Complete Mitochondrial Genome of Decametra tigrina (A.H. Clark, 1907) (Crinoidea, Comatulida, Colobometridae) and Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Storage of Decametra tigrina
2.2. DNA Extraction and Genome Sequencing
2.3. Mitochondrial Genome Annotation and Analysis
2.4. Phylogenetic Analysis
3. Results
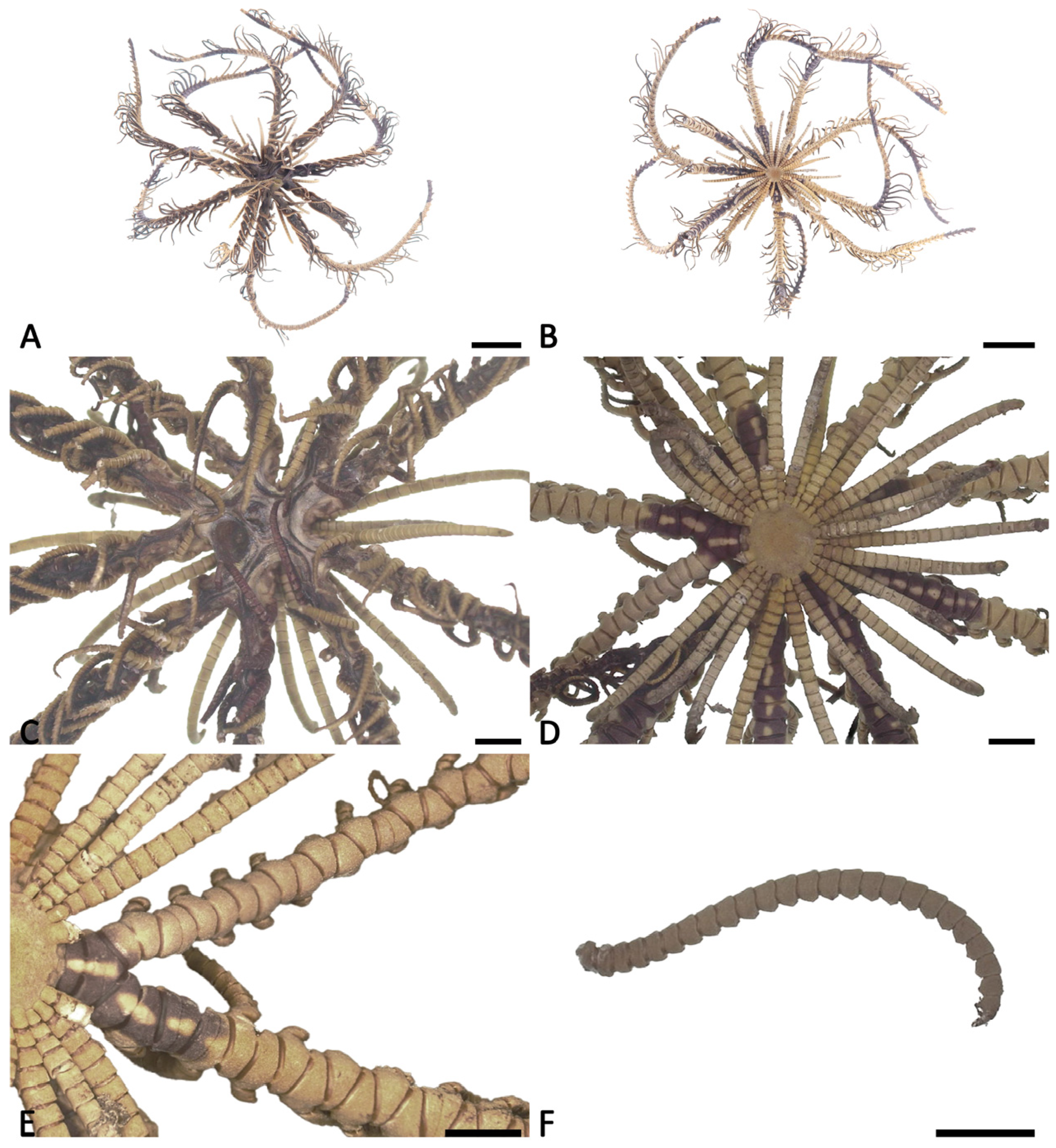
3.1. Morphological Characteristics of Decametra tigrina
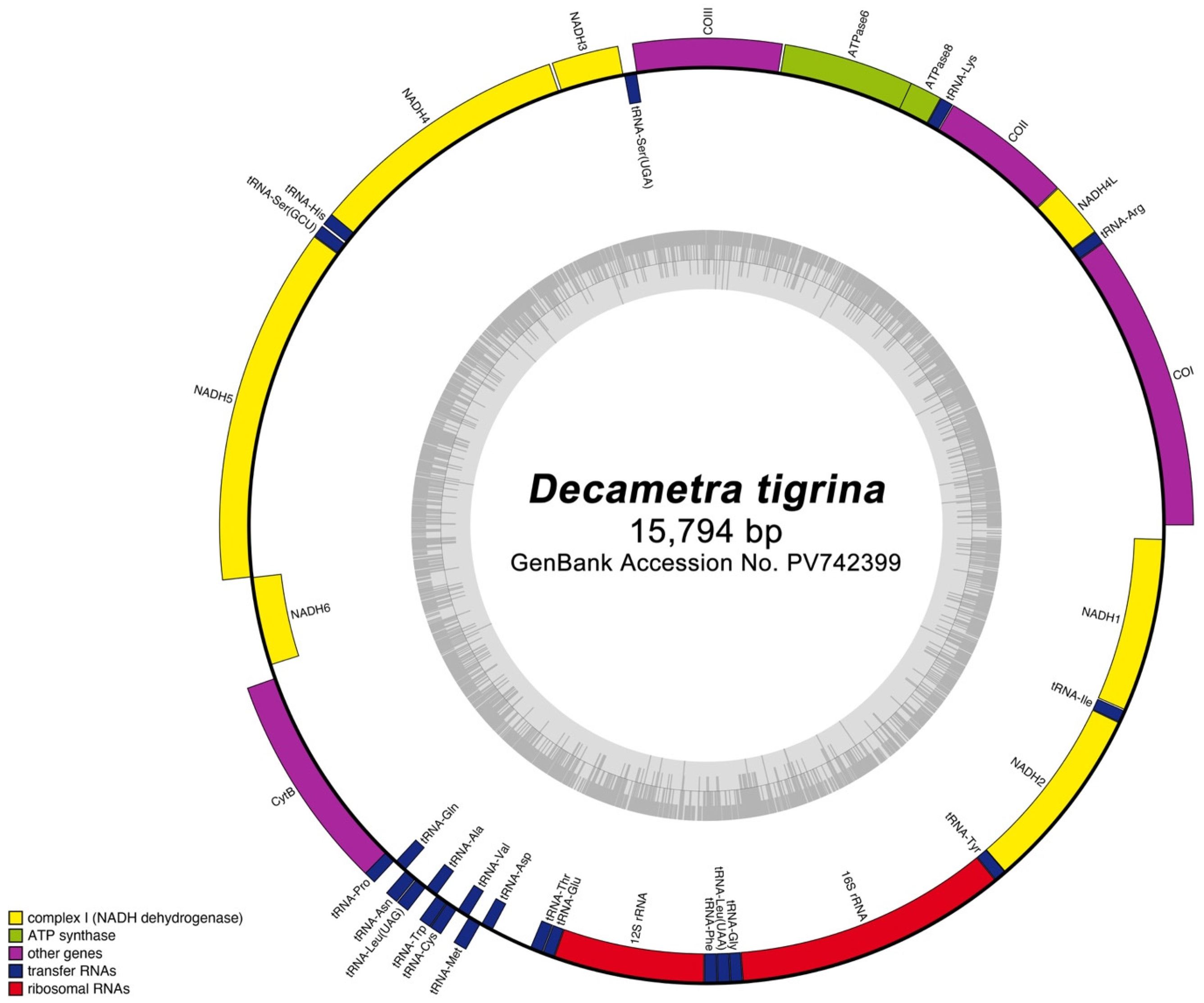
3.2. Gene Arrangement and Characteristics of Decametra tigrina
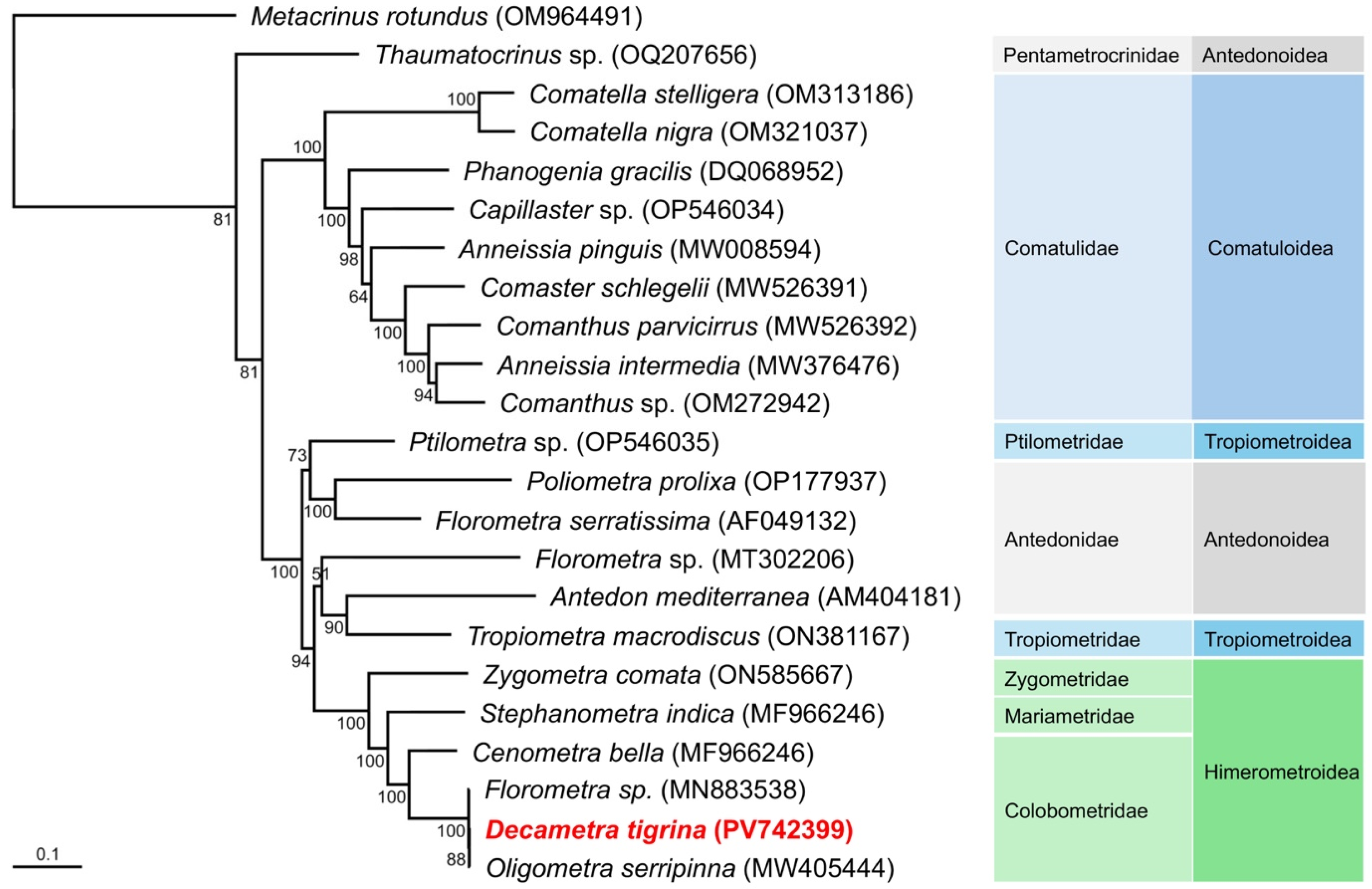
3.3. Phylogenetic Analysis of Comatulids Including Decametra tigrina
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Messing, C.; Gondim, A.I.; Markello, K.; Poatskievick Pierezan, B.; Taylor, K.; Eléaume, M. World List of Crinoidea. Decametra AH Clark, 1911. World Register of Marine Species. Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=204374 (accessed on 12 December 2025).
- Shin, S. Feather stars, basket stars: Echinodermata: Crinozoa: Crinoidea: Comatulida, Asterozoa: Ophiuroidea: Euryalida. In Invertebrate Fauna of Korea, 1st ed.; National Institute of Biological Resources: Incheon, Republic of Korea, 2013; Volume 32, pp. 1–150. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. Mitofish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy Community. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2020, 12, 373–377. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree: Tree Figure Drawing Tool, Version 1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 27 October 2025).
- Perseke, M.; Fritzsch, G.; Ramsch, K.; Bernt, M.; Merkle, D.; Middendorf, M.; Bernhard, D.; Stadler, P.F.; Schlegel, M. Evolution of mitochondrial gene orders in echinoderms. Mol. Phylogenet. Evol. 2008, 47, 855–864. [Google Scholar] [CrossRef]
- Scouras, A.; Smith, M.J. A novel mitochondrial gene order in the crinoid echinoderm Florometra serratissima. Mol. Biol. Evol. 2001, 18, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.-E.; Park, H.S.; Rhee, J.-S. Characterization and phylogenetic analysis of the complete mitochondrial genome of Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA B Resour. 2020, 5, 2010–2011. [Google Scholar] [CrossRef]
- Kwon, H.; Park, H.S.; Rhee, J.S. Complete mitochondrial genome of the crinoid Poliometra prolixa (Crinoidea: Comatulida: Antedonidae). Mitochondrial DNA B Resour. 2023, 8, 927–931. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.M.; Ip, Y.C.A.; Huang, D. Complete mitochondrial genome of the feather star Cenometra bella (Hartlaub, 1890) (Crinoidea: Colobometridae). Mitochondrial DNA B Resour. 2022, 7, 950–952. [Google Scholar] [CrossRef]
- Nam, S.-E.; Park, H.S.; Rhee, J.-S. Complete mitochondrial genome of the crinoid echinoderm, Florometra species (Echinodermata, Crinoidea). Mitochondrial DNA B Resour. 2020, 5, 852–853. [Google Scholar] [CrossRef]
- Kim, P.; Shin, S. The complete mitochondrial genome of Anneissia pinguis (Crinoidea, Articulata, Comatulidae), from South Korea. Mitochondrial DNA B Resour. 2021, 6, 2337–2338. [Google Scholar] [CrossRef]
- Kim, P.; Lee, T.; Shin, S. The complete mitochondrial genome of Anneissia intermedia (Crinoidea: Comatulida: Comatulidae). Mitochondrial DNA B Resour. 2021, 6, 1777–1778. [Google Scholar] [CrossRef]
- Xu, Q.; Lu, M.; Sun, Y.; Li, Z.; Li, Y.; Dong, Y.; Hu, X.; Zhang, Q.; Liu, B.; He, X. Structural features and phylogenetic implications of crinoid echinoderms based on thirteen novel mitochondrial genomes. J. Mar. Sci. Eng. 2024, 12, 361. [Google Scholar] [CrossRef]
- Scouras, A.; Smith, M.J. The complete mitochondrial genomes of the sea lily Gymnocrinus richeri and the feather star Phanogenia gracilis: Signature nucleotide bias and unique nad4L gene rearrangement within crinoids. Mol. Phylogenet. Evol. 2006, 39, 323–334. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, H.; Wang, X.; Yin, J.; Shen, P.; Lin, Q. Characterization and phylogenetic analysis of the complete mitochondrial genome of Stephnometra indica (Pelmatozoa: Crinoidea). Mitochondrial DNA B Resour. 2019, 4, 2283–2284. [Google Scholar] [CrossRef]
- Sun, Y.; Liao, X.; Dong, Y.; Xia, S.; Xu, Q. The complete mitochondrial genome of Comaster schlegelii (Crinoidea, Comatulida). Mitochondrial DNA B Resour. 2022, 7, 314–316. [Google Scholar] [CrossRef]
- Rouse, G.W.; Jermiin, L.S.; Wilson, N.G.; Eeckhaut, I.; Lanterbecq, D.; Oji, T.; Young, C.M.; Browning, T.; Cisternas, P.; Helgen, L.E.; et al. Fixed, free, and fickle: The phylogeny of extant Crinoidea and their Permian–Triassic origin. Mol. Phy. Evol. 2013, 66, 161–181. [Google Scholar] [CrossRef]
- Hemery, L.G.; Roux, M.; Ameziane, N.; Eleaume, M. High-resolution crinoid phyletic inter-relationships derived from molecular data. Cah. Biol. Mar. 2013, 54, 511–523. [Google Scholar]
- Quek, Z.B.R.; Chang, J.J.M.; Ip, Y.C.A.; Chan, Y.K.S.; Huang, D. Mitogenomes reveal alternative initiation codons and lineage-specific gene order conservation in echinoderms. Mol. Biol. Evol. 2021, 38, 981–985. [Google Scholar] [CrossRef]
- Clark, A.H. A monograph of the existing crinoids. Bull. U.S. Natl. Mus. 1915, 1, 1–406. [Google Scholar]
| Order | Family | Species | Accession Number in GenBank | Reference |
|---|---|---|---|---|
| Comatulida | Antedonidae | Antedon mediterranea | AM404181 | [13] |
| Florometra serratissima | AF049132 | [14] | ||
| Florometra sp. | MT302206 | [15] | ||
| Poliometra prolixa | OP177937 | [16] | ||
| Colobometridae | Cenometra bella | OK509084 | [17] | |
| Decametra tigrina | PV742399 | This study | ||
| Florometra sp. | MN883538 | [18] | ||
| Oligometra serripinna | MW405444 | Unpublished | ||
| Comatulidae | Anneissia pinguis | MW008594 | [19] | |
| Anneissia intermedia | MW376476 | [20] | ||
| Capillaster sp. | OP546034 | [21] | ||
| Comanthus parvicirrus | MW526392 | [21] | ||
| Comenthus sp. | OM272942 | [21] | ||
| Comaster schlegelii | MW526391 | [22] | ||
| Comatella nigra | OM321037 | [21] | ||
| Comatella stelligera | OM313186 | [21] | ||
| Phanogenia gracilis | DQ068952 | [22] | ||
| Mariametridae | Stephanometra indica | MF966246 | [23] | |
| Pentametrocrinidae | Thaumatocrinus sp. | OQ207656 | [21] | |
| Ptilometridae | Ptilometra sp. | OP546035 | [21] | |
| Tropiometridae | Tropiometra macrodiscus | ON381167 | [21] | |
| Zygometridae | Zygometra comata | ON585667 | [21] | |
| Isocrinida | Isocrinidae | Metacrinus rotundus | OM964491 | [21] |
| Name | Site | Length (bp) | Strand | Start Codon | Stop Codon | Anti-Codon |
|---|---|---|---|---|---|---|
| COI | 1–1557 | 1557 | H | ATG | TAA | |
| tRNA-Arg | 1558–1625 | 68 | H | UCG | ||
| NADH4L | 1626–1922 | 297 | H | GTG | TAA | |
| COII | 1924–2613 | 690 | H | ATG | TAG | |
| tRNA-Lys | 2616–2685 | 70 | H | CUU | ||
| ATPase8 | 2686–2859 | 174 | H | ATG | TAA | |
| ATPase6 | 2853–3542 | 690 | H | ATG | TAA | |
| COIII | 3552–4334 | 783 | H | ATG | TAA | |
| tRNA-Ser | 4333–4403 | 71 | L | UGA | ||
| NADH3 | 4407–4760 | 354 | H | ATG | TAA | |
| NADH4 | 4773–6164 | 1392 | H | ATG | TAA | |
| tRNA-His | 6155–6223 | 69 | H | GUG | ||
| tRNA-Ser | 6233–6300 | 68 | H | GCU | ||
| NADH5 | 6301–8181 | 1881 | H | GTG | TAA | |
| NADH6 | 8182–8676 | 495 | L | ATG | TAA | |
| CytB | 8751–9902 | 1152 | H | ATG | TAG | |
| tRNA-Pro | 9893–9961 | 69 | H | UGG | ||
| tRNA-Gln | 9967–10,038 | 72 | L | UUG | ||
| tRNA-Asn | 10,042–10,114 | 73 | H | GUU | ||
| tRNA-Leu | 10,117–10,187 | 71 | H | UAG | ||
| tRNA-Ala | 10,189–10,259 | 71 | L | UGC | ||
| tRNA-Trp | 10,262–10,329 | 68 | H | UCA | ||
| tRNA-Cys | 10,330–10,396 | 67 | H | GCA | ||
| tRNA-Val | 10,401–10,470 | 70 | L | UAC | ||
| tRNA-Met | 10,474–10,548 | 75 | H | CAU | ||
| tRNA-Asp | 10,552–10,621 | 70 | L | GUC | ||
| tRNA-Thr | 10,851–10,922 | 72 | L | UGU | ||
| tRNA-Glu | 10,924–10,991 | 68 | L | UUC | ||
| 12S rRNA | 10,992–11,832 | 841 | L | |||
| tRNA-Phe | 11,833–11,905 | 73 | L | GAA | ||
| tRNA-Leu | 11,906–11,977 | 72 | L | UAA | ||
| tRNA-Gly | 11,978–12,046 | 69 | L | UCC | ||
| 16S rRNA | 12,047–13,565 | 1519 | L | |||
| tRNA-Tyr | 13,566–13,636 | 71 | L | GUA | ||
| NADH2 | 13,635–14,675 | 1041 | L | ATG | TAA | |
| tRNA-Ile | 14,676–14,746 | 71 | L | GAU | ||
| NADH1 | 14,751–15,719 | 969 | L | ATG | TAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Kim, G.; Choi, Y.; Kwon, S.; Lee, T. Complete Mitochondrial Genome of Decametra tigrina (A.H. Clark, 1907) (Crinoidea, Comatulida, Colobometridae) and Phylogenetic Analysis. Taxonomy 2026, 6, 2. https://doi.org/10.3390/taxonomy6010002
Kim G, Choi Y, Kwon S, Lee T. Complete Mitochondrial Genome of Decametra tigrina (A.H. Clark, 1907) (Crinoidea, Comatulida, Colobometridae) and Phylogenetic Analysis. Taxonomy. 2026; 6(1):2. https://doi.org/10.3390/taxonomy6010002
Chicago/Turabian StyleKim, Gilpyo, Yujin Choi, Soyeon Kwon, and Taekjun Lee. 2026. "Complete Mitochondrial Genome of Decametra tigrina (A.H. Clark, 1907) (Crinoidea, Comatulida, Colobometridae) and Phylogenetic Analysis" Taxonomy 6, no. 1: 2. https://doi.org/10.3390/taxonomy6010002
APA StyleKim, G., Choi, Y., Kwon, S., & Lee, T. (2026). Complete Mitochondrial Genome of Decametra tigrina (A.H. Clark, 1907) (Crinoidea, Comatulida, Colobometridae) and Phylogenetic Analysis. Taxonomy, 6(1), 2. https://doi.org/10.3390/taxonomy6010002






