1. Introduction
Rapid DNA analysis systems, such as RapidHIT ID
®, are revolutionizing forensic workflows by enabling the generation of genetic profiles in just 90 min [
1]. This technology is especially useful in situations where time is critical, such as criminal investigations that need fast DNA results. Unlike traditional laboratory methods, which can take days or even weeks to produce results, RapidHIT ID
® allows rapid testing, even in decentralized locations, enabling forensic operators to work in areas with minimal infrastructure [
2]. Recent reviews, including the study by Bruijns et al. [
3], emphasize how much these tools have improved the speed and efficiency of forensic DNA profiling.
The Forensic Science Laboratory of the French Gendarmerie (IRCGN) processes over 140,000 individual samples and 70,000 casework analyses annually, ensuring that analyses are completed within the legal time limits for police custody (24 h) in both metropolitan France and Corsica. However, due to logistical challenges, this rapid processing is not possible in France’s overseas territories. To overcome this issue, the French Gendarmerie has introduced RapidHIT ID
® systems in four key locations: New Caledonia, Mayotte, Guadeloupe, and French Guyana. In these areas, samples are tested on site, with the results securely transmitted to the IRCGN lab in Pontoise for final validation [
4]. Over the past two years, more than 400 samples, including blood, cigarette butts, and cadaver remains, have been successfully processed using this decentralized system, achieving impressive success rates [
5,
6,
7].
While the RapidHIT ID
® system offers many benefits, it does have one significant drawback: it uses the entire sample, such as swabs, during analysis [
8]. This poses a challenge if additional testing or reanalysis is needed, as there is no remaining biological material to work with. One exception has been made for cigarette butts, where a protocol allows part of the sample to be kept for potential retesting if the initial results are negative.
To address this issue with blood samples [
9], we have developed a subsampling method that preserves part of the original swab for future testing. The RapidINTEL™ Plus sample cartridge manual does mention a subsampling method but recommends using a flexible swab, which is difficult to handle for this process [
10]. In our study, we first tested different flocked swabs with rigid heads in RapidHIT ID
®, starting from the same blood volume, to compare the quality of DNA profiles obtained. Then, we opted for the 4N6FLOQSwabs
® Subungual Shape code 40U022D (Copan Italia S.p.A.) [
11] to perform subsampling. It has a smaller, rigid head that is ideal and more practical for precise subsampling. We tested this approach on blood samples from four donors, with three different operators conducting the subsampling. The use of four donors was selected as a balanced approach for this preliminary study, as it provided sufficient variability to assess the subsampling method’s effectiveness across different genetic profiles. This study’s primary aim was to evaluate the feasibility and consistency of the subsampling technique, which also involved testing across multiple operators to assess reproducibility and operator effect. All the tests were carried out on a single RapidHIT ID
® machine to avoid variability between machines [
12].
2. Materials and Methods
2.1. Sample Collection
This study follows all the recommendations issued by the ethics commission of the ‘Pôle Judiciaire de la Gendarmerie Nationale’ (PJGN) in good practice for the ethical classification of biological data and the recovery of biological material and in accordance with the Helsinki Accords (1975) and the French National Charter of Research Integrity [
4]. Blood samples were collected from 4 volunteers after obtaining informed consent. After collection, all blood samples were extracted using Crime Prep Adem-Kit (Ademtech, Pessac, France). The DNA extracts were quantified by qPCR using the Investigator
® Quantiplex PRO kit (QIAGEN, Hilden, Germany) [
13] on a real-time PCR ABI 7500 instrument (Thermo Fisher Scientific, MA, USA) to evaluate their DNA concentration.
2.2. Study Design
For all of this study, one RapidHIT ID® instrument (software v1.3.3) (Thermo Fisher Scientific) was used with the RapidINTELTM sample cartridges. We chose to use a single RapidHIT ID® instrument to avoid potential biases that could arise from minor inter-instrument variability. Using a single device allowed us to focus on assessing the subsampling method’s effectiveness without introducing additional variables.
To test different types of flocked swabs, blood samples from 2 donors were used, and three types of swabs were tested: the Chemunex flocked swab (A43212C), the Copan regular flocked swab (4N6FLOQSwabs® code 4504 C– Copan Italia S.p.A), and the Copan mini flocked swab (4N6FLOQSwabs® Subungual Shape code 40U022D–Copan Italia S.p.A). For each type of swab and each blood source, a volume of 10 μL of blood was deposited on 2 swab replicates and then dried for 24 h according to the respective manufacturer’s recommendations.
For the subsampling study, blood samples were drawn from 4 different donors, identified as A, B, C, and D. For each donor, a volume of 10 μL or 150 μL of blood was deposited directly on 7 regular flocked swabs (4N6FLOQSwabs® code 4504 C– Copan) for each volume tested and then dried for 24 h by the active drying system in the cap of the device.
For each volume tested, an entire regular flocked swab was analyzed in the RapidHIT ID® instrument to obtain a reference genetic profile. The other 6 swabs were subsampled by 3 different operators (2 swabs per operator) using a mini flocked swab (4N6FLOQSwabs® Subungual Shape code 40U022D – Copan Italia S.p.A). For this purpose, the mini flocked swab was moistened with 15 μL of ultra-pure DNA-free water and then applied by rolling and tapping on the regular swab until the entire surface of the mini swab head was stained red. This mini swab was then directly analyzed on the RapidHIT ID® instrument.
For all experiments, short-term storage (up to one week) of the samples was conducted at refrigerated temperatures, and long-term storage, when required, was performed in a freezer to preserve sample integrity.
2.3. Primary and Secondary Rapid DNA Analysis
The primary analysis was automatically performed on RapidHIT ID
®, returning information on the quality of the results [
14]. This is often described as rapid DNA analysis that is fully automated. Upon completion of the primary analysis, the RapidHIT ID
® system provides one of three status results: a green checkmark (indicating that all system threshold criteria are met), a yellow checkmark (indicating that one or more system threshold criteria are not met and a manual review is required), or a red ‘X’ (indicating that no result was generated due to a failed run).
Regardless of the quality flag, all results underwent manual review during a secondary analysis using GeneMapper™ ID-X Software v1.6 (Thermo Fisher Scientific) [
15,
16]. The results were analyzed using a simplified pipeline with a specific threshold for allele detection and calls and for heterozygous balance examination.
2.4. Statistical Analysis
The various results obtained are presented as the mean of different independent experiments. Several types of statistical analyses were used to compare the signal intensities from each marker individually. To check whether the distribution was normal, a Shapiro–Wilk normality test was performed for all experiments. Subsequently, a one-way analysis of variance (ANOVA) was used to assess differences in the different types of flocked swabs tested and among the operators’ results. Then, a t-test was performed to compare the intensity (allelic peak heights expressed in RFU) of the genetic profiles obtained with the different subsampling tests for each donor.
4. Discussion
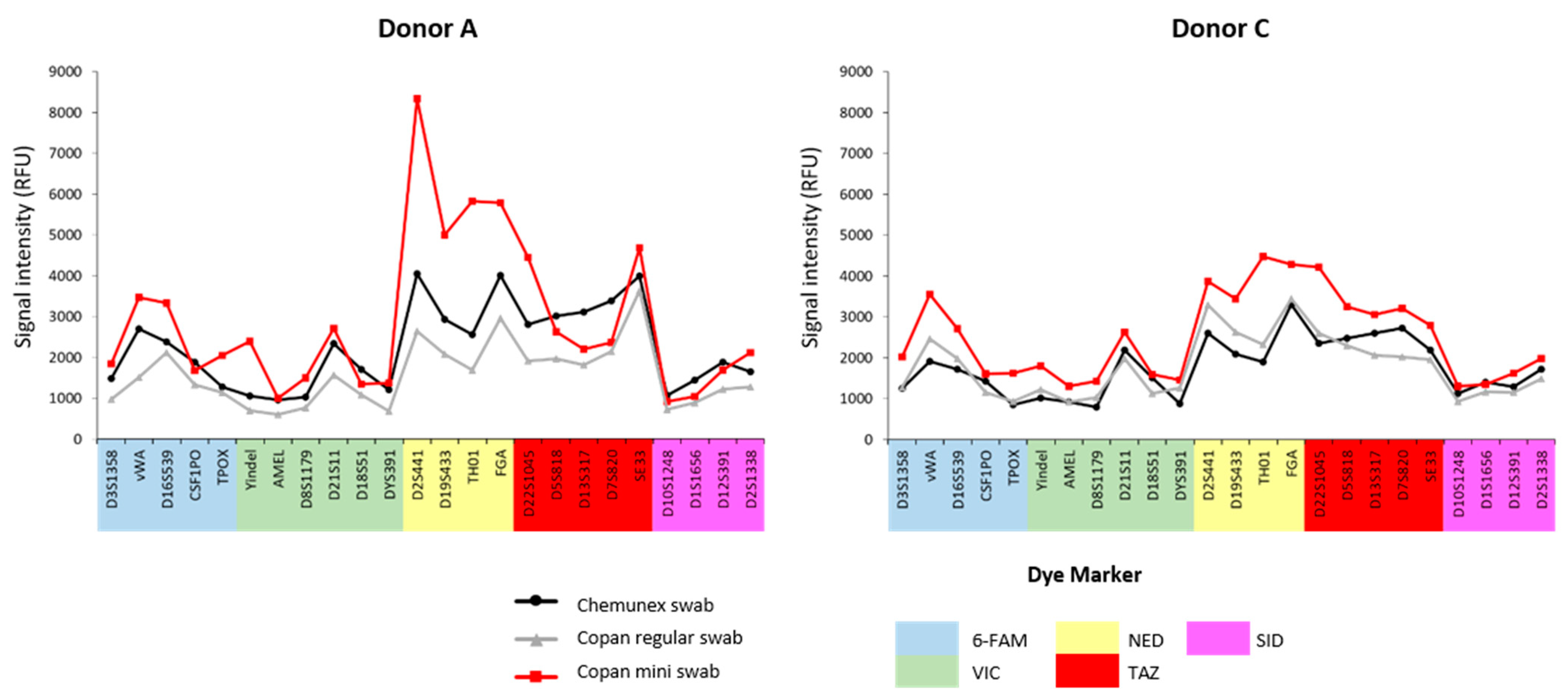
This study explored the potential influence of different types of flocked swabs on profile intensity with the same volume of blood deposited (10 µL). Regarding the device choice, the French Gendarmerie has previously used Chemunex flocked swabs for casework trace sampling and analyses. Recently, we have transitioned to using the Copan regular flocked swab for this purpose. Finally, we studied the quality of the results obtained with a Copan mini flocked swabs to validate its relevance for the rest of this study. All the analyses showed no significant differences in the completeness or intensity of the profiles, regardless of the swab used (
Figure 1).
Based on these results, the Copan mini flocked swab was selected to fulfill the primary objective of this study, which was to evaluate the effectiveness of a subsampling procedure for RapidHIT ID
® analysis, while addressing the challenge of complete sample consumption by the system [
17]. Moreover, the selected subsampling device features a rigid shaft to facilitate sample collection and a smaller tip size that better fits the instrument cartridge compared to the regular swab configuration. The results demonstrate the success of this approach, offering several important insights for forensic DNA analysis in decentralized environments, such as French overseas territories.
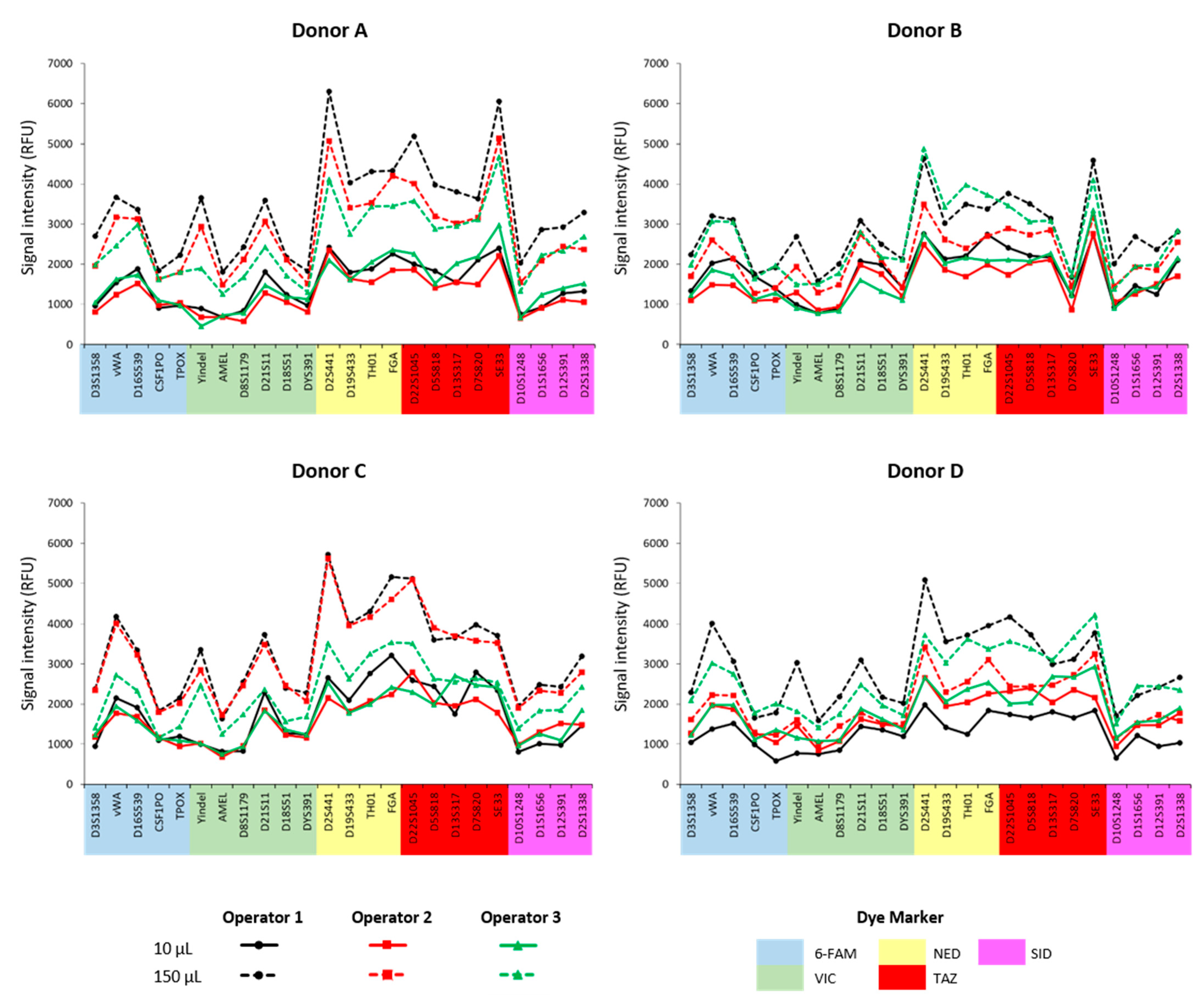
Firstly, the subsampling technique proved to be highly effective, consistently generating complete and usable genetic profiles from as little as 10 µL of blood. This was confirmed across all operators, as indicated by the ANOVA test (p-value = 0.01), which showed no statistically significant differences among the operators’ results. Operators were chosen, as they had never tested this protocol before and had to follow the guidelines provided at the time of the experiment. This suggests that the guideline is clear and that the subsampling method is robust enough to be reliably performed by different individuals, regardless of their prior experience with the procedure.
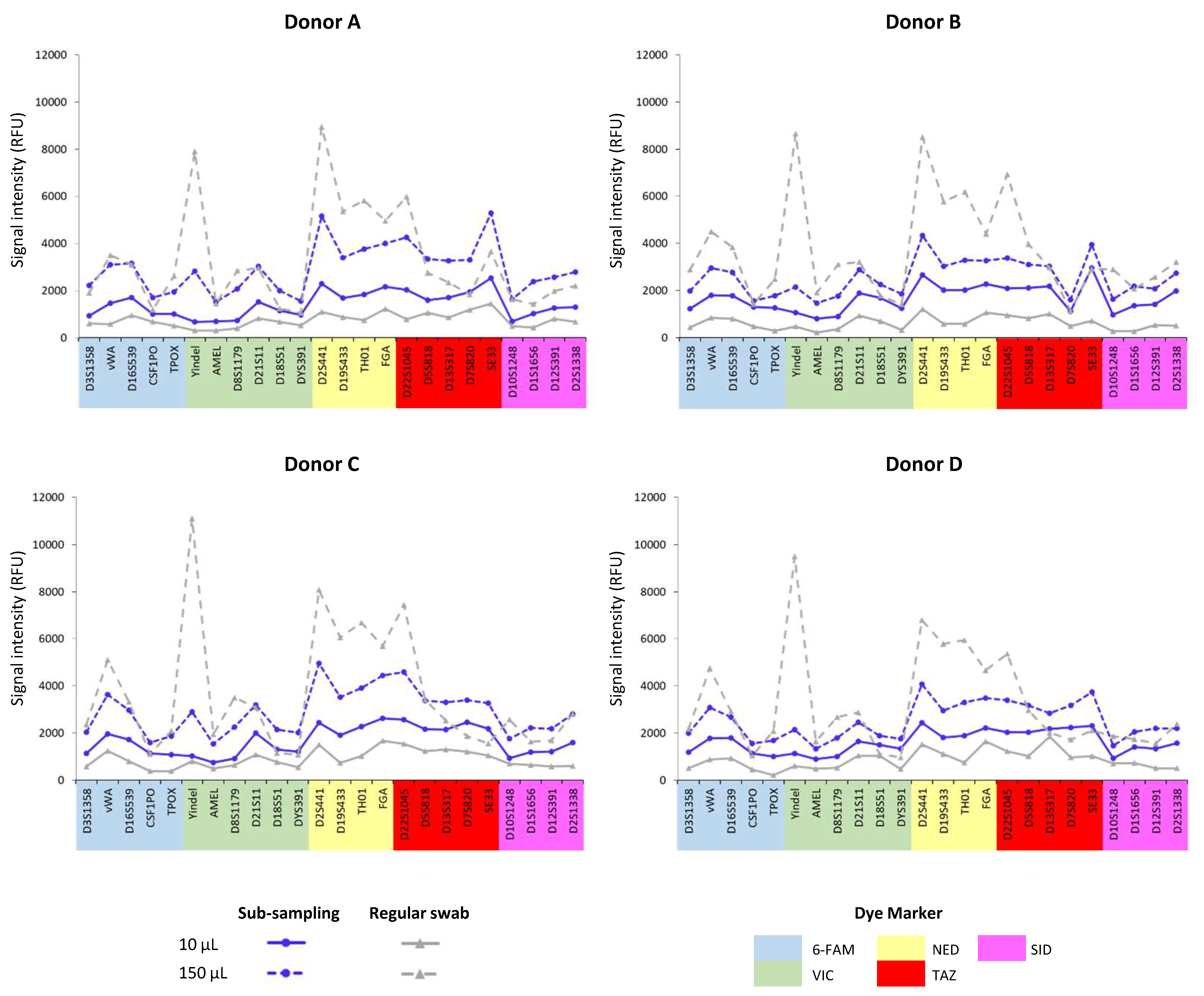
Secondly, analysis of the subsampling results based on different initial blood volumes (10 µL and 150 µL) revealed significant differences in the intensity of the genetic profiles (
t-test,
p-value = 0.05). This finding highlights the impact of initial sample volume on the intensity of the DNA profile. While larger blood volumes generally produce stronger signals, the generation of complete profiles from small volumes on the initial swab (10 µL) demonstrates the robustness and practicality of the subsampling technique, especially when starting biological material is limited (
Figure 2 and
Figure 3).
The method of subsampling described in this study offers a significant advantage in forensic casework. It preserves a portion of the original sample for future analysis [
18]. The subsampling method can be validated with different swab designs, other than the tested mini flocked swab, adding flexibility to its application in forensic workflows. This also highlights the innovative approach of using swabs that were not initially intended for this specific purpose. It demonstrates that such adaptations can still yield reliable results.
Looking ahead, this technique has broad potential for use in the next generation of RapidHIT ID
® systems. The anticipated release of RapidHIT ID
® V2, with enhanced sensitivity and specialized protocols [
19], is likely to produce even better results when paired with the subsampling method. Additionally, subsampling could be extended to buccal swabs from individuals, allowing for analysis with RapidINTEL
TM or RapidINTEL
TM Plus cartridges instead of the ACE cartridges currently in use. This would standardize the amount of biological material analyzed, improving consistency and reducing the need to manage multiple types of cartridges. Such a shift could streamline stock management, particularly within the French Gendarmerie’s strategy of using only RapidINTEL
TM or RapidINTEL
TM Plus cartridges.
The findings and inferences from this study can serve as the foundation for developing practical guidelines to support subsampling protocols (
Supplementary Material Figure S1). These guidelines would ensure that the technique is consistently and effectively applied in routine forensic operations, minimizing the risk of sample loss and maximizing the reliability of RapidHIT ID
® analyses. Therefore, these results suggest that the proposed subsampling method is a practical and reliable solution for saving forensic samples during RapidHIT ID
® analysis. This is especially relevant in remote or resource-limited settings, where preserving sample material for potential reanalysis is critical. Moreover, the method’s consistency across different operators and swab designs bolsters its potential for widespread implementation in forensic laboratories. Researchers may focus on optimizing the protocol for use with different biological samples (e.g., buccal swabs) and testing its compatibility with the next-generation RapidHIT ID
® systems, which may offer even greater sensitivity and accuracy.
5. Conclusions
This study successfully demonstrates the utility and efficiency of a subsampling technique for forensic DNA analysis using RapidHIT ID® technology. By preserving part of the original biological sample, this method addresses a key limitation of RapidHIT ID® systems, which consume the entire swab during analysis, leaving no material for potential retesting. Our findings indicate that subsampling from as little as 10 µL of blood can yield complete and reliable genetic profiles, with no significant variation among operators. This highlights the robustness of the technique, making it an ideal solution for use in decentralized or resource-limited settings, such as French overseas territories.
The results also suggest that the initial sample volume plays a role in the intensity of the genetic profiles obtained. While larger volumes provide stronger signals, subsampling from small volumes still produces complete profiles, supporting the method’s flexibility in situations where sample material is scarce. Moreover, while achieving notable success with the flocked subungual mini swab, the technique can be compatible with different swab head designs and potentially adaptable across various forensic workflows.
Beyond its immediate application in blood sample analysis, this method holds the potential for broader use. It could be extended to buccal swabs in combination with RapidINTEL
TM or RapidINTEL
TM Plus cartridges [
10], offering an opportunity to standardize biological sample volumes in RapidHIT ID
® workflows. Such standardization could streamline forensic operations, particularly in institutions like the French Gendarmerie, where cartridge management is critical.
Importantly, the ability to preserve part of the primary sample for potential reanalysis mitigates the risks associated with carry-over contamination, a known issue in RapidHIT ID® systems. This safeguard enhances the reliability of forensic casework, particularly in high-stakes investigations where evidence re-examination may be necessary. Looking forward, the subsampling protocol described here could be further optimized with the advent of next-generation RapidHIT ID® V2 instruments, which are expected to offer even greater sensitivity and accuracy. The development of practical guidelines for the standardized application of this method will ensure its widespread adoption and consistent use in forensic laboratories worldwide. This solution mitigates the current limitations in RapidHIT ID® technology and paves the way for future advancements in forensic science. This study presents a novel, effective, and adaptable subsampling method that has the potential to significantly improve forensic DNA analysis, particularly in remote or decentralized environments. As these findings demonstrate the feasibility of the subsampling technique in producing viable DNA profiles, validation using ground truth samples could be a valuable next step in future studies to confirm the method’s effectiveness and further establish its reliability in forensic applications.