Innovations in Forensic Sciences for Human Identification by DNA in the French Gendarmerie during the Last 10 Years
Abstract
1. Introduction
1.1. Genetic Profiling in Forensics and DNA Database in France
1.2. Forensic Genetic Laboratories in the French Gendarmerie
2. Innovations Made in the French Gendarmerie
2.1. Setup of a New Protocol for Disaster Victim Identification and Isolated Cadavers
2.1.1. Statement
- As a corpse or body part is an inert biological material containing the potentially abundant genetic material, the FTA® card could be used to recover biological samples, depending on the level of decomposition of the corpse or the body element [30];
- Transmission of the practice is facilitated as all Gendarmerie units have sampling kits;
- Laboratory analysis can be accelerated thanks to the properties of the FTA® card (saving of 2 to 4 h for extraction), and automation allows mass analysis of samples in the event of a multitude of victims or specimens [31];
- For laboratory technicians, the sample is dehumanized (a simple piece of paper), greatly reducing the psychological trauma inflicted during the treatment of a multitude of samples or body elements (morphologically identifiable or not) [32].
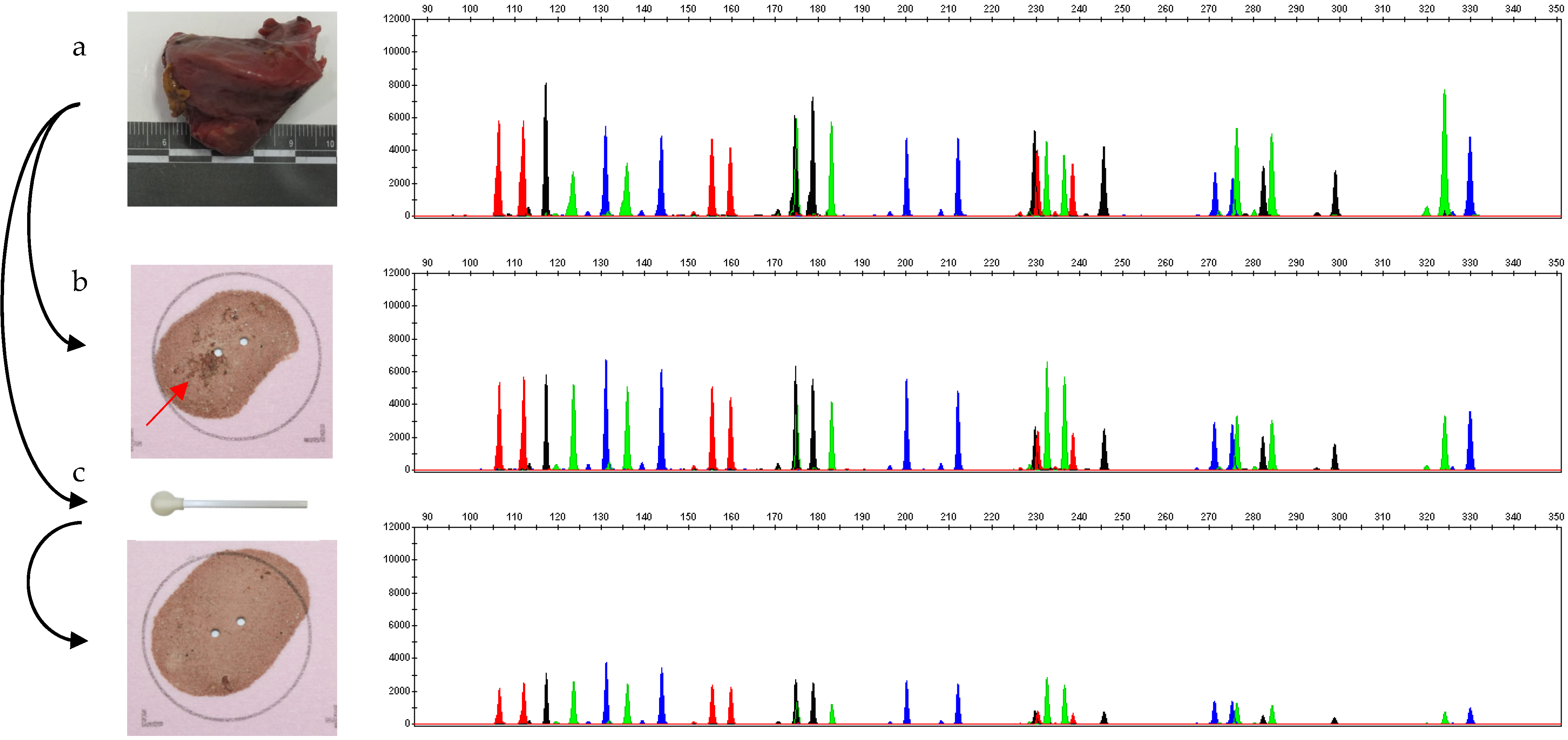
2.1.2. Initial Applications and Basics for the Standardized Cadaver Sampling Protocol
- A piece of muscle analyzed by the method of digestion and DNA extraction;
- A transfer of organic matter via direct affixing of the piece of muscle onto an FTA® card, which was then analyzed;
- Wiping the piece of muscle using the sampling ‘lollipop’ from the standardized ‘individual’ kit, previously moistened with sterile pyrogen-free water, then transferring the sample by applying the lollipop to an FTA card ® for analysis.
2.1.3. Protocol Optimization
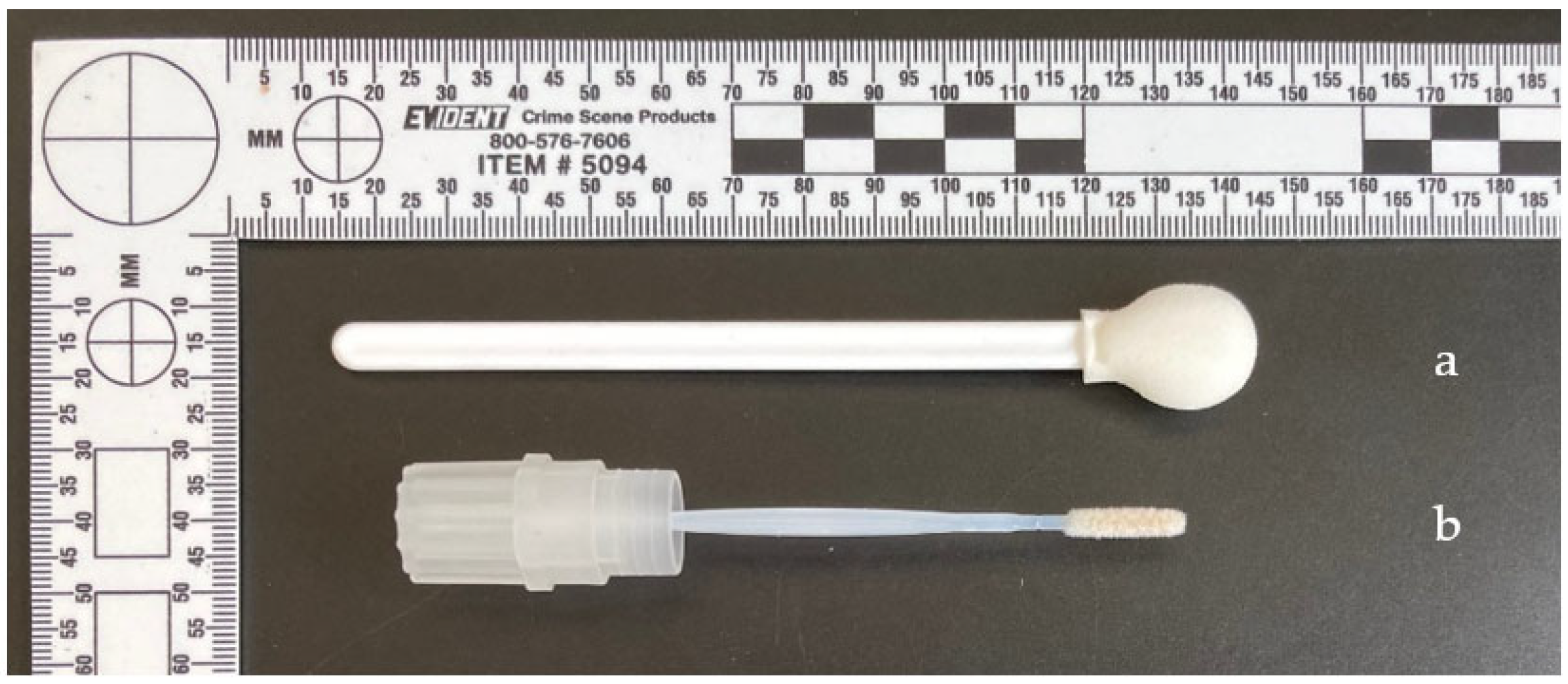
2.2. Design of a New Collection Tool: The MicroFloq®
2.3. Design of the First Mobile Lab for Genetic Analysis
- -
- The versatility of the missions carried out in the vehicle. It is necessary to have a mobile structure dedicated to genetic analyses in order to limit contamination and, on the other hand, to design a specific environment for each analytical step [1] (before and after PCR amplification);
- -
- The size of the vehicle makes it difficult to transport on narrow roads. The same is true for parking, which requires a location and sufficient surface area for deployment and installation [27]. Finally, it is a vehicle requiring a specific driving license, limiting the number of potential drivers.
2.4. Design of a New Swab Sampling Automaton
3. New Innovative Technologies Adapted for Forensic Sciences
3.1. The Revolution of Microfluidic Technologies: The Rapid HIT™ Technology
3.2. NGS Applied for Forensic Analysis
3.3. The Microbial Communities: New Perspectives in Forensic Sciences
4. Conclusions
5. Patents
- DISPOSITIF DE STOCKAGE À TEMPÉRATURE AMBIANTE DE MATÉRIELS BIOLOGIQUES FR2019000016 HUBAC, Sylvain
- DISPOSITIF DE COLLECTION DE MATÉRIEL BIOLOGIQUE À PARTIR D’UNE TRACE BIOLOGIQUE FR2016000026 HUBAC, Sylvain
- LABORATOIRE MOBILE POUR ANALYSE GÉNÉTIQUE FR2017000083 HUBAC, Sylvain
- BOÎTIER D’ÉCHANTILLONNAGE ET AUTOMATE MODULAIRE COMPRENANT CE BOITIER D’ÉCHANTILLONNAGE FR2021050910 PUSSIAU, Amaury, HUBAC, Sylvain
- DISPOSITIF DE STOCKAGE À TEMPÉRATURE AMBIANTE DE MATÉRIELS BIOLOGIQUES EP19708587 HUBAC, Sylvain
- LABORATOIRE MOBILE POUR ANALYSE GÉNÉTIQUE EP17725653 HUBAC, Sylvain
- DISPOSITIF DE COLLECTION DE MATÉRIEL BIOLOGIQUE À PARTIR D’UNE TRACE BIOLOGIQUE EP16722264 HUBAC, Sylvain
- DISPOSITIF POUR ANALYSER DES ELEMENTS SOLIDES BIOLOGIQUES ET DISPOSITIF POUR SA MISE EN OEUVRE EP2021052117 PUSSIAU AMAURY, HUBAC, Sylvain
- DISPOSITIF POUR ANALYSER DES ELEMENTS SOLIDES BIOLOGIQUES ET DISPOSITIF POUR SA MISE EN OEUVRE EP21702660 PUSSIAU Amaury, HUBAC, Sylvain
- DISPOSITIF DE STOCKAGE A TEMPERATURE AMBIANTE DE MATERIELS BIOLOGIQUES FR1870121 SYLVAIN HUBAC
- DISPOSITIF DE COLLECTTION DE MATERIEL BIOLOGIQUE A PARTIR D’UNE TRACE BIOLOGIQUE FR1670047 SYLVAIN HUBAC
- LABORATOIRE MOBILE POUR ANALYSE GENETIQUE FR1670215 SYLVAIN HUBAC
- DISPOSITIFS DE COLLECTION POUR L’ANALYSE RAPIDE DIRECTE ET INDIRECTE D’ACIDES NUCLEIQUES A PARTIR DE TRACES BIOLOGIQUES FR1500350 SYLVAIN HUBAC
- DISPOSITIF DE PRELEVEMENT DE MATIERE OSSEUSE FR2103443 Julien MARTY
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bukyya, J.L.; Tejasvi, M.L.A.; Avinash, A.; Chanchala, H.P.; Talwade, P.; Afroz, M.M.; Pokala, A.; Neela, P.K.; Shyamilee, T.K.; Srisha, V. DNA Profiling in Forensic Science: A Review. Glob. Med. Genet. 2021, 08, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Jeffreys, A.J.; Werrett, D.J. Forensic application of DNA ‘fingerprints’. Nature 1985, 318, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Chu, J.-Y. A Brief Review of Short Tandem Repeat Mutation. Genom. Proteom. Bioinform. 2007, 5, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Yang, Y.W.; Cao, Y.Y.; Zhu, Q.; Huang, Y.G.; Hu, Y.H.; Zhou, Y.J.; Li, X.; Wei, Y.F.; Shu, P.Y.; et al. Construction of SNP-STR Multiplex Amplification System with Genetic Markers and Its Forensic Application. Fa Yi Xue Za Zhi 2020, 36, 316-315. [Google Scholar]
- Wei, T.; Liao, F.; Wang, Y.; Pan, C.; Xiao, C.; Huang, D. A novel multiplex assay of SNP-STR markers for forensic purpose. PLoS ONE 2018, 13, e0200700. [Google Scholar] [CrossRef]
- Parson, W. Age Estimation with DNA: From Forensic DNA Fingerprinting to Forensic (Epi)Genomics: A Mini-Review. Gerontology 2018, 64, 326–332. [Google Scholar] [CrossRef]
- Arenas, M.; Pereira, F.; Oliveira, M.; Pinto, N.; Lopes, A.M.; Gomes, V.; Carracedo, A.; Amorim, A. Forensic genetics and genomics: Much more than just a human affair. PLoS Genet. 2017, 13, e1006960. [Google Scholar] [CrossRef]
- Article A38—Code de Procédure Pénale—Légifrance. Available online: https://www.legifrance.gouv.fr/codes/article_lc/LEGIARTI000031094125/ (accessed on 26 June 2022).
- Leake, S.L.; Pagni, M.; Falquet, L.; Taroni, F.; Greub, G. The salivary microbiome for differentiating individuals: Proof of principle. Microbes Infect. 2016, 18, 399–405. [Google Scholar] [CrossRef]
- Article 706-54—Code de Procédure Pénale—Légifrance. Available online: https://www.legifrance.gouv.fr/codes/article_lc/LEGIARTI000006577698/1998-06-18 (accessed on 26 June 2022).
- Goodwin, W.H. The use of forensic DNA analysis in humanitarian forensic action: The development of a set of international standards. Forensic Sci. Int. 2017, 278, 221–227. [Google Scholar] [CrossRef]
- Kimmelman, J. Risking Ethical Insolvency: A Survey of Trends in Criminal DNA Databanking. J. Law Med. Ethics 2000, 28, 209–221, discussion 222–223. [Google Scholar] [CrossRef]
- Lynch, L.; Gamblin, A.; Vintiner, S.; Simons, J.L. STR profiling of epithelial cells identified by X/Y-FISH labelling and laser microdissection using standard and elevated PCR conditions. Forensic Sci. Int. Genet. 2015, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dumache, R.; Ciocan, V.; Muresan, C.; Enache, A. Molecular DNA Analysis in Forensic Identification. Clin. Lab. 2016, 62, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Haddrill, P.R. Developments in forensic DNA analysis. Emerg. Top. Life Sci. 2021, 5, 381–393. [Google Scholar] [PubMed]
- Johnson, B.T.; Riemen, J.A.J.M. Digital capture of fingerprints in a disaster victim identification setting: A review and case study. Forensic Sci. Res. 2019, 4, 293–302. [Google Scholar] [CrossRef]
- Bécue, A.; Eldridge, H.; Champod, C. Interpol review of fingermarks and other body impressions 2016–2019. Forensic Sci. Int. Synergy 2020, 2, 442–480. [Google Scholar] [CrossRef]
- Mohammed, F.; Fairozekhan, A.T.; Bhat, S.; Menezes, R.G. Forensic Odontology. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: http://www.ncbi.nlm.nih.gov/books/NBK540984/ (accessed on 26 June 2022).
- Prajapati, G.; Sarode, S.C.; Sarode, G.S.; Shelke, P.; Awan, K.H.; Patil, S. Role of forensic odontology in the identification of victims of major mass disasters across the world: A systematic review. PLoS ONE 2018, 13, e0199791. [Google Scholar] [CrossRef]
- Radhika, T.; Jeddy, N.; Ravi, S. Current trends in forensic odontology. J. Forensic Dent. Sci. 2017, 9, 115–119. [Google Scholar]
- Watherston, J.; McNevin, D.; Gahan, M.; Bruce, D.; Ward, J. Current and emerging tools for the recovery of genetic information from post mortem samples: New directions for disaster victim identification. Forensic Sci. Int. Genet. 2018, 37, 270–282. [Google Scholar] [CrossRef]
- Ziętkiewicz, E.; Witt, M.; Daca, P.; Żebracka-Gala, J.; Goniewicz, M.; Jarząb, B.; Witt, M. Current genetic methodologies in the identification of disaster victims and in forensic analysis. J. Appl. Genet. 2012, 53, 41–60. [Google Scholar] [CrossRef]
- McCord, B.R.; Gauthier, Q.; Cho, S.; Roig, M.N.; Gibson-Daw, G.C.; Young, B.; Taglia, F.; Zapico, S.C.; Mariot, R.F.; Lee, S.B.; et al. Forensic DNA Analysis. Anal. Chem. 2019, 91, 673–688. [Google Scholar] [CrossRef]
- Butler, J.M. The future of forensic DNA analysis. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140252. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, L.; Liò, P. Forensic DNA and bioinformatics. Brief. Bioinform. 2007, 8, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Epplen, J.T.; Melmer, G.; Schmidt, P.; Roewer, L.; Hundrieser, J.; Epplen, C.; Buitkamp, J. On the potential of simple repetitive DNA for fingerprinting in clinical, forensic, and evolutionary dynamic studies. J. Mol. Med. 1992, 70, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Turnbough, M.; Eisenberg, A.; Shade, L.L.; Shewale, J.G. Training of Forensic DNA Scientists—A Commentary. Forensic Sci. Rev. 2012, 24, 143–150. [Google Scholar] [PubMed]
- Budowle, B.; Bieber, F.R.; Eisenberg, A.J. Forensic aspects of mass disasters: Strategic considerations for DNA-based human identification. Leg. Med. 2005, 7, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, M.-L.; Buchard, A.; Børsting, C.; Morling, N. High-throughput sequencing of forensic genetic samples using punches of FTA cards with buccal swabs. Biotechniques 2016, 61, 149–151. [Google Scholar] [CrossRef]
- Rahikainen, A.-L.; Palo, J.U.; de Leeuw, W.; Budowle, B.; Sajantila, A. DNA quality and quantity from up to 16 years old post-mortem blood stored on FTA cards. Forensic Sci. Int. 2016, 261, 148–153. [Google Scholar] [CrossRef]
- Raziel, A.; Dell’Ariccia-Carmon, A.; Zamir, A. Reduction of Powerplex® Y23 Reaction Volume for Genotyping Buccal Cell Samples on FTA TM Cards. J. Forensic Sci. 2015, 60, 152–156. [Google Scholar] [CrossRef]
- McNevin, D. Preservation of and DNA Extraction from Muscle Tissue. In Forensic DNA Typing Protocols; Goodwin, W., Ed.; Springer: New York, NY, USA, 2016; Volume 1420, pp. 43–53. [Google Scholar]
- Green, H.; Tillmar, A.; Pettersson, G.; Montelius, K. The use of FTA cards to acquire DNA profiles from postmortem cases. Int. J. Leg. Med. 2019, 133, 1651–1657. [Google Scholar] [CrossRef]
- Ciampolini, R.; Cecchi, F.; Spinetti, I.; Rocchi, A.; Biscarini, F. The use of genetic markers to estimate relationships between dogs in the course of criminal investigations. BMC Res. Notes 2017, 10, 414. [Google Scholar] [CrossRef]
- Mundorff, A.Z.; Amory, S.; Huel, R.; Bilić, A.; Scott, A.L.; Parsons, T.J. An economical and efficient method for postmortem DNA sampling in mass fatalities. Forensic Sci. Int. Genet. 2018, 36, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.S.; Roman, M.G.; Hughes-Stamm, S. In-field collection and preservation of decomposing human tissues to facilitate rapid purification and STR typing. Forensic Sci. Int. Genet. 2018, 36, 124–129. [Google Scholar] [CrossRef]
- Hubac, S. Device for Collecting Biological Material from a Biological Trace. WO2016132028A1, 25 August 2016. Available online: https://patents.google.com/patent/WO2016132028A1/en (accessed on 31 July 2020).
- Ambers, A.; Wiley, R.; Novroski, N.; Budowle, B. Direct PCR amplification of DNA from human bloodstains, saliva, and touch samples collected with microFLOQ® swabs. Forensic Sci. Int. Genet. 2018, 32, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Touron, P.; Siatka, C.; Pussiau, A.; Follot, S.; Fritz, T.; Petit, M.; Latifa, N.; Herrmann, J.-L.; Rottman, M.; Lemoine, A.; et al. A mobile DNA laboratory for forensic science adapted to coronavirus SARS-CoV-2 diagnosis. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 40, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Hubac, S. Laboratoire Mobile Pour Analyse Génétique. FR Patent EP3454986A1, 20 March 2019. [Google Scholar]
- Holland, M.; Wendt, F. Evaluation of the RapidHIT™ 200, an automated human identification system for STR analysis of single source samples. Forensic Sci. Int. Genet. 2015, 14, 76–85. [Google Scholar] [CrossRef]
- Buscaino, J.; Barican, A.; Farrales, L.; Goldman, B.; Klevenberg, J.; Kuhn, M.; Lin, F.; Nguyen, P.; Salceda, S.; Schueren, R.; et al. Evaluation of a rapid DNA process with the RapidHIT® ID system using a specialized cartridge for extracted and quantified human DNA. Forensic Sci. Int. Genet. 2018, 34, 116–127. [Google Scholar] [CrossRef]
- Shackleton, D.; Pagram, J.; Ives, L.; Vanhinsbergh, D. Development and validation of the RapidHIT™ 200 utilising NGMSElect™ Express for the processing of buccal swabs. Forensic Sci. Int. 2018, 289, 244–252. [Google Scholar] [CrossRef]
- Romsos, E.L.; French, J.L.; Smith, M.; Figarelli, V.; Harran, F.; Vandegrift, G.; Moreno, L.I.; Callaghan, T.F.; Brocato, J.; Vaidyanathan, J.; et al. Results of the 2018 Rapid DNA Maturity Assessment. J. Forensic Sci. 2020, 65, 953–959. [Google Scholar] [CrossRef]
- Sun, S.; Tian, Y.M.; Tang, J.M.; Zhang, Q.X.; Hu, Y.L.; Wang, P.F.; Xue, L.Y.; Liu, L.; Tang, H. Application of RapidHITTM 200 System in Forensic Medicine]. Fa Yi Xue Za Zhi 2018, 34, 161–164. [Google Scholar]
- Sherier, A.J.; Kieser, R.E.; Novroski, N.M.; Wendt, F.R.; King, J.L.; Woerner, A.E.; Ambers, A.; Garofano, P.; Budowle, B. Copan microFLOQ® Direct Swab collection of bloodstains, saliva, and semen on cotton cloth. Int. J. Leg. Med. 2020, 134, 45–54. [Google Scholar] [CrossRef]
- Bruijns, B.; Tiggelaar, R.M.; Gardeniers, J. Massively parallel sequencing techniques for forensics: A review. Electrophoresis 2018, 39, 2642–2654. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, C.; Vandewoestyne, M.; Van Criekinge, W.; Deforce, D.; Van Nieuwerburgh, F. My-Forensic-Loci-queries (MyFLq) framework for analysis of forensic STR data generated by massive parallel sequencing. Forensic Sci. Int. Genet. 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Parson, W.; Ballard, D.; Budowle, B.; Butler, J.M.; Gettings, K.B.; Gill, P.; Gusmão, L.; Hares, D.R.; Irwin, J.A.; King, J.L.; et al. Massively parallel sequencing of forensic STRs: Considerations of the DNA commission of the International Society for Forensic Genetics (ISFG) on minimal nomenclature requirements. Forensic Sci. Int. Genet. 2016, 22, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Chow, H.Y.; Siegel, D.; Wurmbach, E. Qualitative and quantitative assessment of Illumina’s forensic STR and SNP kits on MiSeq FGx™. PLoS ONE 2017, 12, e0187932. [Google Scholar] [CrossRef] [PubMed]
- Børsting, C.; Morling, N. Next generation sequencing and its applications in forensic genetics. Forensic Sci. Int. Genet. 2015, 18, 78–89. [Google Scholar] [CrossRef]
- Grandell, I.; Samara, R.; Tillmar, A.O. A SNP panel for identity and kinship testing using massive parallel sequencing. Int. J. Leg. Med. 2016, 130, 905–914. [Google Scholar] [CrossRef]
- Mehta, B.; Daniel, R.; Phillips, C.; McNevin, D. Forensically relevant SNaPshot® assays for human DNA SNP analysis: A review. Int. J. Leg. Med. 2017, 131, 21–37. [Google Scholar] [CrossRef]
- Aly, S.M.; Sabri, D.M. Next generation sequencing (NGS): A golden tool in forensic toolkit. Arch. Forensic Med. Criminol. 2015, 4, 260–271. [Google Scholar] [CrossRef]
- Ansorge, W.J. Next-generation DNA sequencing techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Nicolas, T.; Nolot, F.; Pussiau, A.; Guyomarc, P.; Mazevet, M. Forensic Science and “The Duty of Memory”: The Face of Verdun. Forensic Identif. 2020, 70, 1–15. [Google Scholar]
- Vidaki, A.; Kayser, M. Recent progress, methods and perspectives in forensic epigenetics. Forensic Sci. Int. Genet. 2018, 37, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef] [PubMed]
- Kader, F.; Ghai, M. DNA methylation and application in forensic sciences. Forensic Sci. Int. 2015, 249, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Poussard, A.; Curci, J.-Y.; Siatka, C.; Hermitte, F.; Pussiau, A.; Singla-Sanchez, H.; Hubac, S. Evaluation of DNA Methylation-Based Age-Prediction Models from Saliva and Buccal Swab Samples Using Pyrosequencing Data. Forensic Sci. 2023, 3, 192–204. [Google Scholar] [CrossRef]
- Heidegger, A.; Xavier, C.; Niederstätter, H.; de la Puente, M.; Pośpiech, E.; Pisarek, A.; Kayser, M.; Branicki, W.; Parson, W. Development and optimization of the VISAGE basic prototype tool for forensic age estimation. Forensic Sci. Int. Genet. 2022, 48, 102322. [Google Scholar] [CrossRef]
- Woźniak, A.; Heidegger, A.; Piniewska-Róg, D.; Pośpiech, E.; Xavier, C.; Pisarek, A.; Kartasińska, E.; Boroń, M.; Freire-Aradas, A.; Wojtas, M.; et al. Development of the VISAGE enhanced tool and statistical models for epigenetic age estimation in blood, buccal cells and bones. Aging 2021, 13, 6459–6484. [Google Scholar] [CrossRef]
- Giampaoli, S.; Alessandrini, F.; Frajese, G.V.; Guglielmi, G.; Tagliabracci, A.; Berti, A. Environmental microbiology: Perspectives for legal and occupational medicine. Leg. Med. 2018, 35, 34–43. [Google Scholar] [CrossRef]
- Gouello, A.; Dunyach-Remy, C.; Siatka, C.; Lavigne, J.-P. Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics 2021, 12, 1. [Google Scholar] [CrossRef]
- Tuccia, F.; Zurgani, E.; Bortolini, S.; Vanin, S. Experimental evaluation on the applicability of necrobiome analysis in forensic veterinary science. MicrobiologyOpen 2019, 8, e828. [Google Scholar] [CrossRef]



| Type of Catastrophe | Number of Victims | FTA® Samples Analyzed | Success (%) | Swabs Analyzed | Success (%) |
|---|---|---|---|---|---|
| Air crash AH5017 (Mali, 2014) | 115 | 40 | 93 | 130 | 76 |
| Air crash 4U9525 (France, 2015) | 150 | 2508 | 93 | 222 | 100 |
| Road accident (France, 2015) | 43 | 70 | 100 | 0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sauvagère, S.; Pussiau, A.; Hubac, S.; Gouello, A.; Poussard, A.; Lavigne, J.-P.; Larnane, A.; Siatka, C.; Hermitte, F. Innovations in Forensic Sciences for Human Identification by DNA in the French Gendarmerie during the Last 10 Years. Forensic Sci. 2023, 3, 316-329. https://doi.org/10.3390/forensicsci3020024
Sauvagère S, Pussiau A, Hubac S, Gouello A, Poussard A, Lavigne J-P, Larnane A, Siatka C, Hermitte F. Innovations in Forensic Sciences for Human Identification by DNA in the French Gendarmerie during the Last 10 Years. Forensic Sciences. 2023; 3(2):316-329. https://doi.org/10.3390/forensicsci3020024
Chicago/Turabian StyleSauvagère, Stéphane, Amaury Pussiau, Sylvain Hubac, Audrey Gouello, Alexandre Poussard, Jean-Philippe Lavigne, Amel Larnane, Christian Siatka, and Francis Hermitte. 2023. "Innovations in Forensic Sciences for Human Identification by DNA in the French Gendarmerie during the Last 10 Years" Forensic Sciences 3, no. 2: 316-329. https://doi.org/10.3390/forensicsci3020024
APA StyleSauvagère, S., Pussiau, A., Hubac, S., Gouello, A., Poussard, A., Lavigne, J.-P., Larnane, A., Siatka, C., & Hermitte, F. (2023). Innovations in Forensic Sciences for Human Identification by DNA in the French Gendarmerie during the Last 10 Years. Forensic Sciences, 3(2), 316-329. https://doi.org/10.3390/forensicsci3020024










