Role of microRNA in Oxidative Stress
Abstract
1. Introduction
2. miRNA Biology: Biogenesis and Action
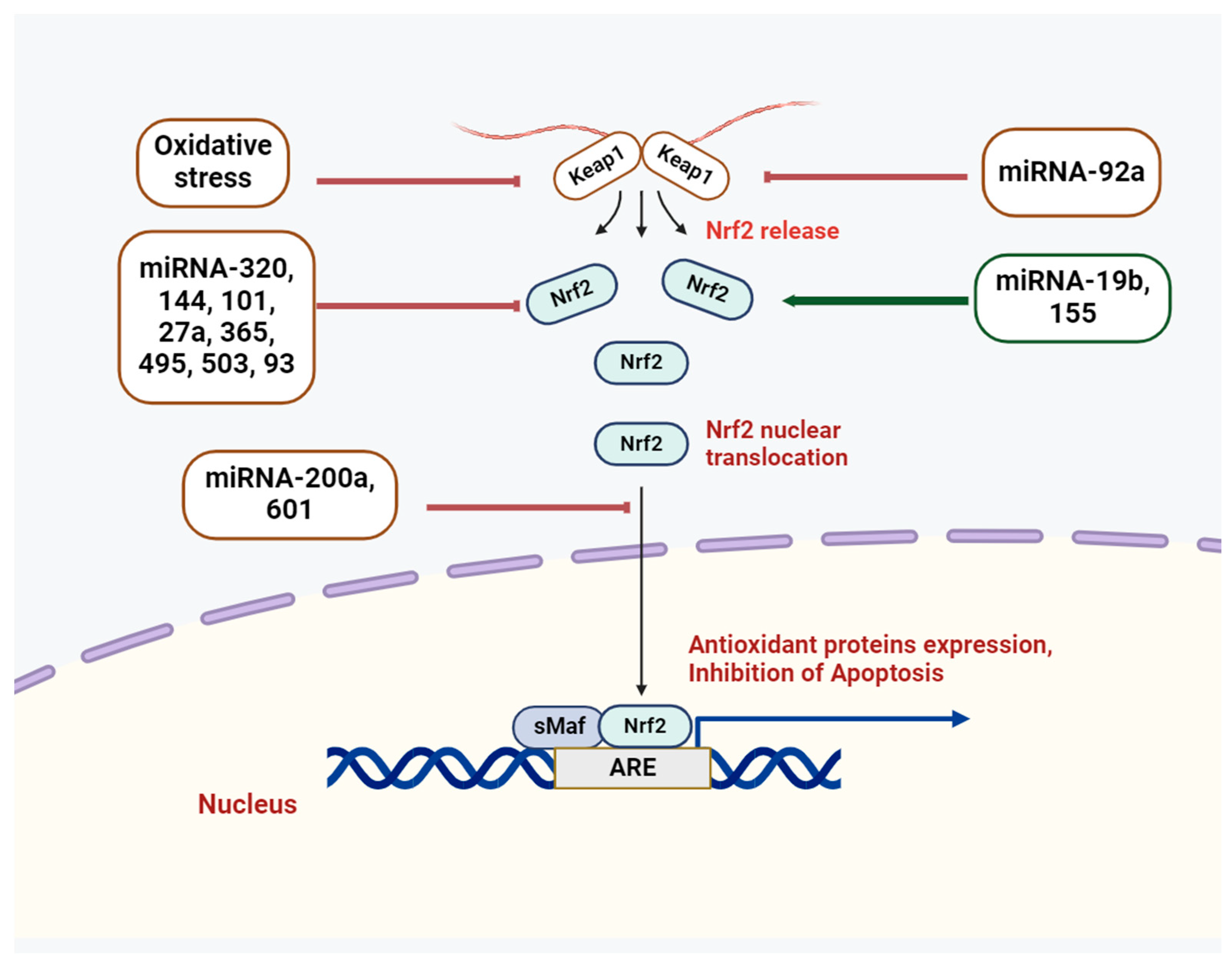
3. Cross-Talk of miRNAs and Nrf2
4. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Profumo, E.; Tucci, P.; Saso, L. A Perspective on Nrf2 Signaling Pathway for Neuroinflammation: A Potential Therapeutic Target in Alzheimer’s and Parkinson’s Diseases. Front. Cell. Neurosci. 2022, 15, 787258. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Saha, S.; Buttari, B.; Profumo, E.; Saso, L. Activation of Nrf2 signaling pathway by natural and synthetic chalcones: A therapeutic road map for oxidative stress. Expert Rev. Clin. Pharmacol. 2021, 14, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A. The Keap1-Nrf2pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Panieri, E.; Pinho, S.A.; Afonso, G.J.M.; Oliveira, P.J.; Cunha-Oliveira, T.; Saso, L. Nrf2 and Mitochondrial Function in Cancer and Cancer Stem Cells. Cells 2022, 11, 2401. [Google Scholar] [CrossRef] [PubMed]
- Boutten, A.; Goven, D.; Artaud-Macari, E.; Bonay, M. Protective role of Nrf2 in the lungs against oxidative airway diseases. Med. Sci. 2011, 27, 966–972. [Google Scholar]
- Gan, L.; Johnson, J.A. Oxidative damage and the Nrf2-ARE pathway in neurodegenerative diseases. Biochim. Biophys. Acta 2014, 1842, 1208–1218. [Google Scholar] [CrossRef]
- Howden, R. Nrf2 and cardiovascular defense. Oxidative Med. Cell. Longev. 2013, 2013, 104308. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Jiang, B.H. Interplay between reactive oxygen species and microRNAs in cancer. Curr. Pharmacol. Rep. 2016, 2, 82–90. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Bushell, M. The complexity of miRNA mediated repression. Cell Death Diff. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Liu, Y.; Qiang, W.; Xu, X.; Dong, R.; Karst, A.M.; Liu, Z.; Kong, B.; Drapkin, R.I.; Wei, J.J. Role of miR-182 in response to oxidative stress in the cell fate of human fallopian tube epithelial cells. Oncotarget 2015, 6, 38983–38998. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, S.; Martinez-Zamora, A.; Garcia-Arumi, E.; Andreu, A.L.; Armengod, M.E. The ROS-sensitive microRNA-9/9* controls the expression of mitochondrial tRNA-modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum. Mol. Genet. 2015, 24, 167–184. [Google Scholar] [CrossRef] [PubMed]
- La Sala, L.; Mrakic-Sposta, S.; Micheloni, S.; Prattichizzo, F.; Ceriello, A. Glucose-sensing microRNA-21 disrupts ROS homeostasis and impairs antioxidant responses in cellular glucose variability. Cardiovasc. Diabetol. 2018, 17, 105. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Kozak, J.; Jonak, K.; Maciejewski, R. The function of miR-200 family in oxidative stress response evoked in cancer chemotherapy and radiotherapy. Biomed. Pharmacother. 2020, 125, 110037. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem. 2010, 148, 381–392. [Google Scholar]
- Li, S.C.; Chan, W.C.; Hu, L.Y.; Lai, C.H.; Hsu, C.N.; Lin, W.C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2010, 96, 1–9. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Lizano, E.; Houben, A.J.; Bezdan, D.; Banez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [PubMed]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Li, M.; Vilborg, A.; Lee, N.; Shu, M.D.; Yartseva, V.; Šestan, N.; Steitz, J.A. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 2013, 155, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef] [PubMed]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, X.; Cheng, Y.; Yang, J.; Huo, Y.; Zhang, C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J. Biol. Chem. 2009, 284, 7903–7913. [Google Scholar] [CrossRef]
- Seo, J.; Jin, D.; Choi, C.H.; Lee, H. Integration of MicroRNA, mRNA, and protein expression data for the identification of cancer-related MicroRNAs. PLoS ONE 2017, 12, e0168412. [Google Scholar] [CrossRef]
- Chorley, B.N.; Campbell, M.R.; Wang, X.; Karaca, M.; Sambandan, D.; Bangura, F.; Xue, P.; Pi, J.; Kleeberger, S.R.; Bell, D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: Influence on retinoid X receptor alpha. Nucleic Acids Res. 2012, 40, 7416–7429. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Happel, C.; Manna, S.K.; Acquaah-Mensah, G.; Carrerero, J.; Kumar, S.; Nasipuri, P.; Krausz, K.W.; Wakabayashi, N.; Dewi, R.; et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Investig. 2013, 123, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- Kurinna, S.; Schafer, M.; Ostano, P.; Karouzakis, E.; Chiorino, G.; Bloch, W.; Bachmann, A.; Gay, S.; Garrod, D.; Lefort, K.; et al. A novel Nrf2-miR-29-desmocollin-2 axis regulates desmosome function in keratinocytes. Nat. Commun. 2014, 5, 5099. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Yi, Y.; Xia, G.; Zhao, Y.; Yu, Y.; Li, L.; Hua, C.; He, B.; Yang, B.; Yu, C.; et al. Nrf2-miR-129-3p-mTOR Axis Controls an miRNA Regulatory Network Involved in HDACi-Induced Autophagy. Mol. Ther. 2019, 27, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Rice, S.J.; Salzberg, A.C.; Runkle, E.A.; Liao, J.; Zander, D.S.; Mu, D. MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle 2012, 11, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.A.; Cenariu, D.; Irimie, A.; Magdo, L.; Nabavi, S.M.; Atanasov, A.G.; Berindan-Neagoe, I. The role of Nrf2 activity in cancer development and progression. Cancers 2019, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Kyeong-Ah, J.; Dayoung, R.; Geon, K.; Mi-Kyoung, K. 281—Role of NRF2 in MiR-181c/AMP-Activated Protein Kinase Signaling in Cancer. Free Radic. Biol. Med. 2016, 100, S124. [Google Scholar]
- Jung, K.A.; Lee, S.; Kwak, M.K. NFE2L2/NRF2 Activity Is Linked to Mitochondria and AMP-Activated Protein Kinase Signaling in Cancers through miR-181c/Mitochondria-Encoded Cytochrome c Oxidase Regulation. Antioxid. Redox Signal. 2017, 27, 945–961. [Google Scholar] [CrossRef]
- Chen, M.L.; Liang, L.S.; Wang, X.K. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin. Exp. Metastasis 2012, 29, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ohuchida, K.; Mizumoto, K.; Sato, N.; Kayashima, T.; Fujita, H.; Nakata, K.; Tanaka, M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer 2010, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liu, C.; Gao, F.; Mitchel, R.E.; Zhao, L.; Yang, Y.; Lei, J.; Cai, J. MiR-200c enhances radiosensitivity of human breast cancer cells. J. Cell. Biochem. 2012, 114, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Hamano, R.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Hara, J.; ho Moon, J.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M.; et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 2011, 17, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Shen, L.; Yu, X.; Wu, J.; Zha, L.; Xia, Y.; Luo, H. miR-200a attenuated oxidative stress, inflammation, and apoptosis in dextran sulfate sodium-induced colitis through activation of Nrf2. Front. Immunol. 2023, 14, 1196065. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, H.; Wang, Z.; Hu, Z.; Li, L. miR-200a attenuated doxorubicin-induced cardiotoxicity through upregulation of Nrf2 in mice. Oxidative Med. Cell. Longev. 2019, 2019, 1512326. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Wu, J.; Ma, F.; Jiang, J.; Xu, L.; Du, L.; Huang, W.; Wang, Z.; Jia, Y.; Lu, L.; et al. MicroRNA-200a improves diabetic endothelial dysfunction by targeting KEAP1/NRF2. J. Endocrinol. 2020, 24, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.J.; Yu, H.W.; Yang, Y.Z.; Wu, W.Y.; Chen, T.Y.; Jia, K.K.; Kang, L.-L.; Jiao, R.-Q.; Kong, L.-D. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. 2018, 18, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jin, Y.; Li, L.; Xu, L.; Tang, Z.; Qi, Y.; Yin, L.; Peng, J. MicroRNA-128-3p aggravates doxorubicin-induced liver injury by promoting oxidative stress via targeting Sirtuin-1. Pharmacol. Res. 2019, 146, 104276. [Google Scholar] [CrossRef]
- Zhao, L.; Qi, Y.; Xu, L.; Tao, X.; Han, X.; Yin, L.; Peng, J. MicroRNA-140-5p aggravates doxorubicin-induced cardiotoxicity by promoting myocardial oxidative stress via targeting Nrf2 and Sirt2. Redox Biol. 2018, 15, 284–296. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, Y.; Luo, Y.; Wang, Z.; Bi, S.; Song, D.; Dai, Y.; Wang, T.; Qiu, L.; Wen, L.; et al. Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1-Nrf2, Tgfβ1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic. Biol. Med. 2014, 67, 91–102. [Google Scholar] [CrossRef]
- Akdemir, B.; Nakajima, Y.; Inazawa, J.; Inoue, J. miR-432 induces NRF2 stabilization by directly targeting KEAP1. Mol. Cancer Res. 2017, 15, 1570–1578. [Google Scholar] [CrossRef]
- Silva-Palacios, A.; Ostolga-Chavarria, M.; Zazueta, C.; Konigsberg, M. Nrf2: Molecular and epigenetic regulation during aging. Ageing Res. Rev. 2018, 47, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.B.; Lai, X.; Guo, X.F. Activation of Nrf2 by miR-152 Inhibits Doxorubicin-Induced Cardiotoxicity via Attenuation of Oxidative Stress, Inflammation, and Apoptosis. Oxidative Med. Cell. Longev. 2021, 2021, 8860883. [Google Scholar] [CrossRef]
- Zhang, C.; Kong, X.; Ma, D. miR-141-3p inhibits vascular smooth muscle cell proliferation and migration via regulating Keap1/Nrf2/HO-1 pathway. IUBMB Life 2020, 72, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Z.; Tang, Y.; Cheng, L.B.; Yao, J.; Jiang, Q.; Li, K.R.; Zhen, Y.-F. Targeting Keap1 by miR-626 protects retinal pigment epithelium cells from oxidative injury by activating Nrf2 signaling. Free Radic. Biol. Med. 2019, 143, 387–396. [Google Scholar] [CrossRef]
- Sangokoya, C.; Telen, M.J.; Chi, J.-T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 2010, 116, 4338–4348. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yao, Y.; Eades, G.; Zhang, Y.; Zhou, Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res. Treat. 2011, 129, 983–991. [Google Scholar] [CrossRef]
- Singh, B.; Ronghe, A.M.; Chatterjee, A.; Bhat, N.K.; Bhat, H.K. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis 2013, 34, 1165–1172. [Google Scholar] [CrossRef]
- Hussen, B.M.; Abdullah, S.R.; Rasul, M.F.; Jawhar, Z.H.; Faraj, G.S.H.; Kiani, A.; Taheri, M. MiRNA-93: A novel signature in human disorders and drug resistance. Cell Commun. Signal. 2023, 21, 79. [Google Scholar] [CrossRef]
- Zimmerman, A.L.; Wu, S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011, 300, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, M.; Riar, A.K.; Rathinam, M.L.; Vedpathak, D.; Henderson, G.; Mahimainathan, L. Hydrogen peroxide responsive miR153 targets Nrf2/ARE cytoprotection in paraquat induced dopaminergic neurotoxicity. Toxicol. Lett. 2014, 228, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sun, T.; Liu, Z.; Chen, X.; Zhao, L.; Qu, G.; Li, Q. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS ONE 2014, 9, e103948. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011, 132, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, Y.; Qin, J.; Lu, S. The role of miR-34a in the hepatoprotective effect of hydrogen sulfide on ischemia/reperfusion injury in young and old rats. PLoS ONE 2014, 9, e113305. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, N.; Prattichizzo, F.; Martino, E.; Anastasio, C.; Mele, L.; La Grotta, R.; Sardu, C.; Ceriello, A.; Marfella, R.; Paolisso, G.; et al. MiR-27b attenuates mitochondrial oxidative stress and inflammation in endothelial cells. Redox Biol. 2023, 62, 102681. [Google Scholar] [CrossRef]
- Shah, N.M.; Zaitseva, L.; Bowles, K.M.; MacEwan, D.J.; Rushworth, S.A. NRF2-driven miR-125B1 and miR-29B1 transcriptional regulation controls a novel anti-apoptotic miRNA regulatory network for AML survival. Cell Death Differ. 2015, 22, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Joo, M.S.; Lee, C.G.; Koo, J.H.; Kim, S.G. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis. 2013, 4, e899. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wu, L.; Chen, Z.; Yang, J.; Chen, X.; Yu, F.; Zheng, F.; Lin, X. MiR-141 activates Nrf2-dependent antioxidant pathway via down-regulating the expression of keap1 conferring the resistance of hepatocellular carcinoma cells to 5-fluorouracil. Cell. Physiol. Biochem. 2015, 35, 2333–2348. [Google Scholar] [CrossRef]
- Shi, L.; Chen, Z.-G.; Wu, L.-L.; Zheng, J.-J.; Yang, J.-R.; Chen, X.-F.; Chen, Z.-Q.; Liu, C.-L.; Chi, S.-Y.; Zheng, J.-Y.; et al. miR-340 reverses cisplatin resistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Asian Pac. J. Cancer Prev. 2014, 15, 10439–10444. [Google Scholar] [CrossRef]
- Solich, J.; Kolasa, M.; Faron-Górecka, A.; Pabian, P.; Latocha, K.; Korlatowicz, A.; Dziedzicka-Wasylewska, M. Modulating Stress Susceptibility and Resilience: Insights from miRNA Manipulation and Neural Mechanisms in Mice. Mol. Neurobiol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Hsin-Yi, C.; Yu-Min, L.; Hsiang-Ching, C.; Yaw-Dong, L.; Ching-Jung, L.; John, H.; Wei-Chi, W.; Feng-Mao, L.; Zhen, C.; Hsien-Da, H.; et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Canc. Res. 2012, 72, 3631–3641. [Google Scholar]
- Wang, X.; Yuan, B.; Cheng, B.; Liu, Y.; Zhang, B.; Wang, X.; Lin, X.; Yang, B.; Gong, G. Crocin alleviates myocardial ischemia/reperfusion-induced endoplasmic reticulum stress via regulation of miR-34a/Sirt1/Nrf2 pathway. Shock 2019, 51, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, H.; Luo, C.; Du, D.; Huang, J.; Ming, Q.; Jin, F.; Wang, D.; Huang, W. Acetaminophen responsive miR-19b modulates SIRT1/Nrf2 signaling pathway in drug-induced hepatotoxicity. Toxicol. Sci. 2019, 170, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.A.; Gao, L.F.; Zhang, Z.G.; Xiang, D.K. Down-regulation of miR-320 exerts protective effects on myocardial I-R injury via facilitating Nrf2 expression. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1730–1741. [Google Scholar] [PubMed]
- Cookson, M.R. Parkinson’s Disease, Disease-Modifying Targets in Neurodegenerative Disorders; Elsevier: Amsterdam, The Netherlands, 2017; pp. 157–174. [Google Scholar]
- Liao, Y.H.; Xia, N.; Zhou, S.F.; Tang, T.T.; Yan, X.X.; Lv, B.J.; Nie, S.-F.; Wang, J.; Iwakura, Y.; Xiao, H.; et al. Interleukin-17A contributes to myocardial ischemia/reperfusion injury by regulating cardiomyocyte apoptosis and neutrophil infiltration. J. Am. Coll. Cardiol. 2012, 59, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Lu, Z.; Lin, V.; May, A.; Shaw, D.H.; Wang, Z.; Che, B.; Tran, K.; Du, H.; Shaw, P.X. MicroRNA miR-24-3p reduces apoptosis and regulates Keap1-Nrf2 pathway in mouse cardiomyocytes responding to ischemia/reperfusion injury. Oxidative Med. Cell. Longev. 2018, 2018, 7042105. [Google Scholar] [CrossRef]
- Huang, R.; Ma, J.; Niu, B.; Li, J.; Chang, J.; Zhang, Y.; Liu, P.; Luan, X. MiR-34b protects against focal cerebral ischemia-reperfusion (I/R) injury in rat by targeting Keap1. J. Stroke Cerebrovasc. Dis. 2019, 28, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Q.; Zhang, Y.H.; Shang, X.Q.; Zeng, Y.J. Effects of miR-101 on the proliferation and apoptosis of gastric mucosal epithelial cells via Nrf2/ARE signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5187–5194. [Google Scholar]
- Wu, L.L.; Cai, W.P.; Lei, X.; Shi, K.Q.; Lin, X.Y.; Shi, L. NRAL mediates cisplatin resistance in hepatocellular carcinoma via miR-340-5p/Nrf2 axis. J. Cell Commun. Signal. 2019, 13, 99–112. [Google Scholar] [CrossRef]
- Geng, J.F.; Liu, X.; Zhao, H.B.; Fan, W.F.; Geng, J.J.; Liu, X.Z. LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int. J. Biochem. Cell Biol. 2018, 99, 133–139. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, W.; Jiao, G.; Li, C.; Liu, H. MiR-455-3p activates Nrf2/ARE signaling via HDAC2 and protects osteoblasts from oxidative stress. Int. J. Biol. Macromol. 2018, 107, 2094–2101. [Google Scholar] [CrossRef]
- Kang, J.S.; Cho, D.; Kim, Y.I.; Hahm, E.; Kim, Y.S.; Jin, S.N.; Na Kim, H.; Kim, D.; Hur, D.; Park, H.; et al. Sodium ascorbate (vitamin C) induces apoptosis in melanoma cells via the down-regulation of transferrin receptor dependent iron uptake. J. Cell. Physiol. 2005, 204, 192–197. [Google Scholar] [CrossRef]
- Talasila, K.M.; Soentgerath, A.; Euskirchen, P.; Rosland, G.V.; Wang, J.; Huszthy, P.C.; Prestegarden, L.; Skaftnesmo, K.O.; Sakariassen, P.; Eskilsson, E.; et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013, 125, 683–698. [Google Scholar] [CrossRef]
- Xu, D.; Zhu, H.; Wang, C.; Zhu, X.; Liu, G.; Chen, C.; Cui, Z. microRNA-455 targets cullin 3 to activate Nrf2 signaling and protect human osteoblasts from hydrogen peroxide. Oncotarget 2017, 8, 59225. [Google Scholar] [CrossRef][Green Version]
- Qu, J.; Zhang, L.; Li, L.; Su, Y. miR-148b functions as a tumor suppressor by targeting endoplasmic reticulum metallo protease 1 in human endometrial cancer cells. Oncol. Res. 2018, 27, 81–88. [Google Scholar] [CrossRef]
- Zhang, X.; Ng, W.L.; Wang, P.; Tian, L.; Werner, E.; Wang, H.; Doetsch, P.; Wang, Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012, 72, 4707–4713. [Google Scholar] [CrossRef]
- Karshovska, E.; Wei, Y.; Subramanian, P.; Mohibullah, R.; Geißler, C.; Baatsch, I.; Popal, A.; Campos, J.C.; Exner, N.; Schober, A. HIF-1α (Hypoxia-Inducible Factor-1α) Promotes Macrophage Necroptosis by Regulating miR-210 and miR-383. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 583–596. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, S. Role of microRNA in Oxidative Stress. Stresses 2024, 4, 269-281. https://doi.org/10.3390/stresses4020016
Saha S. Role of microRNA in Oxidative Stress. Stresses. 2024; 4(2):269-281. https://doi.org/10.3390/stresses4020016
Chicago/Turabian StyleSaha, Sarmistha. 2024. "Role of microRNA in Oxidative Stress" Stresses 4, no. 2: 269-281. https://doi.org/10.3390/stresses4020016
APA StyleSaha, S. (2024). Role of microRNA in Oxidative Stress. Stresses, 4(2), 269-281. https://doi.org/10.3390/stresses4020016





