusan-Llijas, K.; Grohovac, D.; Grahovac, B.; Jonjic, N. JAK2-V617F Mutation is associated with clinical and laboratory features of myeloproliferative neoplasms. Coll Antropol. 2012, 36, 859–865. [Google Scholar] [PubMed]
- Kucine, N. Myeloproliferative Neoplasms in Children, Adolescents, and Young Adults. Curr. Hematol. Malign. Rep. 2020, 15, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Barbui, T. Polycythemia vera and essential thrombocythemia: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, N.T.; Hau, B.B.; Vuong, N.B.; Xuan, N.T. JAK2 rs10974944 is associated with both V617F-positive and negative myeloproliferative neoplasms in a Vietnamese population: A potential genetic marker. Mol. Genet. Genom. Med. 2022, 10, e2044. [Google Scholar] [CrossRef]
- Trifa, A.P.; Cucuianu, A.; Petrov, L.; Urian, L.; Militaru, M.S.; Dima, D.; Pop, I.V.; Popp, R.A. The G allele of the JAK2 rs10974944 SNP, part of JAK2 46/1 haplotype, is strongly associated with JAK2 V617F-positive myeloproliferative neoplasms. Ann. Hematol. 2010, 89, 979–983. [Google Scholar] [CrossRef]
- Harrison, C.N.; Schaap, N.; Mesa, R.A. Management of myelofibrosis after ruxolitinib failure. Ann. Hematol. 2020, 99, 1177–1191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, J.G.; Chen, X.; Emm, T.; Scherle, P.A.; McGee, R.F.; Lo, Y.; Landman, R.R.; McKeever, E.G.; Punwani, N.G.; Williams, W.V.; et al. The Effect of CYP3A4 inhibition or induction on the pharmacokinetics and pharmacodynamics of orally administered ruxolitinib (INCB018424 Phosphate) in healthy volunteers. J. Clin. Pharmacol. 2012, 52, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Appeldoorn, T.Y.J.; Munnink, T.H.O.; Morsink, L.M.; Hooge, M.N.L.-D.; Touw, D.J. Pharmacokinetics and Pharmacodynamics of Ruxolitinib: A Review. Clin. Pharmacokinet. 2023, 62, 559–571. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schepers, K.; Campbell, T.B.; Passegué, E. Normal and leukemic stem cell niches: Insights and therapeutic opportunities. Cell Stem Cell 2015, 16, 254–267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rumi, E.; Cazzola, M. Diagnosis, risk stratification, and response evaluation in classical myeloproliferative neoplasms. Blood 2017, 129, 680–692. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Patnaik, M.M.; Finke, C.M.; Hussein, K.; Hogan, W.J.; Elliott, M.A.; Litzow, M.R.; Hanson, C.A.; Pardanani, A. JAK2 germline genetic variation affects disease susceptibility in primary myelofibrosis regardless of V617F mutational status: Nullizygosity for the JAK2 46/1 haplotype is associated with inferior survival. Leukemia 2009, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]

| Genotype | MPN Patients Positive for JAK2V617F Mutation (n = 50) | ||
|---|---|---|---|
| PMF n = 18 | PV n = 19 | ET n = 13 | |
| CC (n = 18) | 5/18 (28%) | 7/19 (37%) | 6/13 (46%) |
| CG (n = 29) | 11/18 (61%) | 12/19 (63%) | 6/13 (46%) |
| GG (n = 3) | 2/18 (11%) | 0/19 (0%) | 1/6 (8%) |
| Genotype | No Therapy n = 8 | p-Value | Therapy n = 25 | p-Value | Therapy Changed n = 17 | p-Value |
|---|---|---|---|---|---|---|
| CC | 4/8 (50%) | 0.4357 | 9/25 (36%) | 1 | 5/17 (29%) | 0.7568 |
| CG | 4/8 (50%) | 0.7058 | 15/25 (60%) | 1 | 10/17 (58%) | 0.7635 |
| GG | 0 | 1 | 1/25 (4%) | 1 | 2/17 (11%) | 1 |
| Genotype | Onco-Drug Resistance Symptoms | |||
|---|---|---|---|---|
| Splenomegaly n = 17 | Thrombocytosis n = 17 | Leukocytosis n = 17 | Hepatomegaly n = 17 | |
| CC | 4/5 (80%) | 2/5 (40%) | 3/5 (60%) | 1/5 (20%) |
| CG | 4/10 (40%) | 5/10 (50%) | 5/10 (50%) | 4/10 (40%) |
| GG | 1/2 (50%) | 2/2 (100%) | 2/2 (100%) | 1/2 (50%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrone, M.; Sergio, S.; Tarantino, A.; Loglisci, G.; Matera, R.; Seripa, D.; Maffia, M.; Di Renzo, N. Association of JAK2 Haplotype GGCC_46/1 with the Response to Onco-Drug in MPNs Patients Positive for JAK2V617F Mutation. Onco 2024, 4, 241-256. https://doi.org/10.3390/onco4030018
Perrone M, Sergio S, Tarantino A, Loglisci G, Matera R, Seripa D, Maffia M, Di Renzo N. Association of JAK2 Haplotype GGCC_46/1 with the Response to Onco-Drug in MPNs Patients Positive for JAK2V617F Mutation. Onco. 2024; 4(3):241-256. https://doi.org/10.3390/onco4030018
Chicago/Turabian StylePerrone, Michela, Sara Sergio, Amalia Tarantino, Giuseppina Loglisci, Rosella Matera, Davide Seripa, Michele Maffia, and Nicola Di Renzo. 2024. "Association of JAK2 Haplotype GGCC_46/1 with the Response to Onco-Drug in MPNs Patients Positive for JAK2V617F Mutation" Onco 4, no. 3: 241-256. https://doi.org/10.3390/onco4030018
APA StylePerrone, M., Sergio, S., Tarantino, A., Loglisci, G., Matera, R., Seripa, D., Maffia, M., & Di Renzo, N. (2024). Association of JAK2 Haplotype GGCC_46/1 with the Response to Onco-Drug in MPNs Patients Positive for JAK2V617F Mutation. Onco, 4(3), 241-256. https://doi.org/10.3390/onco4030018







