Ester Production Using the Lipid Composition of Coffee Ground Oil (Coffea arabica): A Theoretical Study of Eversa® Transform 2.0 Lipase as an Enzymatic Biocatalyst
Abstract
1. Introduction
2. Materials and Methods
2.1. Homology Modeling
2.1.1. Identification and selection of protein-fold
2.1.2. Alignment of Target and Mold Sequences
2.1.3. Model Construction and Optimization
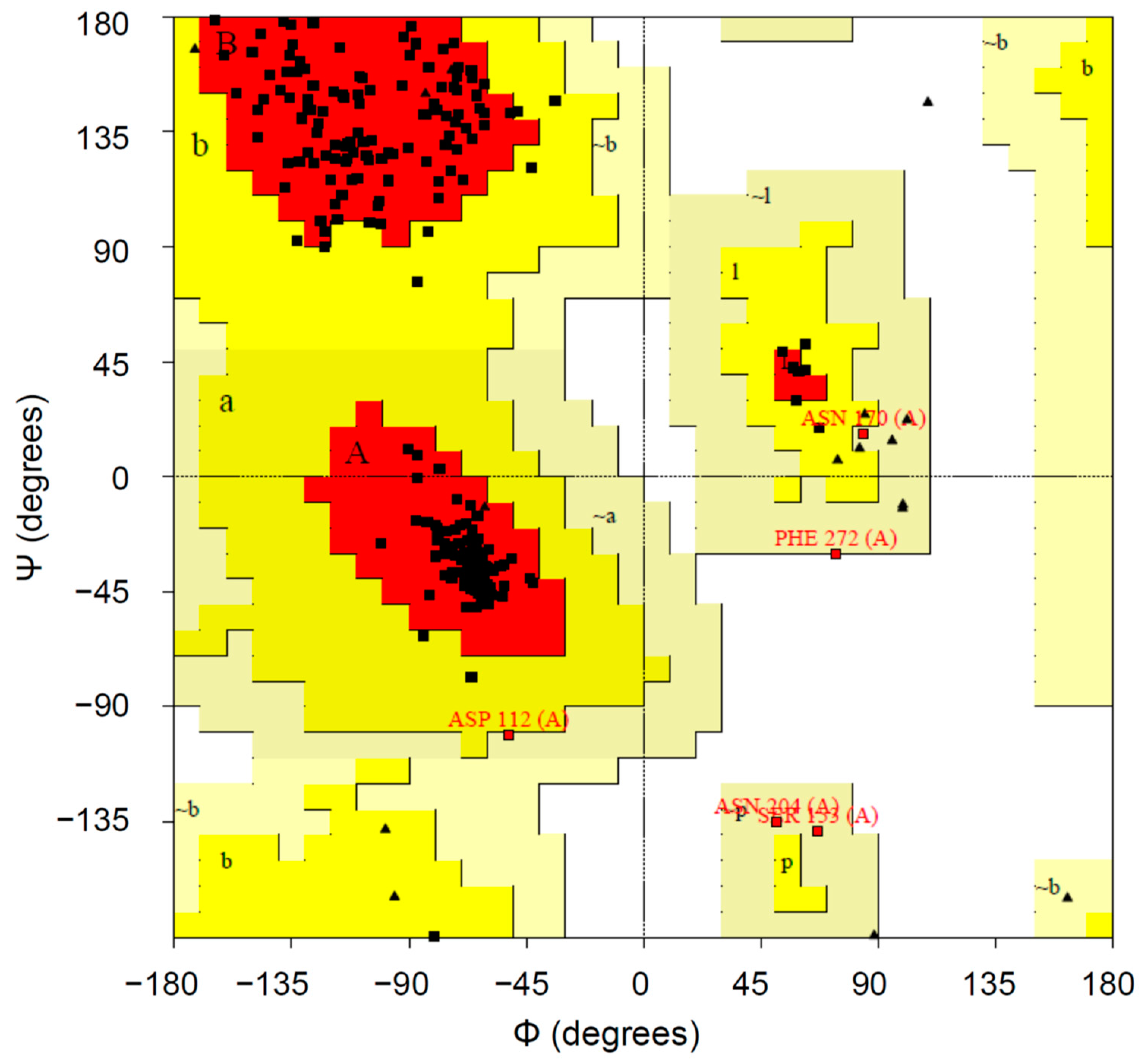
2.1.4. Protein Validation
2.2. Protein Preparation
2.3. Obtaining the Ligand
2.4. Molecular Docking and Visualization of Calculations
2.5. Molecular Dynamic
- Potential energy (kcal/mol) [45];
- Protein–ligand interaction energy (kcal/mol);
- The root mean square deviation of the atomic positions of proteins, binders, and the distances between them (RMSD, Å), and the root mean square deviation of the atomic positions of proteins, ligands, and the distances between them (RMSD, Å);
- Hydrogen bonds were evaluated using Visual Molecular Dynamics (VMD) [46];
- The mean square fluctuation of the minimum distances between proteins and ligands was observed in MD (RMSF, Å) [47]. The plots were generated using the Qtrace program.
- In this study, MD simulations were used to evaluate the stability of a viral protease enzyme with various ligands containing different amounts of α-helix and β-sheets [48]. The long-range interactions were calculated using the SPME method and a Langevin thermal bath at 310 K. The conformational changes of the protein during the MD simulations were described using root mean square deviations (RMSD).
MM/GBSA Calculations
3. Results and Discussion
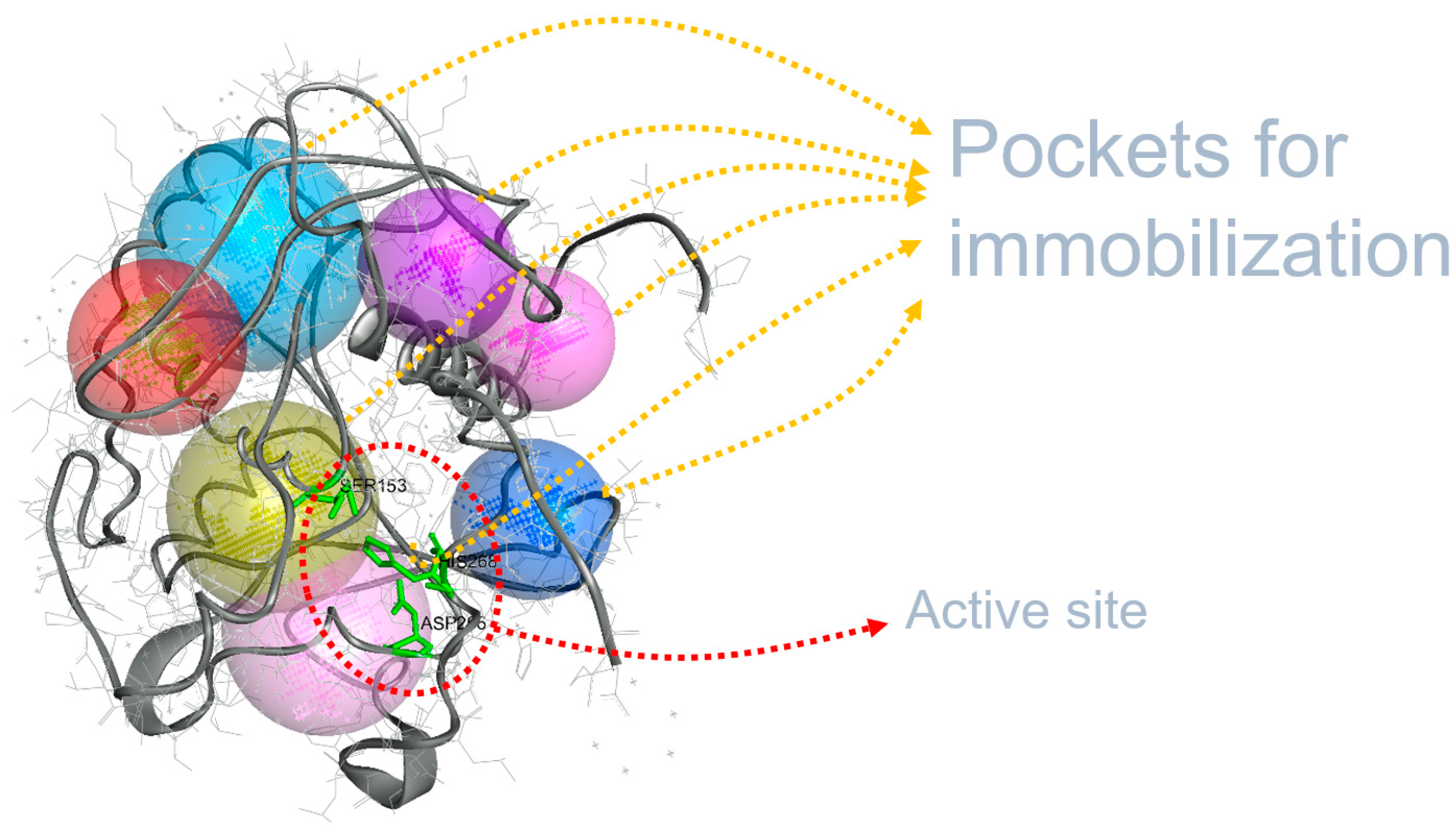
3.1. Immobilization Locations
3.2. Protein Modeling
3.3. Interaction between Substrate and Lipase
3.4. Molecular Dynamics
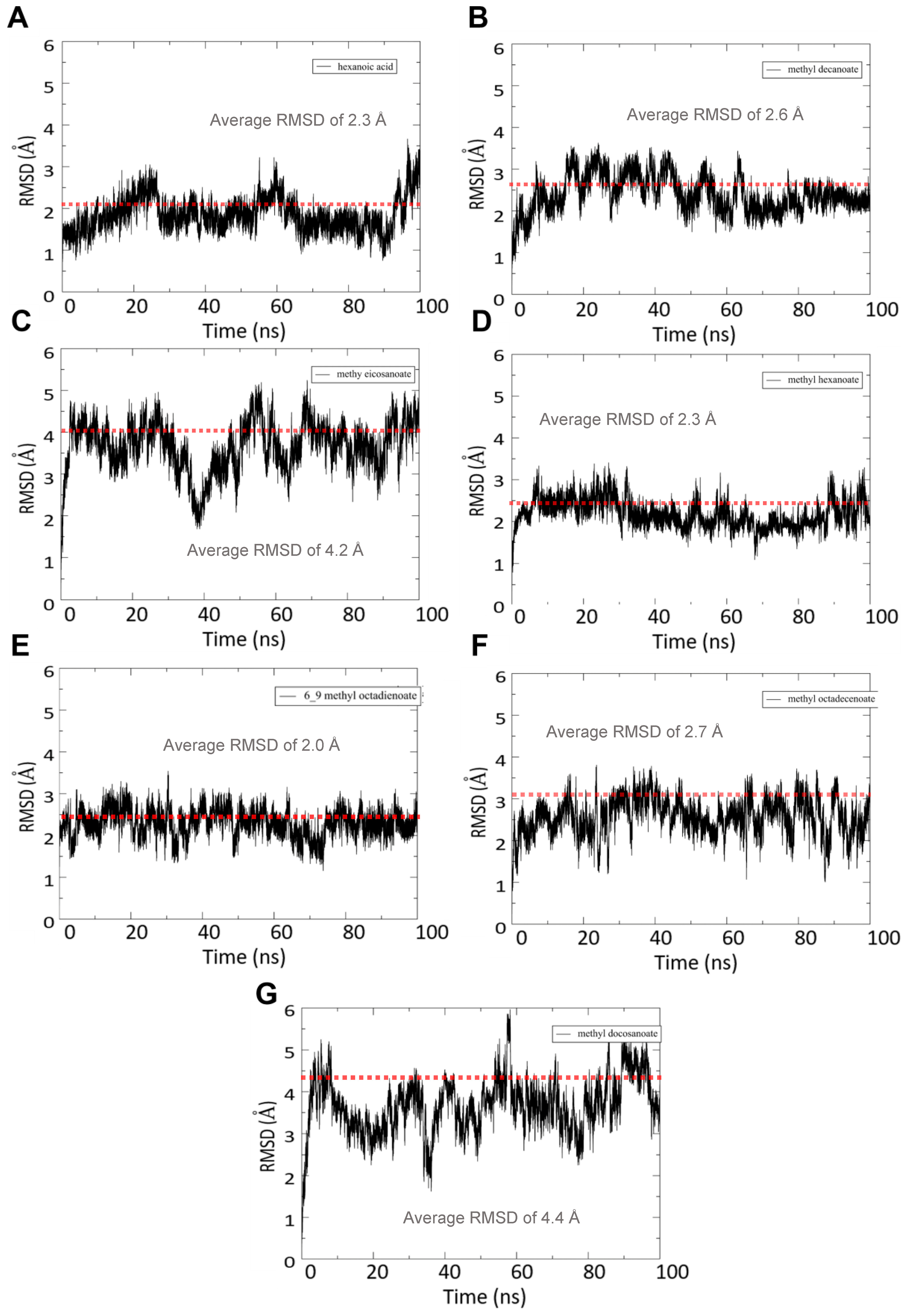
3.4.1. RMSD Analysis
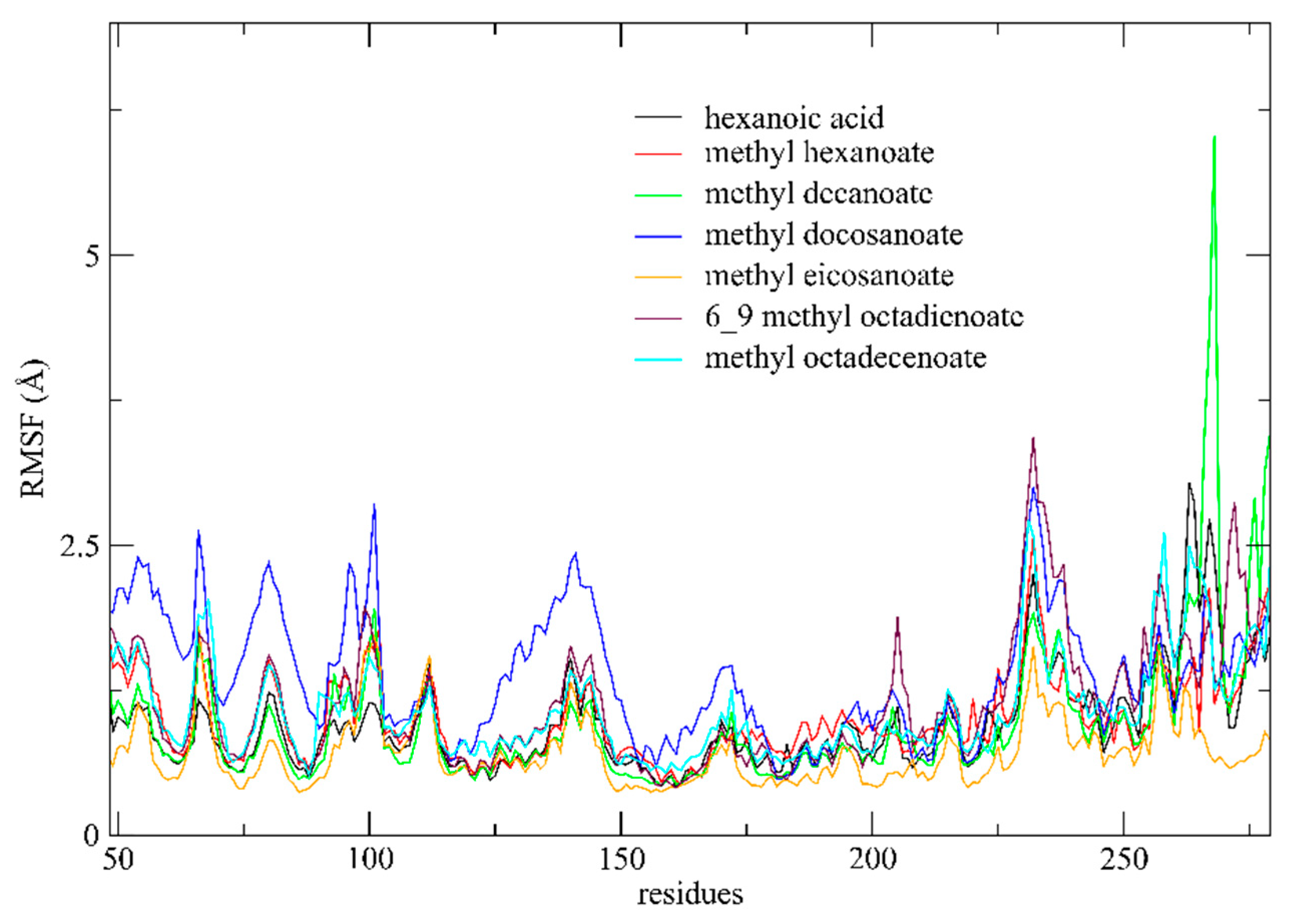
3.4.2. RMSF Analysis
3.4.3. H-bonds Analysis
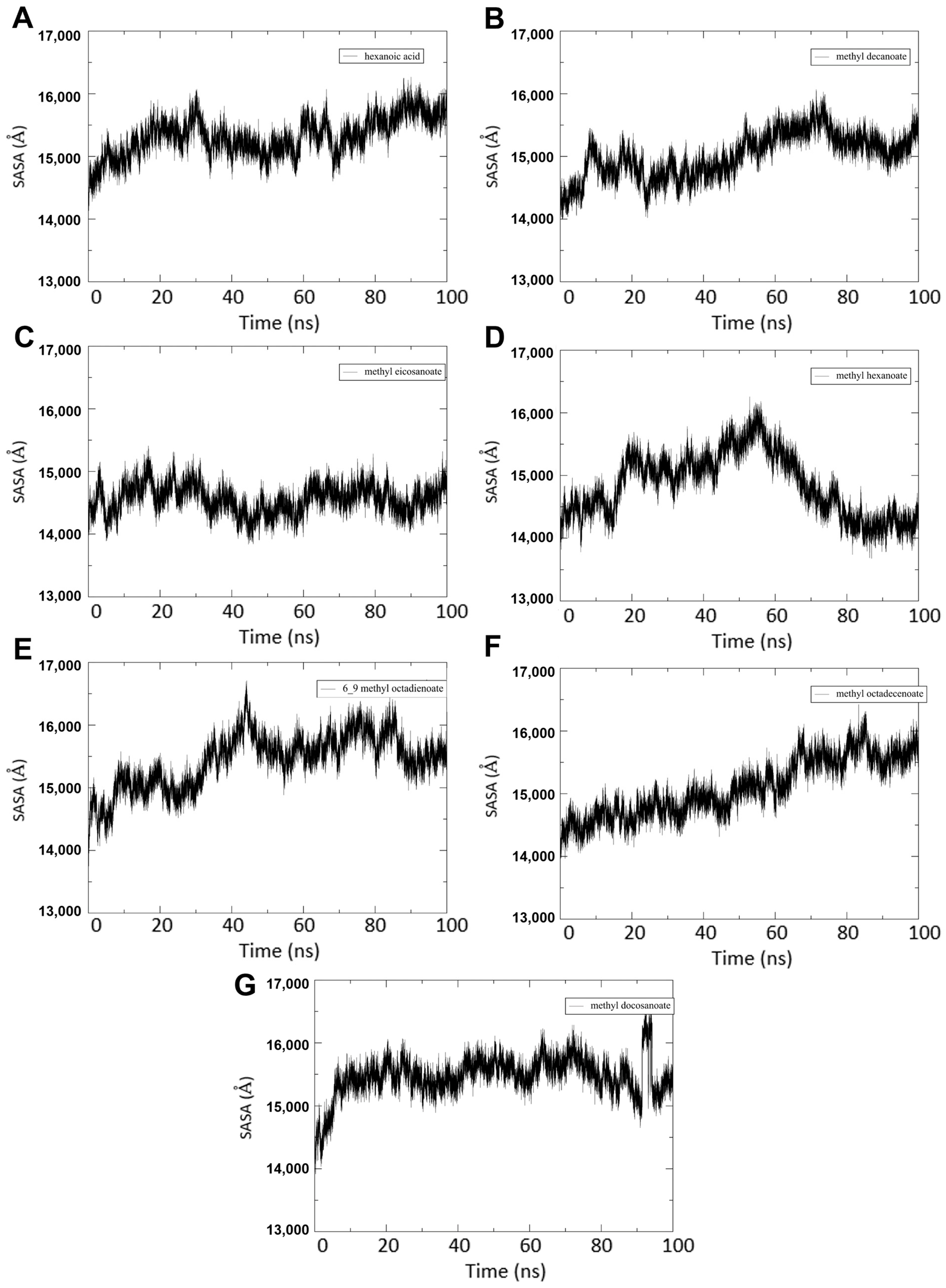
3.4.4. SASA Calculations
3.4.5. MM/GBSA Calculations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asensio-Ramos, M.; D’Orazio, G. Capillary electromigration techniques: Application to coffee analysis—A review. J. Chromatogr. Open 2023, 3, 100083. [Google Scholar] [CrossRef]
- Battista, F.; Zuliani, L.; Rizzioli, F.; Fusco, S.; Bolzonella, D. Biodiesel, Biogas and Fermentable Sugars Production from Spent Coffee Grounds: A Cascade Biorefinery Approach. Bioresour. Technol. 2021, 342, 125952. [Google Scholar] [CrossRef] [PubMed]
- Sarno, M.; Iuliano, M. Active Biocatalyst for Biodiesel Production from Spent Coffee Ground. Bioresour. Technol. 2018, 266, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Mueanmas, C.; Nikhom, R.; Petchkaew, A.; Iewkittayakorn, J.; Prasertsit, K. Extraction and Esterification of Waste Coffee Grounds Oil as Non-Edible Feedstock for Biodiesel Production. Renew. Energy 2019, 133, 1414–1425. [Google Scholar] [CrossRef]
- Bitencourt, R.G.; Mello, F.M.P.A.; Cabral, F.A.; Meirelles, A.J.A. High-Pressure Fractionation of Spent Coffee Grounds Oil Using Green Solvents. J. Supercrit. Fluids 2020, 157, 104689. [Google Scholar] [CrossRef]
- Anika, O.C.; Nnabuife, S.G.; Bello, A.; Okoroafor, E.R.; Kuang, B.; Villa, R. Prospects of Low and Zero-Carbon Renewable Fuels in 1.5-Degree Net Zero Emission Actualisation by 2050: A Critical Review. Carbon Capture Sci. Technol. 2022, 5, 100072. [Google Scholar] [CrossRef]
- Rahman, M.N.; Wahid, M.A. Renewable-Based Zero-Carbon Fuels for the Use of Power Generation: A Case Study in Malaysia Supported by Updated Developments Worldwide. Energy Rep. 2021, 7, 1986–2020. [Google Scholar] [CrossRef]
- Singh, P.; Bishnoi, S.K. Modified Moth-Flame Optimization for Strategic Integration of Fuel Cell in Renewable Active Distribution Network. Electr. Power Syst. Res. 2021, 197, 107323. [Google Scholar] [CrossRef]
- Chang, M.Y.; Chan, E.S.; Song, C.P. Biodiesel Production Catalysed by Low-Cost Liquid Enzyme Eversa® Transform 2.0: Effect of Free Fatty Acid Content on Lipase Methanol Tolerance and Kinetic Model. Fuel 2021, 283, 119266. [Google Scholar] [CrossRef]
- Salih, N. A Review on Eco-Friendly Green Biolubricants from Renewable and Sustainable Plant Oil Sources. Biointerface Res. Appl. Chem. 2021, 11, 13303–13327. [Google Scholar]
- dos Santos, J.C.S.; Garcia-Galan, C.; Rodrigues, R.C.; de Sant’ Ana, H.B.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Improving the Catalytic Properties of Immobilized Lecitase via Physical Coating with Ionic Polymers. Enzyme Microb. Technol. 2014, 60, 1–8. [Google Scholar] [CrossRef]
- da Fonseca, A.M.; de Freitas, Í.B.; Soares, N.B.; de Araújo, F.A.M.; Gaieta, E.M.; Dos Santos, J.C.S.; Sobrinho, A.C.N.; Marinho, E.S.; Colares, R.P. Synthesis, Biological Activity, and in Silico Study of Bioesters Derived from Bixin by the Calb Enzyme. Biointerface Res. Appl. Chem. 2022, 12, 5901–5917. [Google Scholar] [CrossRef]
- Garcia-galan, C.; Barbosa, O.; Hernandez, K.; Santos, J.C.S.; Rodrigues, R.C.; Fernandez-lafuente, R. Evaluation of Styrene-Divinylbenzene Beads as a Support to Immobilize Lipases. Molecules 2014, 19, 7629–7645. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.R.; Bhanderi, K.K.; Patel, J.V. A Review on Bio-Lubricants from Non-Edible Oils-Recent Advances, Chemical Modifications and Applications. J. Indian Chem. Soc. 2023, 100, 100849. [Google Scholar] [CrossRef]
- Parandi, E.; Safaripour, M.; Abdellattif, M.H.; Saidi, M.; Bozorgian, A.; Rashidi, H.; Rezania, S. Biodiesel Production from Waste Cooking Oil Using a Novel Biocatalyst of Lipase Enzyme Immobilized Magnetic Nanocomposite. Fuel 2022, 313, 123057. [Google Scholar] [CrossRef]
- Silva, J.M.F.; dos Santos, K.P.; dos Santos, E.S.; Rios, N.S.; Gonçalves, L.R.B. Immobilization of Thermomyces Lanuginosus Lipase on a New Hydrophobic Support (Streamline PhenylTM): Strategies to Improve Stability and Reusability. Enzyme Microb. Technol. 2023, 163, 110166. [Google Scholar] [CrossRef]
- Remonatto, D.; Jr, R.H.M.; Monti, R.; Bassan, J.C.; Paula, A.V. De Applications of Immobilized Lipases in Enzymatic Reactors: A Review. Process Biochem. 2022, 114, 1–20. [Google Scholar] [CrossRef]
- Sampaio, C.S.; Angelotti, J.A.F.; Fernandez-lafuente, R.; Hirata, D.B. International Journal of Biological Macromolecules Lipase Immobilization via Cross-Linked Enzyme Aggregates: Problems and Prospects—A Review. Int. J. Biol. Macromol. 2022, 215, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, C.E.C.; Neto, F.S.; De Castro Bizerra, V.; Do Nascimento, J.G.A.; Valério, R.B.R.; De Sousa Junior, P.G.; De Sousa Braz, A.K.; Melo, R.L.F.; De França Serpa, J.; De Lima, R.K.C.; et al. Production sustainable bioethanol from first- and second-generation sugar-based feedstocks: Advanced bibliometric analysis. Bioresour. Technol. Rep. 2023, 23, 101543. [Google Scholar] [CrossRef]
- Feedstocks, L.A.; Vieira, A.C.; Ana, B.; Guimar, R.; Marquettotti, A.; Vieira, S.; Fernandez-lafuente, R.; Tardioli, P.W. Performance of Liquid Eversa on Fatty Acid Ethyl Esters Production by Simultaneous Esterification/Transesterification of Low-to-High Acidity Feedstocks. Catalysts 2021, 11, 1486. [Google Scholar]
- Bresolin, D.; Hawerroth, B.; de Oliveira Romera, C.; Sayer, C.; de Araújo, P.H.H.; de Oliveira, D. Immobilization of Lipase Eversa Transform 2.0 on Poly(Urea–Urethane) Nanoparticles Obtained Using a Biopolyol from Enzymatic Glycerolysis. Bioprocess Biosyst. Eng. 2020, 43, 1279–1286. [Google Scholar] [CrossRef]
- Bonazza, H.L.; Manzo, R.M.; dos Santos, J.C.S.; Mammarella, E.J. Operational and Thermal Stability Analysis of Thermomyces Lanuginosus Lipase Covalently Immobilized onto Modified Chitosan Supports. Appl. Biochem. Biotechnol. 2018, 184, 182–196. [Google Scholar] [CrossRef]
- De Sousa, I.G.; Chaves, A.V.; Barros, A.L.; Oliveira, D.; Moreira, S.; Gonçalves, P.; Junior, D.S.; Neto, F.S.; Cristina, S.; Carvalho, F.D.; et al. A Novel Hybrid Biocatalyst from Immobilized Eversa Transform 2.0 Lipase and Its Application in Biolubricant Synthesis. Biocatal. Biotransform. 2022, 1–22. [Google Scholar] [CrossRef]
- Kato, K.; Nakayoshi, T.; Kurimoto, E.; Oda, A. Molecular Dynamics Simulations for the Protein–Ligand Complex Structures Obtained by Computational Docking Studies Using Implicit or Explicit Solvents. Chem. Phys. Lett. 2021, 781, 139022. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Cavalcante, A.L.G.; Chaves, A.V.; Fechine, P.B.A.; Holanda Alexandre, J.Y.N.; Freire, T.M.; Davi, D.M.B.; Neto, F.S.; de Sousa, I.G.; da Silva Moreira, K.; de Oliveira, A.L.B.; et al. Chemical Modification of Clay Nanocomposites for the Improvement of the Catalytic Properties of Lipase A from Candida Antarctica. Process Biochem. 2022, 120, 1–14. [Google Scholar] [CrossRef]
- Rodrigues, A.F.S.; Ananias, F.; Francisco, L.B.; Kaiany, M.; De Oliveira, M.P.; Nobre, M.M.R.; Catumba, B.D.; Sales, M.B.; Braz, S.; Cavalcante, A.L.G.; et al. A Scientometric Analysis of Research Progress and Trends in the Design of Laccase Biocatalysts for the Decolorization of Synthetic Dyes. Process Biochem. 2023, 126, 272–291. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinforma. 2016, 54, 1–37. [Google Scholar] [CrossRef]
- Bedoya, O.F.; Tischer, I. Detección de Homología Remota de Proteínas Usando Modelos 3D Enriquecidos Con Propiedades Fisicoquímicas. Ing. Y Compet. 2015, 17, 75–84. [Google Scholar] [CrossRef]
- Ramachandran, G.N.R.C.; Sasisekharan, V. Stereochemistry of Polypeptide Chain Conformations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Ahmadi, M.; Jahed Motlagh, M.; Rahmani, A.T.; Zolfagharzadeh, M.M.; Shariatpanahi, P.; Chermack, T.J.; Coons, L.M.; Cotter, J.; Eyiah-Donkor, E.; Poti, V.; et al. Chem3D 15.0 User Guide. Macromolecules 2005, 24, 1–61. [Google Scholar]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Cen, Y.; Singh, W.; Arkin, M.; Moody, T.S.; Huang, M.; Zhou, J.; Wu, Q.; Reetz, M.T. Artificial Cysteine-Lipases with High Activity and Altered Catalytic Mechanism Created by Laboratory Evolution. Nat. Commun. 2019, 10, 3198. [Google Scholar] [CrossRef]
- Ben Hlima, H.; Dammak, M.; Karray, A.; Drira, M.; Michaud, P.; Fendri, I.; Abdelkafi, S. Molecular and Structural Characterizations of Lipases from Chlorella by Functional Genomics. Mar. Drugs 2021, 19, 70. [Google Scholar] [CrossRef]
- PyMOL Molecular Graphics System. Version 1.3r1. Schrödinger, LLC: New York, NY, USA, 2010. Available online: https://pymol.org/ (accessed on 12 March 2023).
- Shityakov, S.; Förster, C. In Silico Predictive Model to Determine Vector-Mediated Transport Properties for the Blood-Brain Barrier Choline Transporter. Adv. Appl. Bioinforma. Chem. 2014, 7, 23–36. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Farago, O. Langevin Thermostat for Robust Configurational and Kinetic Sampling. Phys. A Stat. Mech. its Appl. 2019, 534, 122210. [Google Scholar] [CrossRef]
- Diez, M.; Petuya, V.; Martínez-Cruz, L.A.; Hernández, A. Insights into Mechanism Kinematics for Protein Motion Simulation. BMC Bioinform. 2014, 15, 1–14. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Arshia, A.H.; Shadravan, S.; Solhjoo, A.; Sakhteman, A.; Sami, A. De Novo Design of Novel Protease Inhibitor Candidates in the Treatment of SARS-CoV-2 Using Deep Learning, Docking, and Molecular Dynamic Simulations. Comput. Biol. Med. 2021, 139, 104967. [Google Scholar] [CrossRef] [PubMed]
- Gohlke, H.; Case, D.A. Converging Free Energy Estimates: MM-PB(GB)SA Studies on the Protein-Protein Complex Ras-Raf. J. Comput. Chem. 2004, 25, 238–250. [Google Scholar] [CrossRef]
- Gohlke, H.; Kiel, C.; Case, D.A. Insights into Protein-Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras-Raf and Ras-RalGDS Complexes. J. Mol. Biol. 2003, 330, 891–913. [Google Scholar] [CrossRef]
- DasGupta, D.; Mandalaparthy, V.; Jayaram, B. A Component Analysis of the Free Energies of Folding of 35 Proteins: A Consensus View on the Thermodynamics of Folding at the Molecular Level. J. Comput. Chem. 2017, 38, 2791–2801. [Google Scholar] [CrossRef]
- Pal, G.; Araújo, L.S.; Vidal, M.S.; Baldani, J.I.; Henrique, C.; Gadelha, S. Modelagem Molecular de Proteínas: O Caso de Uma Glucoronosiltransferase (GumK) de Gluconacetobacter Diazotrophicus PAL5. J. Biol. Pharm. Agric. Manag. 2014, 10, 1–6. [Google Scholar]
- Chaturvedi, S.K.; Zaidi, N.; Alam, P.; Khan, J.M.; Qadeer, A.; Siddique, I.A.; Asmat, S.; Zaidi, Y.; Khan, R.H. Unraveling Comparative Anti-Amyloidogenic Behavior of Pyrazinamide and D-Cycloserine: A Mechanistic Biophysical Insight. PLoS ONE 2015, 10, 1–21. [Google Scholar] [CrossRef]
- Silvestrini, L.; Cianci, M. Principles of Lipid–Enzyme Interactions in the Limbus Region of the Catalytic Site of Candida Antarctica Lipase B. Int. J. Biol. Macromol. 2020, 158, 358–363. [Google Scholar] [CrossRef]
- Hari Krishna, S.; Karanth, N.G. Lipases and Lipase-Catalyzed Esterification Reactions in Nonaqueous Media. Catal. Rev. Sci. Eng. 2002, 44, 499–591. [Google Scholar] [CrossRef]
- Stergiou, P.Y.; Foukis, A.; Filippou, M.; Koukouritaki, M.; Parapouli, M.; Theodorou, L.G.; Hatziloukas, E.; Afendra, A.; Pandey, A.; Papamichael, E.M. Advances in Lipase-Catalyzed Esterification Reactions. Biotechnol. Adv. 2013, 31, 1846–1859. [Google Scholar] [CrossRef] [PubMed]
- Corici, L.; Pellis, A.; Ferrario, V.; Ebert, C.; Cantone, S.; Gardossi, L. Understanding Potentials and Restrictions of Solvent-Free Enzymatic Polycondensation of Itaconic Acid: An Experimental and Computational Analysis. Adv. Synth. Catal. 2015, 357, 1763–1774. [Google Scholar] [CrossRef]
- Struchtrup, H. Entropy and the Second Law of Thermodynamics-The Nonequilibrium Perspective. Entropy 2020, 22, 793. [Google Scholar] [CrossRef]
- Beretta, G.P. The Fourth Law of Thermodynamics: Steepest Entropy Ascent. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2020, 378, 1–17. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Byléhn, F.; Menéndez, C.A.; Perez-Lemus, G.R.; Alvarado, W.; De Pablo, J.J. Modeling the Binding Mechanism of Remdesivir, Favilavir, and Ribavirin to SARS-CoV-2 RNA-Dependent RNA Polymerase. ACS Cent. Sci. 2021, 7, 164–174. [Google Scholar] [CrossRef]
- Cavallari, M.; Ghio, C.; Monti, S.; Ferrario, M.; Maritan, A.; Carloni, P. Partially Folded States of HIV-1 Protease: Molecular Dynamics Simulations and Ligand Binding. J. Mol. Struct. THEOCHEM 2006, 769, 111–121. [Google Scholar] [CrossRef]
- Vorobjev, Y.N. Free Energies of Protein Decoys Provide Insight into Determinants of Protein Stability. Protein Sci. 2001, 10, 2498–2506. [Google Scholar] [CrossRef]
- Qin, X.; Zhong, J.; Wang, Y. A Mutant T1 Lipase Homology Modeling, and Its Molecular Docking and Molecular Dynamics Simulation with Fatty Acids. J. Biotechnol. 2021, 337, 24–34. [Google Scholar] [CrossRef]
- Roe, D.R.; Brooks, B.R. A Protocol for Preparing Explicitly Solvated Systems for Stable Molecular Dynamics Simulations. J. Chem. Phys. 2020, 153, 1–9. [Google Scholar] [CrossRef]
- Thirumalai, D.; Lorimer, G.H.; Hyeon, C. Iterative Annealing Mechanism Explains the Functions of the GroEL and RNA Chaperones. Protein Sci. 2020, 29, 1–18. [Google Scholar] [CrossRef]
- Mascoli, V.; Liguori, N.; Cupellini, L.; Elias, E.; Mennucci, B.; Croce, R. Uncovering the Interactions Driving Carotenoid Binding in Light-Harvesting Complexes. Chem. Sci. 2021, 12, 5113–5122. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, A.; Malathi, K.; Anbarasu, A. MurB as a Target in an Alternative Approach to Tackle the Vibrio Cholerae Resistance Using Molecular Docking and Simulation Study. J. Cell. Biochem. 2018, 119, 1726–1732. [Google Scholar] [CrossRef]
- Mazola, Y.; Guirola, O.; Palomares, S.; Chinea, G.; Menéndez, C.; Hernández, L.; Musacchio, A. A Comparative Molecular Dynamics Study of Thermophilic and Mesophilic β-Fructosidase Enzymes. J. Mol. Model. 2015, 21, 1–11. [Google Scholar] [CrossRef]
- Street, A.G.; Mayo, S.L. Pairwise Calculation of Protein Solvent-Accessible Surface Areas. Fold. Des. 1998, 3, 253–258. [Google Scholar] [CrossRef]
- Durham, E.; Dorr, B.; Woetzel, N.; Staritzbichler, R.; Meiler, J. Solvent Accessible Surface Area Approximations for Rapid and Accurate Protein Structure Prediction. J. Mol. Model. 2009, 15, 1093–1108. [Google Scholar] [CrossRef]
- Marsh, J.A.; Teichmann, S.A. Relative Solvent Accessible Surface Area Predicts Protein Conformational Changes upon Binding. Structure 2011, 19, 859–867. [Google Scholar] [CrossRef]
- Novotny, M.; Seibert, M.; Kleywegt, G.J. On the Precision of Calculated Solvent-Accessible Surface Areas. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 270–274. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar]
- Wright, D.W.; Hall, B.A.; Kenway, O.A.; Jha, S.; Coveney, P.V. Computing Clinically Relevant Binding Free Energies of HIV-1 Protease Inhibitors. J. Chem. Theory Comput. 2014, 10, 1228–1241. [Google Scholar] [CrossRef]
- Ben-Tal, N.; Honig, B.; Bagdassarian, C.K.; Ben-Shaul, A. Association Entropy in Adsorption Processes. Biophys. J. 2000, 79, 1180–1187. [Google Scholar] [CrossRef]
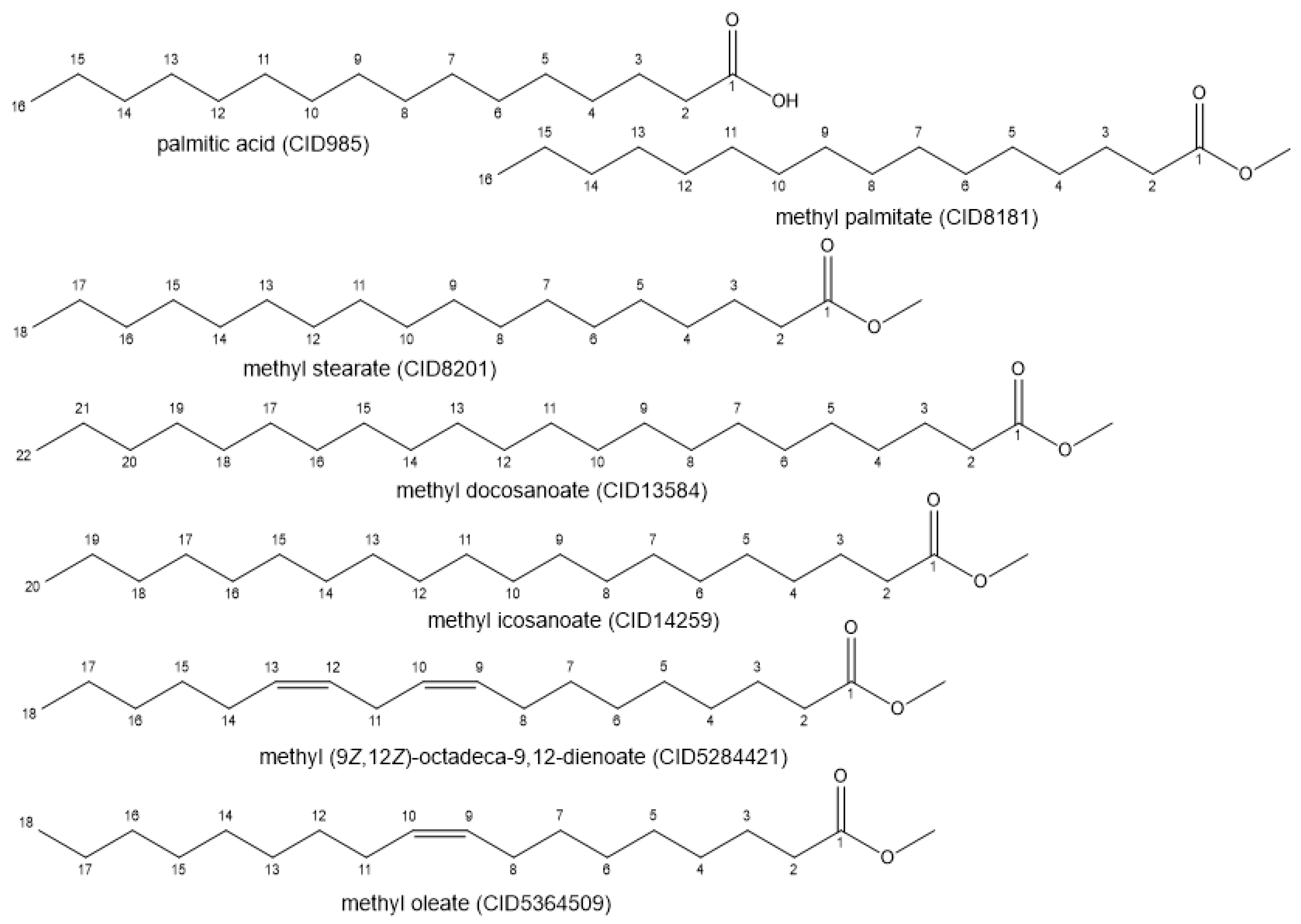
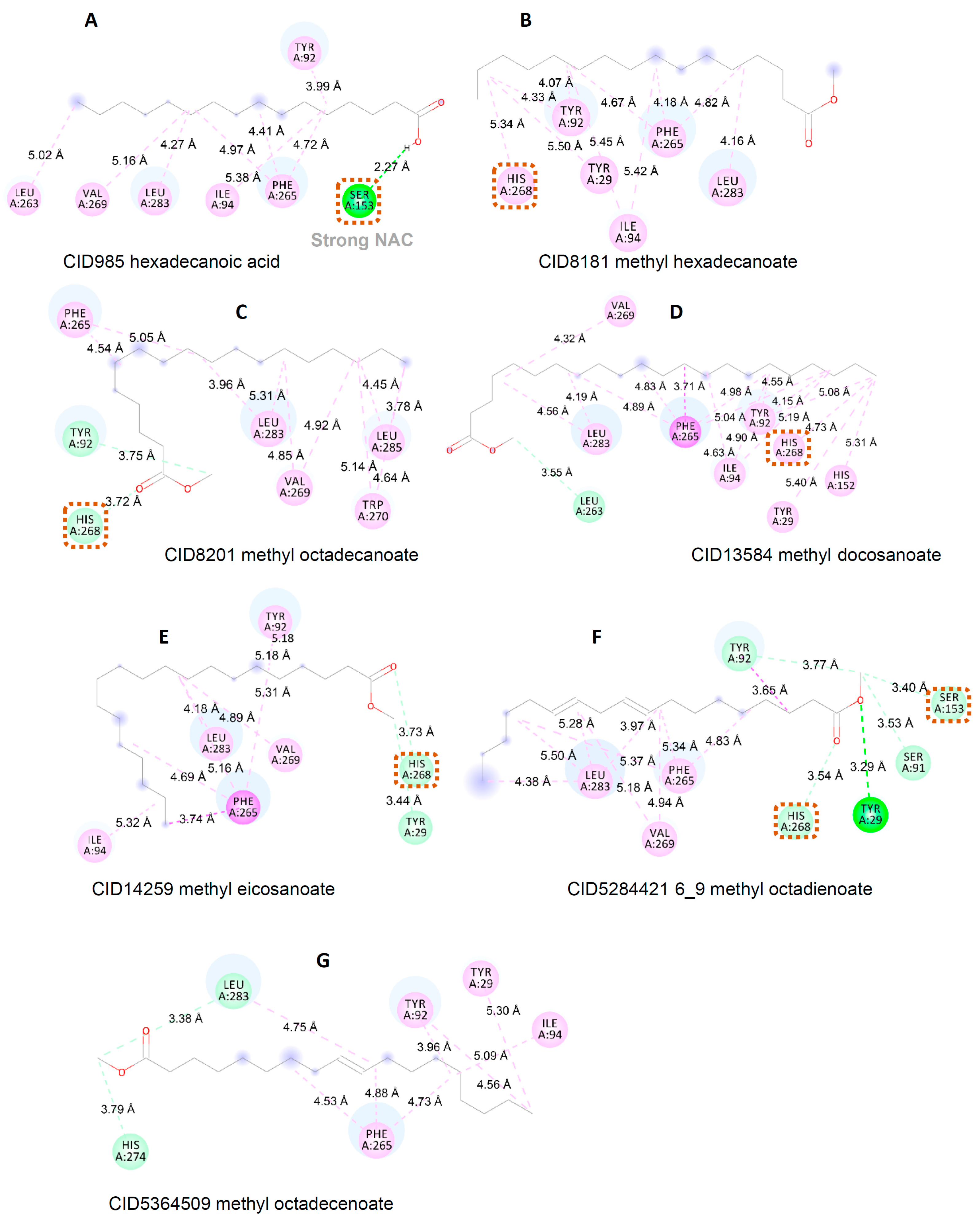
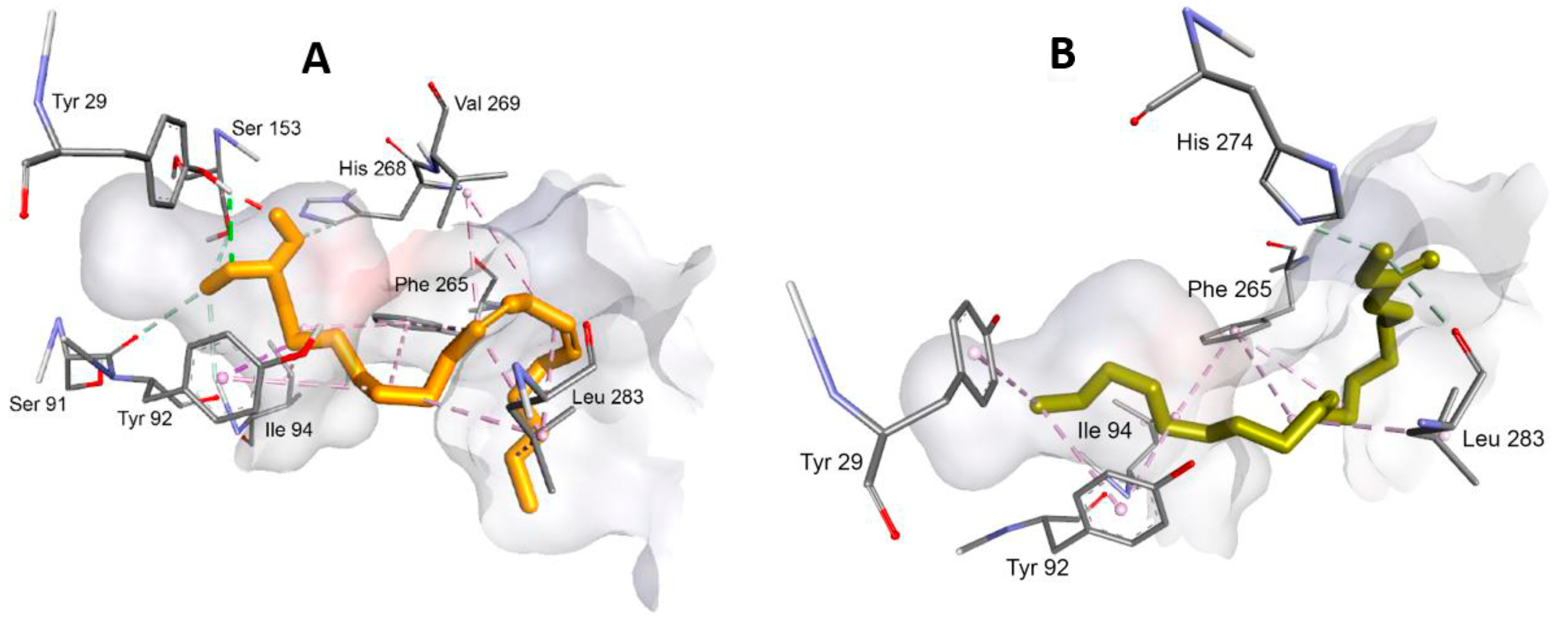


| Sample | Energy (kcal/mol) |
|---|---|
| CID985 hexadecanoic acid | −5.6 |
| CID8181 methyl hexadecanoate | −5.4 |
| CID8201 methyl octadecanoate | −5.6 |
| CID13584 methyl docosanoate | −5.4 |
| CID14259 methyl eicosanoate | −5.7 |
| CID5284421 6,9-methyl octadienoate | −6.1 |
| CID5364509 methyl octadecenoate | −5.7 |
| Sample | Residue | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tyr 29 | Tyr 92 | Ile 94 | His 152 | Ser 153 | Leu 263 | Phe 265 | His 268 | Val 269 | Trp 270 | His 274 | Leu 283 | Leu 285 | |
| CID985 | 3.99 (HI) | 5.38 (HI) | 2.27 (HB) | 5.02 (HI) | 4.41 (HI) 4.72 (HI) 4.97 (HI) | 5.16 (HI) | 4.27 (HI) | ||||||
| CID8181 | 5.50 (HI) | 4.33 (HI) 4.07 (HI) | 5.42 (HI) 5.45 (HI) | 4.82 (HI) 4.18 (HI) 4.67 (HI) | 5.34 (HI) | 4.16 (HI) | |||||||
| CID8201 | 3.75(CH) | 4.54 (HI) 5.05 (HI) | 3.72 (CH) | 4.85 (HI) 4.92 (HI) | 4.64(HI) 5.14(HI) | 3.96 (HI) 5.31 (HI) | 3.78 4.45 | ||||||
| CID13584 | 5.40 (HI) | 4.15 (HI) 4.55 (HI) 5.08 (HI) | 4.63 (HI) 4.55 (HI) 5.19 (HI) | 5.31(HI) | 3.55(CH) | 3.71 (PA) 4.83 (HI) 4.89 (HI) 4.98 (HI) 5.04 (HI) | 4.73 (HI) | 4.32 (HI) | 4.19 (HI) 4.56 (HI) | ||||
| CID14259 | 3.44 | 5.32 | 3.74 4.96 5.16 5.31 | 3.73 | 4.89 | ||||||||
| CID5284421 | 3.29 | 3.65 | 4.83 5.34 5.18 | 3.54 | 4.94 5.18 | 3.97 4.38 5.28 5.50 | |||||||
| CID5364509 | 5.30 | 3.96 | 5.09 | 4.53 4.73 4.88 | 3.79 | 4.75 | |||||||
| Complex | ∆Eele + ∆Gsol | ∆Evdw | ∆Gbind (kcal/mol) | Standard Deviation |
|---|---|---|---|---|
| (kcal/mol) | (kcal/mol) | |||
| hexadecanoic acid/Eversa | 18.43 | −29.14 | −13.27 | +/− 0.052 |
| methyl hexadecanoate/Eversa | 17.67 | −33.93 | −16.26 | +/− 0.023 |
| methyl octadecanoate/Eversa | 10.33 | −37.19 | −26.86 | +/− 0.027 |
| methyl docosanoate/Eversa | 20.76 | −39.79 | −19.03 | +/− 0.024 |
| methyl eicosanoate/Eversa | 13.75 | −37.37 | −23.62 | +/− 0.027 |
| 6,9-methyl octadienoate/Eversa | 11.97 | −35.38 | −23.41 | +/− 0.026 |
| methyl octadecenoate/Eversa | 19.56 | −35.03 | −15.47 | +/− 0.026 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nobre, M.M.R.; Silva, A.F.d.; Menezes, A.M.; Silva, F.L.B.d.; Lima, I.M.; Colares, R.P.; Souza, M.C.M.d.; Marinho, E.S.; Melo, R.L.F.; Santos, J.C.S.d.; et al. Ester Production Using the Lipid Composition of Coffee Ground Oil (Coffea arabica): A Theoretical Study of Eversa® Transform 2.0 Lipase as an Enzymatic Biocatalyst. Compounds 2023, 3, 411-429. https://doi.org/10.3390/compounds3030031
Nobre MMR, Silva AFd, Menezes AM, Silva FLBd, Lima IM, Colares RP, Souza MCMd, Marinho ES, Melo RLF, Santos JCSd, et al. Ester Production Using the Lipid Composition of Coffee Ground Oil (Coffea arabica): A Theoretical Study of Eversa® Transform 2.0 Lipase as an Enzymatic Biocatalyst. Compounds. 2023; 3(3):411-429. https://doi.org/10.3390/compounds3030031
Chicago/Turabian StyleNobre, Millena Mara Rabelo, Ananias Freire da Silva, Amanda Maria Menezes, Francisco Lennon Barbosa da Silva, Iesa Matos Lima, Regilany Paulo Colares, Maria Cristiane Martins de Souza, Emmanuel Silva Marinho, Rafael Leandro Fernandes Melo, José Cleiton Sousa dos Santos, and et al. 2023. "Ester Production Using the Lipid Composition of Coffee Ground Oil (Coffea arabica): A Theoretical Study of Eversa® Transform 2.0 Lipase as an Enzymatic Biocatalyst" Compounds 3, no. 3: 411-429. https://doi.org/10.3390/compounds3030031
APA StyleNobre, M. M. R., Silva, A. F. d., Menezes, A. M., Silva, F. L. B. d., Lima, I. M., Colares, R. P., Souza, M. C. M. d., Marinho, E. S., Melo, R. L. F., Santos, J. C. S. d., & da Fonseca, A. M. (2023). Ester Production Using the Lipid Composition of Coffee Ground Oil (Coffea arabica): A Theoretical Study of Eversa® Transform 2.0 Lipase as an Enzymatic Biocatalyst. Compounds, 3(3), 411-429. https://doi.org/10.3390/compounds3030031







