Abstract
Advances in forensic biology have increased the options for the collection, sampling, preservation and processing of human remains for DNA-based identification. Combined with a plethora of commercial DNA testing kits that are far more forgiving of inhibited and degraded samples, efficient DNA approaches to post-mortem samples are explored here for DNA-based identification of compromised human remains. Approaches which preserve sample and reduce analytical turnaround times whilst saving resources also have the potential to expedite the identification process, to provide answers to grieving families sooner, or to provide leads in a criminal investigation. Targeting sample types that are minimally-invasive and do not require extensive preparation and testing protocols also has benefit for disaster victim identification (DVI) by facilitating field sampling. We have assessed minimally-invasive and simple to collect sample types compatible with minimal pre-treatment and efficient DNA profiling approaches. Incubating nail, distal phalanges and whole digits in 500 µL of PrepFiler™ Lysis Buffer for 2 h was an efficient and simple method, limiting or removing sample preparation. A reduced 15 min incubation also yielded DNA profiles suggesting a shorter incubation may lyse sufficient DNA. Preservative solutions offer an even simpler process in some cases. Furthermore, the efficient approaches described in this study offer storage solutions and are compatible with backend automated processing. This study will inform further research to develop and optimise efficient protocols. These DNA approaches should not be pursued for every sample; more compromised samples may best be submitted to the laboratory for more effective extraction and genotyping.
1. Introduction
1.1. Compromised Human Remains
The extent of decomposition of human remains can affect DNA recovery because the quality of the genetic material is strongly influenced by time since death and environmental conditions [1]. Optimal post mortem (PM) sample selection can increase the chances of successfully recovering a DNA profile from compromised human remains. Current recommendations suggest the collection of bone is most appropriate due to a higher success rate of DNA recovery from femur shafts and teeth as compared to blood, buccal and tissue samples [1,2,3,4,5,6]. DNA is well preserved in bone cells and teeth [4], making them reliable sources of DNA, particularly in adverse environmental conditions and for long-term sampling [1,7]. However, numerous novel PM sample types have since been reported as successfully yielding DNA (see [8] for a detailed review); amongst these are toenails [9] and small cancellous bones such as distal phalanges of the hands and feet [10,11].
1.2. Disaster Victim Identification (DVI)
The World Health Organisation describes a disaster as ‘a sudden ecological phenomena of sufficient magnitude to require external assistance’ [12]. DNA profiling offers an ability to identify victims, reassociate body parts and assist in the identification of perpetrators [5]. DNA profiling has become the gold standard for the identification of victims in both mass casualty incidents and forensic cases where human remains are highly fragmented and/or degraded, due to a relatively low cost and high degree of discrimination [5]. Conditions associated with a mass disaster often cause severe fragmentation, decomposition and commingling of the remains of victims [5]. Challenges associated with sampling of remains include the number of victims, mechanisms of body destruction and extent of body fragmentation. Body accessibility can also make sample collection difficult [13].
1.3. Efficient PM Protocols
Traditional DVI samples such as femurs are difficult to collect requiring surgical removal from the deceased and several additional processing steps prior to DNA analysis [9]. Recent studies have highlighted the availability of DNA from alternative sample types [8] which may offer time and resource savings by removing or limiting time-consuming sample collection, preparation and extraction [8]. Advances in technology have also increased options for the collection, sampling, preservation and DNA processing of samples [8]. It follows that this study has focused on minimally-invasive nail and distal phalanx sample types.
Nail samples are easily accessible, are associated with a simple and non-invasive collection with fewer health and safety risks [14,15], can be collected with minimal training, and require less storage without the need for refrigeration [14,16]. Both nail clippings and the nail bed offer a source of DNA [16] and the success of recovering DNA from nail has been shown in decomposed remains [14,17]. Toenails have specifically been targeted due to their limited exposure to exogenous DNA sources due to protection by footwear including socks [9]. It has also been suggested that toenails may yield less mixed profiles compared to fingernails [14]; an hypothesis confirmed empirically [18].
Small cancellous bones (e.g., finger, toe and ankle bones) yield more DNA than cortical bones at increasing post mortem intervals (PMIs) [11]. Intra-individual comparison of DNA yields showed that, on average, the small cancellous bones have much higher concentrations of DNA per unit mass, than dense cortical bones such as femur [11]. The phalanges of the hallux have been used for DNA-based identification in the Brazilian floods and mudslides in January 2011, mostly resulting in DNA yields of > 1.0 ng/µL with the added benefit of a simple collection method [10]. Consequently, distal phalanges show promise for rapid identification by the removal of the tip of a finger and/or toe [19].
Time and cost-effective DNA profiling protocols are invaluable for DVI. Additionally, reducing turnaround times for DNA analysis in forensic casework can streamline investigations, provide investigative leads and exclude persons of interest (POIs) for forensic decision-making [20,21]. If body parts are rapidly re-associated, remains can be compiled for reconciliation following their identification [19].
While sample selection and collection can assist in making downstream processing more efficient, there are approaches within the DNA processing workflow that can also assist in making identification faster. Some of these may include reducing or removing sample preparation, lysis and extraction steps [22,23]; or application of a preservative solution which both preserves at room temperature and leaches DNA into solution. Dimethyl sulfoxide (DMSO)—ethylenediaminetetraacetic acid (EDTA)—salt saturated (DESS) solution has been applied to PM tissue [24] and directly to PCR [25].
Due to the findings of recent studies and advances in technology, there is an opportunity for international DVI standards and guidelines to be revised to reflect new findings. However, INTERPOL DVI Guidelines [26] still recommend the collection of invasive samples such as femur and teeth as the most reliable sources of DNA in collection activities spanning weeks or more or in adverse environmental conditions. However, simple, minimally-invasive collection procedures may not jeopardise DNA recovery and can help in removing the practitioner from hostile environments more promptly. Furthermore, while INTERPOL recommend the use of a preservative for tissue and muscle samples collected for DVI [26], they focus on traditional preservatives such as ethanol which preserve but do not leach DNA from the tissue.
Efficiency has been achieved in real-world DVI efforts by focusing on sample selection and collection. During the 9/11 World Trade Center victim identification effort and the Brazilian floods, the value of the small cancellous bones was reported [10,11,27]. Sampling small bones intact with a disposable scalpel eliminates the laborious task of sawing bone whilst decreasing the risk of contamination and harm to the practitioner [10,11]. During the MH17 operation, the successful direct swabbing of exposed bone marrow after removal of a bone tissue wedge was reported [28]. Swabbing exposed muscle tissue following an incision with a disposable scalpel and has also been shown to be an economical and efficient method for DVI, with transfer to FTA® Card (Whatman®) [27], and a similar method was used in the 2004 South East Asia Tsunami [29]. Applying efficient PM protocols that also incorporate an approach to sample preservation, preparation and DNA testing offers an opportunity to expedite the identification process while minimising the resources and equipment required. An efficient PM workflow can offer protocols that combine these efficient approaches.
1.4. Taphonomic Facility
This study was carried out at the Australian Facility for Taphonomic Experimental Research (AFTER), a 12-acre taphonomic facility located at Yarramundi in the lower Blue Mountains, approximately 65 km north-west of the Sydney central business district. The facility itself adjoins the Blue Mountains National Park and other dense bushland and is in close proximity to both the Nepean River and Lynchs Creek. The temperature at Yarramundi ranges from approximately 8–40 °C throughout the year.
1.5. SAim
This study aimed to develop an efficient method of genetic identification for compromised PM samples and DVI with a focus on minimally-invasive sampling approaches by: (1) assessing nail and distal phalanges as a sample type; (2) attempting to recover DNA from non-pulverised skeletonised remains such as whole phalanges and drilling into long bones; (3) trialling samples subject to various environmental insults such as surface and sub-surface decomposition and various PMIs; (4) applying a range of efficient protocols including leaching preservative solutions; and (5) comparing efficient protocols against current standard operating procedures (SOPs). PM samples were collected from experimental scenarios representing DVI, surface decomposition and sub-surface decomposition.
2. Methods
2.1. Ethics & Governance
Ethics approval for this research was granted by the University of Technology Sydney (UTS) Human Research Ethics Committee (HREC), with approval number UTS HREC REF NO. ETH18-2999. Ethics approval was also granted by the Western Sydney Local Health District (WSLHD) HREC under HREC/17/WMEAD/334 and governance approval by WSLHD Research Governance under SSA/17/WMEAD/547. Samples were transported from AFTER to the NSW Health Pathology, Forensic & Analytical Science Service under a material transfer agreement (MTA).
2.2. Donated Cadavers
PM samples were collected from a number of donated cadavers (Table 1). Cadavers were subject to a range of deposition sites and PMIs representing DVI, sub-surface decomposition and short and longer-term surface decomposition.

Table 1.
Details of body cadavers used in this research.
2.2.1. Experimental Setup
A male cadaver (19-01) was laid unclothed on the surface of a plot at AFTER in February 2019 (Australian summer). The cadaver was unclothed and laid in a supine position. The cleared plot was surrounded by sclerophyll trees. The surface of the plot included dirt, grasses and leaves from the surrounding trees.
2.2.2. Sample Collection
Two phalanges from each of the hand and foot were sampled at time intervals of 0, 2, 6, 10 and 14 days, consistent with a DVI timeframe [30]. To account for intra-individual differences and different sizes of the digits of the hand and feet, phalanges were sampled in duplicate (Table 2). At each time point, two distal phalanges were collected by cutting at the first distal joint with secateurs to remove the digit. Collecting the digit whole provided samples of tissue, nail and distal phalanx bone.

Table 2.
Sampling order of distal phalanges. The thumb and big toe are considered the 1st digits and the pinky and small toe are considered the 5th digits.
A nail clipping was removed from all digits using nail clippers and placed into 1.5 mL tubes. Both small toe samples were placed directly into a 15 mL tube. A ~0.5 cm × 0.5 cm area of tissue was excised from each sample (except small toes) using sterile disposable scalpels and placed in a 1.5 mL tube. Nail bed was sampled using a sterile disposable scalpel by cutting half of the nail bed from the distal to proximal end. A scalpel was used to remove as much adhering tissue as possible from the nail bed and it was placed in a 1.5 mL tube.
Right and left big toes were sampled for the 1st distal phalanx bone. Tissue was removed by making an incision vertically on the bottom of the toe and folding tissue away from the bone. Remaining tissue was removed with a scalpel. After sampling and preparation all samples were weighed.
Nail and tissue samples underwent extraction using the PrepFiler™ BTA Forensic DNA Extraction Kit (Thermo Fisher Scientific: TFS—hereafter referred to as PrepFiler™)—Protocol 1 in Table 3. A reagent blank accompanied every batch of 12 samples. PrepFiler™ Lysis Buffer (PLB) was added and samples vortexed prior to incubation on a MixMate® (Eppendorf®) for 2 h at 56 °C whilst shaking at 900 rpm. Following incubation, samples were centrifuged for 3 min to remove condensation before transferring lysate to a new tube for extraction.

Table 3.
Rapid protocols trialled on surface and sub-surface decomposition remains.
Some samples were also subject to minimal preparation procedures (Table 3). Whole small toe and big toe distal phalanx samples in 15 mL tubes had 500 µL PLB added directly and were incubated on the MixMate® for 2 h at 56 °C whilst shaking at 750 rpm (Protocol 2 in Table 3). Following incubation, samples were centrifuged for 3 min to remove condensation before transferring lysate to a new 1.5 mL tube for extraction. Where distinct lipid layers were present following centrifugation, lysate was transferred and centrifuged again. Extraction was carried out on the AutoMate Express™ Forensic DNA Extraction System (TFS—hereafter referred to as AutoMate) as per manufacturer guidelines. After extraction, final elution tubes were stored in the freezer at −20 °C until quantification.
The two middle toes of the foot (6 day PMI) were also immersed in ~5 mL DESS (Protocol 3 in Table 3), made up according to McNevin (2016). This protocol removed all sample cleaning and preparation steps. At 1, 2, 4 and 8 days, a swab was placed into the solution to collect leeched DNA. The swab was then processed using automated PrepFiler™ lysis and extraction. After the eighth day, the preserved whole toe samples were removed from solution and submitted to the 2 h PLB protocol described previously.
2.3. Surface Remains—DVI Exercise (14–17 Days PMI)
2.3.1. Experimental Setup
In February 2020 (Australian summer), a national Australia New Zealand Policing Advisory Agency (ANZPAA) Disaster Victim Identification Committee (ADVIC) capacity building exercise was held at AFTER and the NSW Forensic Medicine & Coroners Court Complex. The exercise involved six cadavers and two fragmented cadavers decomposing for 2–2.5 weeks following a building collapse scenario. Six cadavers were covered in building rubble (e.g., concrete blocks, pipes, bricks and mild steel reinforcement), while fragmented remains from two cadavers were placed in and around an exploded vehicle adjacent to the building collapse.
2.3.2. Sample Collection
Nail and little toe samples were collected from two cadavers in a temporary mortuary set up at the DVI scene (20-02 and 20-03). These cadavers were decomposing under building rubble for approximately 17 and 14 days, respectively.
2.3.3. Sample Preparation/Examination
No sample preparation or cleaning of samples was carried out. Nail and whole little toe samples were weighed and a reagent blank initiated. Samples were submitted to the 2 h PLB approach as described previously (Protocol 2 in Table 3). Distinct soil and lipid layers were observed and the lysates were re-centrifuged as before.
2.4. Surface Remains—Two-Year PMI
2.4.1. Experimental Setup
A female cadaver (18–14) was laid unclothed in the supine position on the surface of a plot at AFTER in May 2018 (Australian autumn).
2.4.2. Sample Collection
Sample collection occurred in July 2020 after the cadaver was subject to approximately two years of surface decomposition. At collection, the remains were covered in grasses. All five mummified and/or disarticulated distal phalanges of the right hand and foot were collected.
2.4.3. Sample Preparation/Examination
Distal phalanges were cleaned with detergent, de-ionised water (>18 MΩ·cm) and 70% ethanol. Remaining tissue was scraped away and the bone allowed to air dry. Approximately 0.28–0.84 g of bone chips for distal phalanges of the hand and ~0.12–1.20 g for those of the foot were generated and subject to three water washes (water added and agitated in a thermomixer for 5 min at room temperature with shaking at 750 rpm) and an ethanol wash. Bone fragments were irradiated with ultra-violet (UV) light for 15 min on each side before bone powder was generated by cryogenic milling in a freezer mill.
Approximately 0.21–0.76 g of bone powder for distal phalanges of the hand and ~0.09–1.01 g for those of the foot was extracted using a total demineralisation buffer (0.5M EDTA, n-Lauroylsarcosine and Proteinase K (Pro K)) followed by a silica-based clean-up using Amicon® Centrifugal Filter Device (Merck Millipore) concentration and QIAquick® PCR Purification Kit (QIAGEN) purification modified from [33,37,38]—Protocol 4 in Table 3. This method of extensive decontamination, milling and total demineralisation followed by a silica-based clean-up is currently considered the gold standard for skeletal remains but is a lengthy and laborious procedure [39,40].
2.5. Surface Remains—Four-Year PMI
2.5.1. Experimental Setup
A male cadaver (16-03) was laid unclothed in the supine position on the surface of a plot at AFTER in February 2016 (Australian summer).
2.5.2. Sample Collection
Sample collection occurred in July 2020 after the cadaver was subject to approximately four years of surface decomposition. At collection, the remains were fully skeletonised and disarticulated. Nine distal phalanges of the feet were collected excluding the 1st distal phalange of the left foot as it was fused with the 1st proximal phalanx.
2.5.3. Sample Preparation/Examination
Soil and moss were cleaned off the distal phalanges with wipes and rinsing in water. Bones were cleaned using 10% bleach, twice with sterile water and then with 100% ethanol. Distal phalanges were then placed into a 15 mL tube whole, or placed within a Day 2 Day Towel (Livingstone) and hit with a hammer 2–3 times before adding bone pieces into a 15 mL tube. Samples had 500 µL PLB added as previously described, except that 15 min and 2 h incubations were trialled—Protocol 5 in Table 3. By applying field-amenable rapid or nil cleaning and preparation steps for bone, combined with an assessment of a 15 min lysis incubation against a standard 2 h incubation, Protocol 5 sought to expedite DNA testing overall. Following lysis, processing was completed by automated extraction and genotyping. Two of the distal phalanges were also subject to the cleaning, milling and total demineralisation protocol as described earlier, allowing for a comparison of the efficient protocol to the current gold standard approach for skeletal remains.
2.6. Sub-Surface Remains
2.6.1. Experimental Setup
Two plots (one cadaver per plot) were allocated at AFTER for a shallow grave study. An excavator was used to clear each plot and machine dig graves to approximate dimensions of 2 m × 0.5 m × 0.5 m, which were later refined using a shovel.
In July 2018 (Australian winter), two cadavers were clothed and their temperatures taken (under the armpit) prior to being placed in the shallow graves. The male cadaver (18-16) wore a short sleeved (cotton) shirt and shorts, and the female cadaver (18-17) wore a long sleeved (cotton) shirt and jeans. The male cadaver (18-16) was measured at 2 °C and female cadaver (18-17) measured 8 °C. The difference in temperature was attributed to cadaver 18-16 being frozen while 18-17 had been refrigerated. Cadavers wore socks and shoes on alternate feet to allow comparison of DNA yield of samples with and without footwear (Figure 1). Cadavers were placed in the shallow graves in the supine position and the graves were filled in completely. The centre point of each grave was marked at the time of burial to facilitate later excavations.
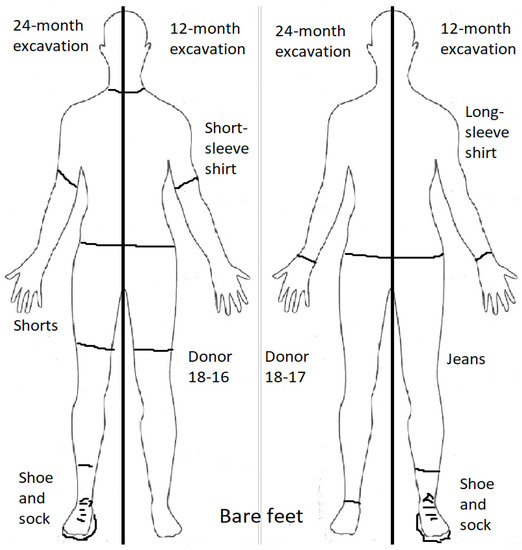
Figure 1.
Experimental setup of sub-surface decomposition in shallow graves. One cadaver was laid in the supine position in each grave. Cadavers were dressed in either short or long clothing, and with and without footwear to provide data for 1 and 2 year excavations.
2.6.2. Grave Excavation
At one and two years, one half of each of the shallow graves was excavated to recover PM samples from the two cadavers. At one year, the excavation was conducted with the assistance of an archaeologist by locating the cut of the grave, exposing the long bones and then the left side of the cadavers. Surrounding soil was sifted for remains and items. At two years the excavation of the other half of each shallow grave was carried out during a NSW Police Force (NSWPF) Crime Scene Section training exercise hosted at AFTER. NSWPF crime scene examiners located cadavers using ground penetrating radar, gridded out the graves, exposed cadavers and sifted surrounding soil for remains and items.
At the year one excavation, differences in decomposition between the two cadavers was observed. The male cadaver (who was frozen before burial) was observed to be mummified with adiopocere present in parts, while the female cadaver (refrigerated) was skeletonised. At the year two excavation, both cadavers were more skeletonised although some tissue did remain, particularly around the male cadaver’s torso area.
2.6.3. Sample Collection
Samples of distal phalanges from the hands, feet and femur were collected (Table 4). Nails were very difficult to locate if not contained in a shoe and therefore deemed unsuitable for collection in sub-surface remains cases with PMI one year or longer.

Table 4.
PM samples collected from sub-surface cadavers in 2019 and 2020.
A large amount of tissue remained attached to both femurs of one cadaver over both years. Consequently, the femur was not collected whole as for the other cadaver. Instead, a ~5 cm × 5 cm window of bone was cut out of the femur using a portable bone saw. At year two, tissue was removed from the proximal area of the femur shaft with a disposable scalpel and the surface wiped with ethanol before drilling into the femur with a portable drill and 6–8 mm drill bit to generate ~100 mg of bone drillings collected on UV-sterilised foil. The femur from the other cadaver was collected whole at the two-year time point. Following each excavation, graves were filled in.
2.6.4. Sample Preparation/Examination
Soil was cleaned off the distal phalanges using detergent wipes and rinsing in water. Bones were then cleaned, prepared and lysed in 500 µL PLB as described previously except that only a 15 min incubation was applied (Protocol 5—Table 3). Additionally, distal phalanges were cleaned and prepared as described for other samples but then placed directly in demineralisation buffer—Protocol 6 in Table 3. Protocol 6 removes extensive decontamination and preparation steps including milling prior to total demineralisation. A 1st distal phalanx from year two remains (18-17) was crushed in a BioPulveriser 59014N (BioSpec) generating ~100 mg of bone powder which was collected in a 1.5 mL microcentrifuge tube.
Drillings were generated from the femur window as well as from year one and year two whole femur shafts (18-17) in the laboratory as per the method used in the field. Approximately 100 mg of femur drillings were transferred into a 15 mL tube and no further cleaning or preparation took place prior to lysis in 500 µL PLB—Protocol 7 in Table 3. Protocol 7 offers an in-field collection technique that also negates the need for bone cutting equipment thus reducing the generation of bone powder by sanding and cutting, and the risk of cross contamination. No further preparation prior to a 15 min incubation in PLB is required. The ~100 mg of year two femur drillings and powdered 1st distal phalanx (18-17) were lysed in 230 µL PLB for 2 h at 56 °C whilst shaking at 1100 rpm.
2.7. Quantification and Genotyping
Samples were quantified using the Quantifiler™ Trio DNA Quantification Kit (TFS: hereafter referred to as Quantifiler™) and subject to quantitative real time PCR on the QuantStudio™ 5 Real-Time PCR System (TFS) according to manufacturer guidelines [41]. Quantifiler™ contains an internal positive control (IPC) system and amplification targets of different sizes to report a Degradation Index (DI), providing an objective measure of PCR inhibition and DNA degradation, respectively [42]. When DNA degrades, the large autosomal (LA) target fragments are selectively depleted, increasing DI [43]. The DI has been shown to effectively characterise degraded samples. With sufficient template, complete or partial STR profiles are expected from mildly degraded (DI < 4) samples, and partial profiles are expected from moderately degraded (DI > 4) samples [44]. Samples with a failed IPC or quantification value in excess of the standards range (0.005–50 ng/µL) were diluted and re-quantified.
Samples were amplified and genotyped using the PowerPlex® 21 System (Promega: hereafter PowerPlex®) according to manufacturer guidelines [45]. A DNA input amount of 0.5 ng is recommended via an input volume of 15 µL [45]. This corresponds with a concentration of ~0.0333 ng/µL although a lower yield may still generate a useful profile. Thermal cycling was carried out on a ProFlex™ PCR System (TFS) and amplified for 29 cycles as per manufacturer guidelines. Capillary electrophoresis (CE) was carried out on a 3500xl Genetic Analyzer (TFS). Fragments were separated according to manufacturer guidelines [46] following the ‘HID’ application type using 36 cm capillaries and POP-4 polymer.
2.8. Data Analysis
Samples were analysed using GeneMapper® ID-X Software (TFS) and a peak height analytical threshold of 175 relative fluorescent units (rfu) and homozygous threshold of 700 rfu was applied. Statistical tests were applied (for n ≥ 5) to determine significant differences in allelic recovery using the Statistical Package for the Social Sciences (SPSS®) (IBM). Statistical tests for normality and variance were performed using Shapiro-Wilk and Wilcoxon Signed Rank tests, respectively.
3. Results
The different PM samples and efficient protocols were compared based on DNA yield according to quantification values and/or allelic recovery in PowerPlex®. A listing of results from all PM samples is provided in Supplemental Table S1. Small autosomal (SA) target, LA target and the DI values from Quantifiler™ were used to assess sample quantity and quality.
3.1. Surface Remains
3.1.1. 0–14 Day PMI
Mummification of the cadaver was observed by day 10 and partial skeletonisation had occurred by day 14. All distal phalanges including nail were present and available for collection at each of the five time points. However, as decomposition progressed, the mass of samples appeared to decrease.
Quantification values ranged from 0.0062–650 ng/µL with sufficient DNA yields recovered across all time points (Figure 2). Due to a large number of samples with high DNA yields (>50 ng/µL), dilution (1:20) and re-quantification was required. Most samples were mildly degraded (i.e., DI < 4), with a few samples exhibiting moderate degradation (i.e., DI 5–8).
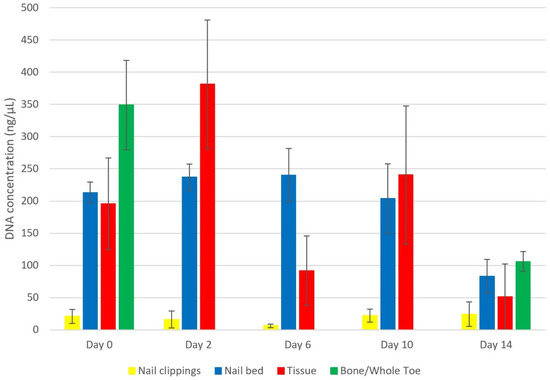
Figure 2.
Average DNA concentration (ng/µL) for nail clippings (n = 20), nail bed (n = 18), tissue (n = 18) and bone/whole toe (n = 4) at 0–14 day time points of summer surface decomposition. Bone and whole toe were subject to a rapid PrepFiler™ lysis (Protocol 2). The other samples were processed using standard PrepFiler™ (Protocol 1). Standard error (SE) bars show the uncertainty of the estimates.
Genotyping was carried out according to DNA input calculated from quantification values of the LA target. All but two samples genotyped (n = 18), yielded full single source profiles, however some were weak with clear signs of degradation. The two day 14 tissue samples yielded 57% and 80% complete profiles. The application of whole toe and distal phalanges to the 2 h PLB with no preparation (Protocol 2) recovered full DNA profiles in PowerPlex® across 0–14 day PMIs.
The DESS solution in which toes were soaked (Protocol 3) yielded complete DNA profiles at all time points; 1, 2, 4 and 8 days after immersion. Quantification values ranged from 2.6–15.2 ng/µL with all DIs <1.5. Generally, an increase in DNA concentration was observed over time in DESS solution but sufficient DNA was still recovered after one day (Figure 3). While one of the day 8 samples resulted in suboptimal amplification, re-amplification of the same dilution and input amount resulted in a complete DNA profile. The first failed amplification was likely due to an extensive dilution being carried out on an automated platform with small input volumes. The two DESS-preserved whole toe samples later applied to the 2 h PLB protocol also yielded complete DNA profiles.
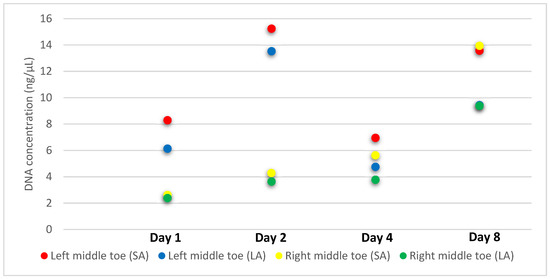
Figure 3.
Small autosomal (SA) and large autosomal (LA) target DNA concentrations (ng/µL) recovered from 6 day post mortem interval toes (n = 2) immersed in leaching preservative solution (Protocol 3) across four time points.
3.1.2. DVI Exercise (14–17 Days PMI)
Nail and whole little toe samples collected during the DVI exercise (Protocol 2) were associated with greater sample degradation with DI values ranging from 3.3–14.4. Samples were also associated with greater sample inhibition with a high IPC threshold cycle (CT > 27) indicating suppressed amplification. Quantification values ranged from 0.0095–0.16 ng/µL, all of which were sufficient for genotyping although one sample required dilution and re-quantification. Allelic recovery ranged from 45–85%.
3.1.3. Two-Year PMI
Milled distal phalange samples yielded 0.21–0.76 g of bone powder from the hand and 0.14–1.01 g of bone powder from the foot. The largest amount of bone powder recovered was from the 1st distal phalanx of the thumb and big toe, respectively.
Following total demineralisation and silica-based clean-up (Protocol 4), quantification values ranged from 15 ng/µL (5th distal phalanx of the foot) to 127 ng/µL (3rd distal phalanx of the hand) (Figure 4). It follows that the 3rd distal phalanx of the hand provided the highest hand DNA yield (despite the 1st distal phalanx yielding the most bone powder), and overall yield, while the 1st distal phalanx of the foot yielded the highest foot DNA concentration (91 ng/µL). DI values ranged from 0.85–1.90. While all DNA concentrations were more than sufficient for genotyping, allelic recovery ranged from 80–100% (Figure 4).
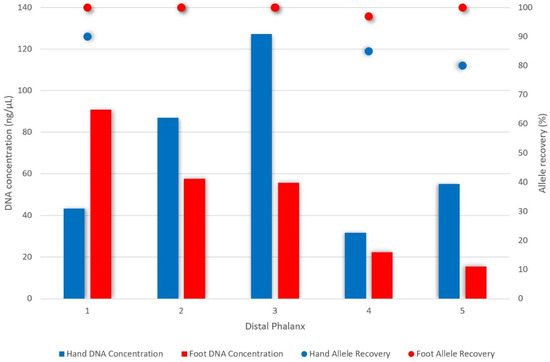
Figure 4.
DNA concentration (ng/µL) and allelic recovery (using the PowerPlex® 21 System) of distal phalanges of the hand and foot from two-year surface remains (Protocol 4, n = 10). Overall, the greatest DNA yield was recovered from the 3rd distal phalanx of the hand (127 ng/µL). The distal phalanx of the big toe gave the highest yield for foot samples (91 ng/µL). Allelic recovery ranged from 80–100% complete profiles with distal phalanges of the foot generally yielding more complete profiles despite higher quantification values recovered for the hand.
3.1.4. Four-Year PMI
The four-year surface distal phalanges weighed from 0.18–2.2 g with the 1st distal phalanx weighing significantly more than other distal phalanges (0.18–0.33 g). Quantification values from milling and total demineralisation (Protocol 4), and whole and crushed distal phalanges subject to 15 min and 2 h PLB incubations (Protocol 5), ranged from 0.0025–3.4 ng/µL (Figure 5), with samples subject to total demineralisation yielding larger concentrations of DNA (2.0 and 3.4 ng/µL). However, genotyping saw most samples generate ~47–92% complete profiles with one sample yielding ~11% complete profile (Figure 6). While demineralised samples consistently yielded more complete profiles (78% and 85%), the whole bone and 15 min incubation gave the highest allelic recovery with a ~92% complete profile.
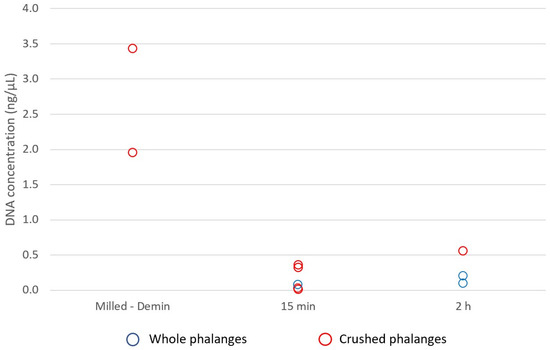
Figure 5.
DNA concentration (ng/µL) of distal phalanges of the feet from four-year surface remains. Samples were subject to a milled preparation followed by total demineralisation (Protocol 4), or a whole or crushed preparation followed by either a 15 min or 2 h PrepFiler™ Lysis Buffer incubation (Protocol 5). Milled and demineralised samples recovered the greatest DNA concentrations.
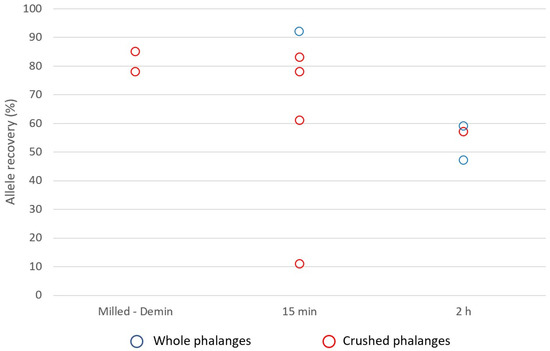
Figure 6.
Allelic recovery of four-year surface distal phalanges of the feet using the PowerPlex® 21 System. Recovery mostly ranged from ~47–92% complete profiles with one sample yielding an ~11% complete profile. Whole bone with a 15 min incubation (Protocol 5) gave the highest recovery with a ~92% complete profile.
3.2. Sub-Surface Remains
3.2.1. Sample Collection
At year one and two excavations, samples of nail, bones of the hands and feet, and femur were successfully recovered. While nail and distal phalanges contained within a shoe were easily collected, several nail samples and distal phalanges were unable to be located within the grave due to detachment and/or disarticulation when shoes were not worn. Disarticulation of the hand was more evident in one of the cadavers. Attempts to recover the nails and distal phalanges by sifting was also unsuccessful and the success of locating nail samples was largely found to be dependent upon finding an attached distal phalanx. Consequently, nails were deemed unsuitable for collection in sub-surface remains cases and were not tested.
The collection of bone drillings was found to be a simple and effective method for in-field and in situ sample collection. While whole femur collection was simple for one of the cadavers, collecting ~100 mg of bone drillings from the other was found to be much simpler than removing the whole femur due to large amounts of attached tissue. Collecting drillings was also simpler than the removal of a femur window, particularly in field conditions. However, drillings were easily generated from the femur window (and femur shaft) in the laboratory.
3.2.2. DNA Testing
The femur drillings and 1st distal phalanx (crushed in the BioPulveriser) subject to 230 µL PLB for 2 h yielded 0.0016 ng/µL and 0.019 ng/µL of DNA, respectively. Genotyping produced a ~35% complete and a complete profile, respectively. Across the one and two-year time points, quantification values ranged from 0.0021–2.2 ng/µL for one-year-old distal phalanges, and an undetectable amount (per µL) to 1.1 ng/µL for two-year-old distal phalanges (Figure 7). One-year-old phalanges yielded an average of 0.44 ng/µL of DNA, while two-year-old phalanges yielded an average 0.11 ng/µL. Quantification values from whole and crushed distal phalanges subject to 15 min PLB incubation (Protocol 5) ranged from an undetectable amount to 1.86 ng/µL (Figure 7). Data for the whole bone and crushed bone protocols both deviated significantly from normal distributions. The average quantification value for the whole bone protocol was 0.21 ng/µL, while the crushed bone protocol recovered an average of 0.20 ng/µL—there was no significant difference using a Wilcoxon signed-rank test (z = −0.663, p > 0.05). The highest quantification value was recovered from a 2nd distal phalanx recovered in a shoe after one year of burial, by submitting the whole distal phalanx for a 15 min incubation.
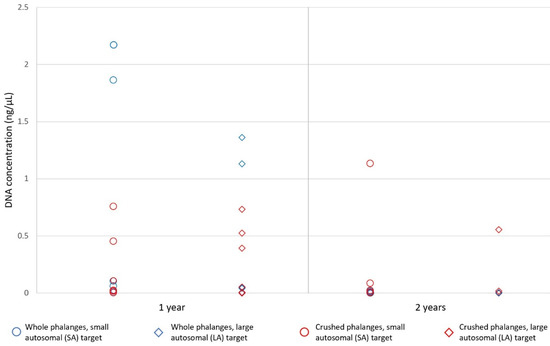
Figure 7.
Regardless of protocol (i.e., 5 or 6), small autosomal (SA) and large autosomal (LA) target quantification values ranged from 0.0021–2.8 ng/µL for one-year-old phalanges, and an undetectable amount (per µL) to 1.1 ng/µL for two year-old distal phalanges. Quantification values (SA and LA targets) from whole and crushed distal phalanges subject to a 15 min PrepFiler™ Lysis Buffer incubation (Protocol 5) ranged from an undetectable amount (per µL) to 1.86 ng/µL.
Overall, DI values ranged from 1.00 (from a one-year crushed foot phalanx) indicating no degradation, to 198 (from a two-year crushed foot phalanx) indicating severe degradation—defined as DI > 10 [44] (Figure 8). Some DIs were unable to be calculated due to an undetectable amount of DNA from the LA target. DIs ranged from 1.0–19.9 for one-year-old phalanges, and 2.0–198 for two-year-old phalanges.
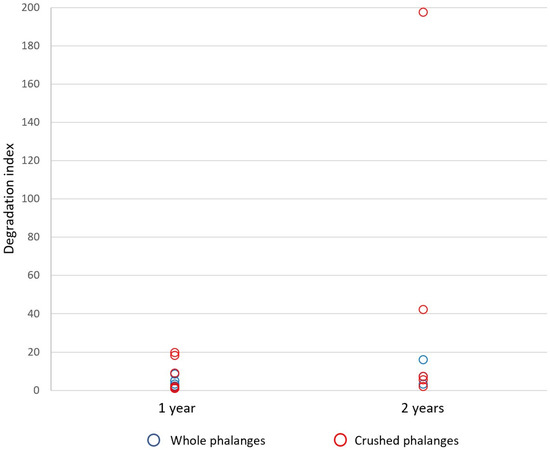
Figure 8.
Degradation Indices (DI) from whole and crushed distal phalanges subject to a 15 min PrepFiler™ Lysis Buffer incubation (Protocol 5). Overall, values ranged from 1.00 indicating no degradation, to 198 indicating severe degradation. DIs ranged from 1.0–19.9 for one-year-old phalanges, and 2.0–198 for two-year-old phalanges.
Genotyping resulted in a range of profiles from 0% to 100% complete and data were not normally distributed (Figure 9). The whole bone protocol recovered an average ~52% complete profiles and the crushed bone protocol recovered ~64% complete profiles but the difference was not significant using a Wilcoxon signed-rank test (z = −0.676, p > 0.05). One-year old distal phalanges ( = ~83%) yielded significantly more complete profiles than two-year old distal phalanges ( = ~32%) using a Wilcoxon signed-rank test (z = −3.062, p < 0.05).
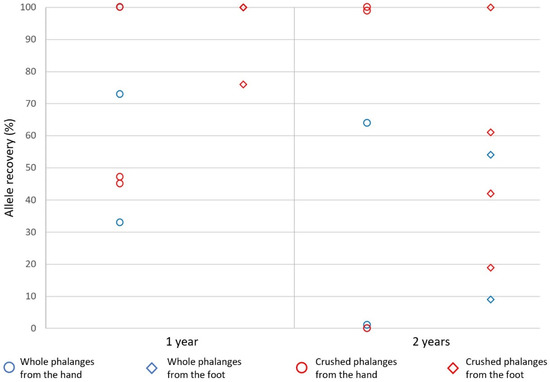
Figure 9.
Allelic recovery of crushed and whole sub-surface distal phalanges of the hands and feet using the PowerPlex® 21 System (Protocol 5). Recovery ranged from 0–100% complete profiles with the whole bone protocol yielding an average ~52% complete profiles and the crushed bone protocol recovering ~64% complete profiles. One-year old distal phalanges ( = ~83%) yielded significantly more complete profiles than two-year old distal phalanges ( = ~32%) using a Wilcoxon signed-rank test (z = −3.062, p < 0.05).
Whole and crushed bone preparation followed by total demineralisation (Protocol 6, n = 4) yielded quantification values ranging from 0.018–2.2 ng/µL and DI values 1.9–18. Genotyping recovered complete profiles from two whole bone applications (one-year PMI). From crushed bone, a complete profile (two-year PMI) and a 76% complete profile (one-year PMI) were recovered.
Adding ~150 mg of femur drillings subject to 15 min PLB (n = 3) yielded 0.011–0.038 ng/µL of DNA with DIs ranging from 1.7–5.1. Genotyping resulted in two complete profiles from different femurs of the same cadaver (one- and two-years post burial) and a ~97% complete profile from a femur (one year post burial).
4. Discussion
The efficient DNA protocols were developed across a number of different PM sample types, PMIs and environmental conditions, and were compared to gold standard extraction methods. By conducting this study at a human taphonomic facility, authentic PM samples in various stages of decomposition could be tested. Consequently, this study offers unique perspectives and directions for future research into the application and optimisation of efficient protocols for the identification of decomposing and skeletonised human remains. Applying efficient PM protocols that also incorporate preservation and minimal preparation of samples offers an opportunity to further expedite identifications and provide early intelligence, while minimising the use of valuable resources and equipment.
4.1. Novelty and Practicality
Several studies have investigated alternative methods for genotyping bone samples including bone powder-free protocols [22,47,48]. Since this research was conducted, another study has shown that PrepFiler™ BTA is often sufficient for genotyping degraded bones after demineralisation of large bone fragments [49]. Additionally, Hasap et al. [50] describe a novel 4 h DNA extraction method for STR typing of casework bone samples. Our protocols are faster, combining and modifying previous work [22,23] to develop a powder-free, rapid 15 min PrepFiler™ Lysis Buffer incubation that is compatible with downstream automated processing; reducing sample preparation and extraction to 2 h. Applying minimally-invasive samples such as nail and distal phalanges further expedites the process. When considering previous DVI efforts that have successfully increased efficiency by simplifying sample selection and collection [10,11,27,28,29], this study offers more efficiency gains by addressing sample preservation, preparation and processing, as well as novel and practical in-field solutions.
4.2. Efficient DNA Approaches
Sample selection and collection can assist in making downstream processing more efficient. With respect to DNA processing, sample preparation can be removed or limited, and lysis buffers can be added directly to whole samples followed by a shorter incubation time [19,22,23]. These crude approaches are likely facilitated by an increased sensitivity and tolerance to inhibitors present in modern commercial multiplexes [51,52,53].
4.2.1. Minimally Invasive Collection
Testing of surface remains showed nail and distal phalange including whole digit samples were simple to collect and minimally-invasive. These samples also require less room for storage which will be an important factor where storage space, especially in a freezer, is limited. This is often the case where temporary facilities may be deployed to remote and hostile locations [54].
In remains with shorter PMIs typically encountered in DVI scenarios, collection of a whole digit offered another simple approach for collecting nail, tissue and bone whilst facilitating retesting. Removing digits was easily achieved by cutting at the first distal joint with secateurs. The use of a cutting implement over a scalpel also reduces health and safety risks but increases contamination risks. Sampling of nail clippings only was proven to be an effective approach, however decomposed remains recovered in the DVI exercise demonstrated that nails could be lost in the field, during transport to the temporary mortuary, or within the body bag or bagged hands. While targeting toenail may reduce the instance of extraneous DNA [9,14], nail selection was limited by availability in a number of scenarios.
In mummified remains, distal phalanges were collected by cutting through mummified tissue with a disposable scalpel. In skeletonised remains, collecting disarticulated distal phalanges by locating them amongst grasses and foliage was often challenging. It should be considered that distal phalanges may not always be present due to environmental factors or animal predation. Their actual presence and/or location in relation to the body may impact the ability to associate samples with that individual and their identification.
Sub-surface remains presented further challenges in recovering nail and distal phalanges. Locating them was largely dependent upon finding them attached to distal phalanges. Therefore, they are not recommended target samples for identification of sub-surface remains although feet in shoes are an exception. In sub-surface scenarios, femurs were easier to locate and drillings were able to be generated in the field and in situ, offering a possible solution in the event distal phalanges are unable to be recovered.
4.2.2. Minimal Preparation
Processing nail and distal phalanges rather than traditionally-targeted tooth and femur sample negates the requirement for laborious and time-consuming cleaning, preparation, sampling and total demineralisation DNA extraction methods. Instead, no or minimal cleaning and preparation can be employed. Pursuing these minimally-invasive sample types also limits the requirement for subsampling. Following collection, whole digits, distal phalanges and femur drillings require no further preparation.
While nail clippings were simple to process requiring no preparation following collection (though commingled remains may warrant decontamination steps prior to testing), sampling nail bed from mummified remains was challenging. Likewise, although greater amounts of DNA were recovered from nail bed, nail clippings yielded sufficient DNA for genotyping over a two-week PMI. Nail samples could also be applied to fully-automated laboratory processing with no cleaning required. During the DVI exercise involving decomposed and commingled remains, samples benefitted from the removal of attached decomposed tissue to mitigate against the increased presence of lipid layers following incubation and subsequent centrifugation. The simple removal of inhibitors such as dirt or soil, decomposition by-products and other inhibitors by rinsing with sterile water could also negate retesting.
Applying whole digits to PLB or DESS solutions was a simple and effective approach with no preparation required. DESS solution has been shown to preserve sample and DNA at room temperature as well as at elevated temperatures associated with mass disaster sites in tropical climates [30]. Collected digits were stored in a 50 mL tube and 5 mL of DESS solution was sufficient for full immersion of a middle toe. No incubation or interaction was required following its addition and DNA was collected by immersing a swab into the solution to collect leeched DNA. This approach offers multiple opportunities for retesting: if the swab is unsuccessful, then nail clipping can be removed, then whole nail and, finally, bone. Alternatively, preserved toe sample can be removed from preservative solution and further lysed by the addition of 500 µL PLB without further preparation.
While a combination of physical and chemical cleaning followed by UV irradiation is recommended as the best way to decontaminate bone [55,56], an efficient decontamination protocol for distal phalanges was applied by cleaning with 10% bleach, twice with sterile water and 100% ethanol. This approach facilitates efficient testing without contamination as evidenced by no extraneous DNA detected in recovered DNA profiles, controls and reagent blanks. Similarly, no extraneous DNA was detected from the femur drilling samples. The main issue for distal phalanges recovered in sub-surface environments is the amount of soil remaining even after cleaning. Optimising an effective and efficient protocol for removing soil prior to DNA testing would assist in their rapid processing.
Placing distal phalanges within a Day 2 Day Towel kept the sample contained while breaking bone with a hammer. This approach was thought to open the bone matrix and facilitate lysis of DNA as it has been reported that fine granulation of bone powder is a crucial step in efficient DNA extraction from bone powder [23]. However, there was no significant difference between DNA yield from whole and crushed bones suggesting that crushing bones was unnecessary. Submitting phalanges whole was a simpler approach further reducing preparation.
4.2.3. Minimal DNA Processing
Incubating nail, distal phalanges and whole digits in 500 µL of PLB for 2 h was an efficient and simple method, limiting or removing sample preparation. The 500 µL volume of PLB was an upper limit imposed by lysate processing on the AutoMate but this was sufficient to submerge most samples. Incubation in the MixMate® likely facilitated coverage of larger samples due to heating and mixing. Genotyping successfully yielded DNA profiles from nail and digit samples subject to up to 2.5 weeks of surface decomposition, and distal phalanx samples up to four years surface decomposition.
Further, a reduced 15 min incubation also yielded DNA profiles suggesting that a shorter incubation may lyse sufficient DNA. DNA profiles were obtained from distal phalanges subject to four years of surface decomposition and up to two years of sub-surface decomposition although sub-surface distal phalanges were inconsistent, particularly at the two-year time point. A significant decrease in allelic recovery from sub-surface distal phalanges at the two-year time point suggests alternative PM samples such as femur drillings should be targeted after one year of burial. After two years of burial, ~100 mg of drillings was sufficient to recover complete DNA profiles. Despite the small sample size (n = 3), femur drillings offer a promising approach particularly where nail and distal phalanges are absent in sub-surface environments.
Adding 15 mL of demineralisation buffer to whole and crushed distal phalanges also produced encouraging results. Three complete profiles and one ~76% complete profile were recovered across one- and two-year distal phalanx samples. Although subsequent demineralisation and silica-based clean-up is laborious; the removal of physical cleaning, chemical cleaning and milling prior to demineralisation offers a significant time reduction, especially where whole bone is applied.
4.2.4. Limitations
Following the PLB incubation and centrifugation to remove condensation prior to lysate transfer, samples with large amounts of tissue such as decomposed nail and whole digits resulted in distinct lipid layers. Similarly, long-PMI surface and sub-surface distal phalanges resulted in distinct soil layers. While this was addressed by pipetting off the lipid layers and re-centrifuging, this required manual intervention and added to processing time.
Many samples yielded a DNA concentration of more than 50 ng/µL DNA which is more than the highest Quantifiler™ standard concentration. Dilution and re-quantification of these samples was required. As retesting adds to processing time, samples that yield sufficient but not excess DNA help to negate workflow intervention and re-testing.
The degraded and inhibited nature of the samples tested in this study often meant partial profiles were recovered. PowerPlex®, a 20-locus multiplex, can provide statistical discrimination in the order of 10−25 for a full profile, depending on population databases and accounting for subpopulation effects [57]. Partial profiles can still provide useful information for identification by offering statistical support in conjunction with other supporting evidence, or by being applied in other ways such as the re-association of remains where discrimination may be less critical. In a closed disaster, statistical discrimination may also be less critical for the DVI effort, particularly where a manifest is available.
4.3. Comparison to Standard Laboratory Typing
Hard tissue cleaning procedures are traditionally utilised for nail samples including 5% Tergazyme® (Alconox) and ultrasonic water baths [58]. In the efficient approach, no cleaning was carried out and processing nail clippings instead of whole nail offered time savings by removing preparation completely. If clippings were collected in the field, no additional intervention would be required following sampling. Despite an average mass ~1/10 of nail bed, nail clippings still gave an average yield of 24 ng/µL after 14 days of summer surface decomposition, and an overall average yield of 18 ng/µL over 0–14 day timeframes (n = 20). However, this may be affected by the scenario as biological fluids from another body such as blood could be present on the nails. Genotyping of our samples (n = 6) yielded complete profiles for day 0 and 14 samples of both the hands and feet with no contamination observed.
Whole nail samples collected from the DVI exercise yielded suboptimal results. Standard testing would remove attached decomposed tissue and utilise hard tissue cleaning procedures [58], removing inhibitory material. Whole toe samples were applied to the 2 h PLB protocol without cleaning and preparation. For these samples, in contrast to the nail samples, the efficient protocol consistently yielded complete profiles without contamination.
Using a gold standard extraction method (i.e., total demineralisation), the utility of distal phalanges as a source of DNA was reinforced for surface remains, even after two years. For skeletal samples, efficient approaches such as whole and crushed phalanges with a 15 min incubation, although recovering less DNA than milling and total demineralisation protocols, provided sufficient DNA for profiling which in some cases gave greater allelic recovery. By applying whole (or crushed) bone to a 15 min PLB incubation instead of bone preparation, milling and a total demineralisation protocol means that a turnaround time of 1.5 days is reduced to approximately 2 h.
4.4. Optimisation of PM Sample Types
4.4.1. Intra and Inter-Individual Differences
Profound differences were observed in the way cadavers decomposed and/or mummified. Not only did this vary based on their environmental exposure, i.e., surface or sub-surface, but there were also differences between cadavers in adjacent plots. Season, deposition environment, clothing, body mass index (BMI), age, sex, pathology, diet and the unique Australian climate may impact the decomposition process. However, these factors seemed to have minimal impact on recovery of DNA from nail and distal phalanx samples.
The DNA yields of nail clippings and nail bed samples were observed to be random across the different digits of the hands and feet, and PMIs. That is, larger digits did not necessarily yield more DNA than smaller digits and this was observed over 0–14 day PMIs. Furthermore, DNA yield from nail clippings and nail bed did not appear to differ between the hands and feet. Consequently, any available nail sample is suitable for collection and testing.
Allelic recovery was higher for distal phalanges from the foot than from the hand after total demineralisation and silica-based clean-up. This was in spite of the fact that DNA quantities were generally lower from the foot phalanges. These results may not necessarily reflect DNA recoveries using other DNA extraction procedures, e.g., PrepFiler™.
4.4.2. Future Studies
Optimising the application of these sample types should be the focus of future studies although more replicates across different scenarios will confirm if success is determined by each sample’s circumstances. This could be achieved by empirically determining optimal input amounts and optimising pre-treatment steps to allow sufficient cleaning and optimal lysis sufficient to eliminate inhibition and contamination. Because the efficient approaches described in this study have been developed to be compatible with backend automated processing, they could also be tested on other commercially-available multiplex kits.
4.5. Recommendations
Several recommendations are offered according to PMI and deposition site (Table 5). For surface remains with a PMI of up to 2.5 weeks, nail clippings could be applied to a fully-automated workflow with no cleaning or preparation. However, this may not be appropriate for commingled remains and/or obvious contamination by body fluids from other bodies which may warrant decontamination steps prior to testing. Alternatively, immersing whole digits in leaching preservative solutions such as DESS is also a viable option with additional benefits for DVI due to its preservation properties and provision of surplus sample for retesting. For surface remains with a PMI up to four years, whole distal phalanges could be applied to a 15 min PLB incubation protocol.

Table 5.
Recommended efficient protocol based on PMI and deposition site.
For sub-surface remains, collection of nail or distal phalanges in shoes should be targeted. Whole distal phalanges could be subject to a 15 min PLB incubation protocol for a rapid solution however, if remains have a PMI greater than a year, alternative samples should be sought. The collection of duplicate samples provides an option for repeat testing in the event the first sample fails. Femur drillings subject to a 15 min PLB incubation offers an effective alternative. However, further research is required to confirm findings found in this study and results may be dependent on a myriad of variables identified earlier.
Disarticulation and/or animal predation studies may highlight limitations with the availability of distal phalanges. It should also be considered that efficient DNA approaches should not be pursued for every sample. For more compromised samples, laboratory submission for more effective extraction and genotyping may be best. This may be more crucial where limited sample is available for identification.
5. Conclusions
While milling of bone followed by total demineralisation may represent the gold standard for DNA identification from skeletonised remains and certainly recovers more DNA, submission of nail and bone fragments to PrepFiler™ may be sufficient in many cases, especially when there is excess tissue available. Application of whole digits to preservative solutions and submission of nail clippings directly to standard, automated laboratory genotyping pipelines offer even simpler processes for shorter PMIs, of the type encountered in DVI scenarios. Faster DNA analysis in forensic casework produces informative results for forensic decision-making [20] and can streamline the investigation process by providing preliminary leads and early exclusionary evidence during the early crucial stages of an investigation [21]. While the collection of ante mortem samples can be a lengthy process itself, generating PM sample profiles for uploading onto databases can assist the DVI effort early.
Implementing efficient approaches to sample selection, collection, preservation, preparation and DNA testing can reduce identification timeframes whilst reducing costs and time-consuming, laborious processes. Furthermore, combining efficient approaches further facilitates rapid identifications. Simple, in-field sample collection can also free up specialist staff for other complex tasks requiring their expertise. Additionally, downstream DNA processing steps for all efficient protocols described in this study are compatible with high-throughput automated DNA laboratory platforms. The approaches described in this study have the potential to expedite the identification process and provide answers to grieving families sooner.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/forensicsci1030014/s1, Table S1: Summary of cadaver information, exposure, sample preparation and DNA results of rapid and traditional approaches.
Author Contributions
Conceptualization, J.W. (Jeremy Watherston), J.W. (Jodie Ward) and D.M.; methodology, J.W. (Jessica Watson), D.B., J.W. (Jodie Ward) and D.M.; formal analysis, J.W. (Jessica Watson); writing—original draft preparation, J.W. (Jeremy Watherston); writing—review and editing, D.B., J.W. (Jodie Ward) and D.M.; supervision, D.B., J.W. (Jodie Ward) and D.M.; funding acquisition, J.W. (Jodie Ward) and D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. NSW Health Pathology, Forensic & Analytical Science Service provided in-kind support for the DNA testing carried out in the study.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of University of Technology Sydney (UTS) Human Research Ethics Committee (HREC) (UTS HREC REF NO. ETH18-2999 date of approval) and the Western Sydney Local Health District (WSLHD) HREC (HREC/17/WMEAD/334 date of approval). Governance approval was granted by WSLHD Research Governance under SSA/17/WMEAD/547.
Informed Consent Statement
Cadavers used in the exercise were donated to the Australian Facility for Taphonomic Experimental Research (AFTER) through the UTS Body Donation Program. Written informed consent was obtained from each donor, or from their legal guardians.
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on application.
Acknowledgments
The authors gratefully thank: The Australian Facility for Taphonomic Experimental Research (AFTER) for facilitating this research. The AFTER donors from the University of Technology (UTS) Body Donation Program involved in this research for their invaluable contribution to forensic science. NSW Health Pathology, Forensic & Analytical Science Service (FASS) for supporting the research and assisting with genotyping and profile analysis. NSW Police Force Crime Scene Section and Eline Schotsmans (University of Bordeaux/University of Wollongong) for assisting with the excavation of buried cadavers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prinz, M.; Carracedo, A.; Mayr, W.; Morling, N.; Parsons, T.; Sajantila, A.; Scheithauer, R.; Schmitter, H.; Schneider, P.M. DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI). Forensic Sci. Int. Genet. 2007, 1, 3–12. [Google Scholar] [CrossRef]
- Andelinović, S.; Sutlović, D.; Ivkosić, I.E.; Skaro, V.; Ivkosić, A.; Paic, F.; Rezić, B.; Definis-Gojanović, M.; Primorac, D. Twelve-year experience in identification of skeletal remains from mass graves. Croat. Med. J. 2005, 46, 530–539. [Google Scholar] [PubMed]
- Milos, A.; Selmanović, A.; Smajlović, L.; Huel, R.; Katzmarzyk, C.; Rizvić, A.; Parsons, T.J. Success Rates of Nuclear Short Tandem Repeat Typing from Different Skeletal Elements. Croat. Med. J. 2007, 48, 486–493. [Google Scholar] [PubMed]
- Montelius, K.; Lindblom, B. DNA analysis in Disaster Victim Identification. Forensic Sci. Med. Pathol. 2012, 8, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Ziętkiewicz, E.; Witt, M.; Daca, P.; Zebracka-Gala, J.; Jarząb, M.G.B. Current genetic methodologies in the identification of disaster victims and in forensic analysis. J. Appl. Genet 2012, 53, 41–60. [Google Scholar] [CrossRef] [Green Version]
- Calacal, G.C.; Apaga, D.L.T.; Salvador, J.M.; Jimenez, J.A.D.; Lagat, L.J.; Villacorta, R.P.F.; Lim, M.C.F.; Fortun, R.D.; Datar, F.A.; De Ungria, M.C.A. Comparing different post-mortem human samples as DNA sources for downstream genotyping and identification. Forensic Sci. Int. Genet. 2015, 19, 212–220. [Google Scholar] [CrossRef]
- Manjunath, B.C.; Chandrashekar, B.R.; Maheshand, M.; Rani, R.M.V. DNA profiling and forensic dentistry--a review of the recent concepts and trends. J. Forensic Leg. Med. 2011, 18, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Watherston, J.; McNevin, D.; Gahan, M.; Bruce, D.; Ward, J. Current and emerging tools for the recovery of genetic information from post mortem samples: New directions for disaster victim identification. Forensic Sci. Int. Genet. 2018, 37, 270–282. [Google Scholar] [CrossRef]
- Schlenker, A.; Grimble, K.; Azim, A.; Owen, R.; Hartman, D. Toenails as an alternative source material for the extraction of DNA from decomposed human remains. Forensic Sci. Int. 2015, 258, 1–10. [Google Scholar] [CrossRef]
- Ferreira, S.T.; Garrido, R.G.; Paula, K.A.; Nogueira, R.C.; Oliveira, E.S.; Moraes, A.V. Cartilage and phalanges from hallux: Alternative sources of samples for DNA typing in disaster victim identification (DVI). A comparative study. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e366–e367. [Google Scholar] [CrossRef]
- Mundorff, A.; Davoren, J.M. Examination of DNA yield rates for different skeletal elements at increasing post mortem intervals. Forensic Sci. Int. Genet 2014, 8, 55–63. [Google Scholar] [CrossRef]
- Noji, E.K. The Public Health Consequences of Disasters. Prehospital Disaster Med. 2000, 15, 21–31. [Google Scholar] [CrossRef]
- Alonso, A.; Martin, P.; Albarrán, C.; Garcia, P.; De Simon, L.F.; Iturralde, M.J.; Fernández-Rodriguez, A.; Atienza, I.; Capilla, J.; García-Hirschfeld, J.; et al. Challenges of DNA profiling in mass disaster investigations. Croat. Med. J. 2005, 46, 540–548. [Google Scholar]
- Allouche, M.; Hamdoum, M.; Mangin, P.; Castella, V. Genetic identification of decomposed cadavers using nails as DNA source. Forensic Sci. Int. Genet. 2008, 3, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Moriyaand, F.; Hashimoto, Y. Effects of environmental conditions to which nails are exposed on DNA analysis of them. Leg. Med. 2003, 5, S194–S197. [Google Scholar] [CrossRef]
- Ottens, R.; Taylorand, D.; Linacre, A. DNA profiles from fingernails using direct PCR. Forensic Sci. Med. Pathol. 2015, 11, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, A.; Cucurachi, N.; Betti, F.; Capra, M.; Coco, S.; D’Avila, F.; Lorenzoni, R.; Lovisolo, A. Forensic DNA typing of human nails at various stages of decomposition. Int. Congr. Ser. 2006, 1288, 586–588. [Google Scholar] [CrossRef]
- Watherston, J.; Bruce, D.; Ward, J.; Gahan, M.E.; McNevin, D. Automating direct-to-PCR for disaster victim identification. Aust. J. Forensic Sci. 2019, 51, S39–S43. [Google Scholar] [CrossRef]
- Watherston, J.; Watson, J.; Bruce, D.; Ueland, M.; McNevinand, D.; Ward, J. An in-field evaluation of rapid DNA instruments for disaster victim identification. Int. J. Legal Med. 2021. under review. [Google Scholar]
- Butler, J.M.; Willis, S. Interpol review of forensic biology and forensic DNA typing 2016–2019. Forensic Sci. Int. 2020, 2, 352–367. [Google Scholar] [CrossRef]
- Gangano, S.; Elliott, K.; Anoruo, K.; Gass, J.; Buscaino, J.; Jovanovich, S.; Harris, D. DNA investigative lead development from blood and saliva samples in less than two hours using the RapidHIT™ Human DNA Identification System. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e43–e44. [Google Scholar] [CrossRef]
- Harrel, M.; Hughes-Stamm, S. A Powder-free DNA Extraction Workflow for Skeletal Samples. J. Forensic Sci. 2020, 65, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Duijs, F.; Sijen, T. A rapid and efficient method for DNA extraction from bone powder. Forensic Sci. Int. Rep. 2020, 2, 100099. [Google Scholar] [CrossRef]
- Sorensen, A.; Rahman, E.; Canela, C.; Gangitano, D.; Hughes-Stamm, S. Preservation and rapid purification of DNA from decomposing human tissue samples. Forensic Sci. Int. Genet. 2016, 25, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, A.; Berry, C.; Bruce, D.; Gahan, M.E.; Hughes, S.; McNevin, D. Direct-to-PCR tissue preservation for DNA profiling. Int. J. Leg. Med. 2015, 130, 607–613. [Google Scholar] [CrossRef] [PubMed]
- INTERPOL. Disaster Victim Identification Guide; INTERPOL: Lyon, France, 2018. [Google Scholar]
- Mundorff, A.Z.; Amory, S.; Huel, R.; Bilić, A.; Scottand, A.L.; Parsons, T.J. An economical and efficient method for postmortem DNA sampling in mass fatalities. Forensic Sci. Int. Genet 2018, 36, 167–175. [Google Scholar] [CrossRef] [PubMed]
- de Boer, H.H.; Maat, G.J.; Kadarmo, D.A.; Widodo, P.T.; Kloosterman, A.D.; Kal, A.J. DNA identification of human remains in Disaster Victim Identification (DVI): An efficient sampling method for muscle, bone, bone marrow and teeth. Forensic Sci. Int. 2018, 289, 253–259. [Google Scholar] [CrossRef]
- Steinlechner, M.; Parson, W.; Rabl, W.; Grubweiserand, P.; Scheithauer, R. Tsunami disaster: DNA typing of Sri Lanka victim samples and related AM matching procedures. Int. Congr. Ser. 2006, 1288, 741–743. [Google Scholar] [CrossRef]
- Allen-Hall, A.; McNevin, D. Human tissue preservation for disaster victim identification (DVI) in tropical climates. Forensic Sci. Int. Genet. 2012, 6, 653–657. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. PrepFiler™ Forensic DNA Extraction Kit; Thermo Fisher Scientific: Waltham, MA, USA, 2008. [Google Scholar]
- McNevin, D. Preservation of and DNA Extraction from Muscle Tissue. In Forensic DNA Typing Protocols; Methods in Molecular Biology; Goodwin, W., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1420, pp. 43–53. [Google Scholar] [CrossRef]
- Loreille, O.M.; Diegoli, T.M.; Irwin, J.A.; Coble, M.; Parsons, T.J. High efficiency DNA extraction from bone by total demineralization. Forensic Sci. Int. Genet. 2007, 1, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Applied BiosystemsTM. AutoMate Express™ Instrument User Guide; Rev. D; Thermo Fisher Scientific: Waltham, MA, USA, 2012. [Google Scholar]
- TECAN. HID EVOlutionTM—Extraction Application Manual; TECAN: Männedorf, Switzerland, 2008. [Google Scholar]
- Yang, D.Y.; Eng, B.; Waye, J.S.; Dudarand, J.C.; Saunders, S.R. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998, 105, 539–543. [Google Scholar] [CrossRef]
- Huel, R.; Amory, S.; Bilić, A.; Vidović, S.; Jasaragićand, E.; Parsons, T.J. DNA extraction from aged skeletal samples for STR typing by capillary electrophoresis. Methods Mol. Biol. 2012, 830, 185–198. [Google Scholar] [PubMed]
- Edson, S.M. Extraction of DNA from Skeletonized Postcranial Remains: A Discussion of Protocols and Testing Modalities. J. Forensic Sci. 2019, 64, 1312–1323. [Google Scholar] [CrossRef]
- Seo, S.B.; Zhang, A.; Kim, H.Y.; Yi, J.A.; Lee, H.Y.; Shin, D.H.; Lee, S.D. Technical note: Efficiency of total demineralization and ion-exchange column for DNA extraction from bone. Am. J. Phys. Anthr. 2009, 141, 158–162. [Google Scholar] [CrossRef]
- Lee, H.Y.; Park, M.J.; Kim, N.Y.; Sim, J.E.; Yang, W.I.; Shin, K.-J. Simple and highly effective DNA extraction methods from old skeletal remains using silica columns. Forensic Sci. Int. Genet. 2010, 4, 275–280. [Google Scholar] [CrossRef]
- Applied Biosystems™. Quantifiler™ HP and Trio DNA Quantification Kits User Guide; Thermo Fisher Scientific: Waltham, MA, USA, 2017. [Google Scholar]
- Ward, J.; Watherston, J. Quantitative and qualitative assessment of DNA recovered from human skeletal remains. In Forensic Genetic Approaches for the Identification of Human Skeletal Remains: Challenges, Best Practices, and Emerging Technologies; Ambers, A., Ed.; Elsevier Academic Press: New York, NY, USA, in press.
- Holt, A.; Wootton, S.C.; Mulero, J.J.; Brzoska, P.M.; Langit, E.; Green, R.L. Developmental validation of the Quantifiler® HP and Trio Kits for human DNA quantification in forensic samples. Forensic Sci. Int. Genet. 2016, 21, 145–157. [Google Scholar] [CrossRef]
- Vernarecci, S.; Ottaviani, E.; Agostino, A.; Mei, E.; Calandro, L.; Montagna, P. Quantifiler ® Trio Kit and forensic samples management: A matter of degradation. Forensic Sci. Int. Genet. 2015, 16, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Promega Corporation. Promega Corporation Technical Manual #TMD034—PowerPlex ® 21 System Technical Manual; Promega Corporation: Madison, WI, USA, 2012. [Google Scholar]
- Thermo Fisher Scientific. 3500/3500xL Genetic Analyzer User Guide—Data Collection Software v3.1.; Thermo Fisher Scientific: Waltham, MA, USA, 2018. [Google Scholar]
- Kitayama, T.; Ogawa, Y.; Fujii, K.; Nakahara, H.; Mizuno, N.; Sekiguchi, K.; Kasai, K.; Yurino, N.; Yokoi, T.; Fukuma, Y.; et al. Evaluation of a new experimental kit for the extraction of DNA from bones and teeth using a non-powder method. Leg. Med. 2010, 12, 84–89. [Google Scholar] [CrossRef]
- Harrel, M.; Mayes, C.; Gangitano, D.; Hughes, S. Evaluation Of A Powder-Free DNA Extraction Method For Skeletal Remains. J. Forensic Sci. 2018, 63, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, H.; Cortellini, V.; Franceschetti, L.; Verzeletti, A. Large fragment demineralization: An alternative pretreatment for forensic DNA typing of bones. Int. J. Leg. Med. 2021, 135, 1417–1424. [Google Scholar] [CrossRef]
- Hasap, L.; Chotigeat, W.; Pradutkanchana, J.; Vongvatcharanon, U.; Kitpipit, T.; Thanakiatkrai, P. A novel, 4-h DNA extraction method for STR typing of casework bone samples. Int. J. Leg. Med. 2020, 134, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Y.Y.; Tan, Y.P.; Ng, S.; Tay, A.S.; Phua, Y.H.; Tan, W.J.; Ong, T.Y.R.; Chua, L.M.; Syn, C.K.C. A preliminary evaluation study of new generation multiplex STR kits comprising of the CODIS core loci and the European Standard Set loci. J. Forensic Leg. Med. 2017, 52, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.W.; Liand, C.; Ip, S.C.Y. A selection guide for the new generation 6-dye DNA profiling systems. Forensic Sci. Int. Genet 2017, 30, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Elwick, K.; Mayesand, C.; Hughes-Stamm, S. Comparative sensitivity and inhibitor tolerance of GlobalFiler® PCR Amplification and Investigator® 24plex QS kits for challenging samples. Leg. Med. 2018, 32, 31–36. [Google Scholar] [CrossRef]
- INTERPOL Tsunami Evaluation Working Group. The DVI Response to the South East Asian Tsunami between December 2004 and February 2006; INTERPOL: Lyon, France, 2010. [Google Scholar]
- Goodwin, W.H. The use of forensic DNA analysis in humanitarian forensic action: The development of a set of international standards. Forensic Sci. Int. 2017, 278, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Ward, J. Best practice recommendations for the establishment of a national DNA identification program for missing persons: A global perspective. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e43–e45. [Google Scholar] [CrossRef]
- Bright, J.-A.; Allen, C.; Fountain, S.; Gray, K.; Grover, D.; Neville, S.; Poy, A.L.; Taylor, D.; Turbett, G.; Wilson-Wilde, L. Australian population data for the twenty Promega PowerPlex 21 short tandem repeat loci. Aust. J. Forensic Sci. 2014, 46, 442–446. [Google Scholar] [CrossRef]
- Cline, R.E.; Laurentand, N.M.; Foran, D.R. The fingernails of Mary Sullivan: Developing reliable methods for selectively isolating endogenous and exogenous DNA from evidence. J. Forensic Sci. 2003, 48, 328–333. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).