Taxonomic Notes on Lerniana Delicado et Hauffe, 2022, Trichonia Schütt, 1980 (Truncatelloidea: Hydrobiidae: Horatiinae) and Allied Taxa †
Abstract
1. Introduction
2. Material and Methods
3. Results
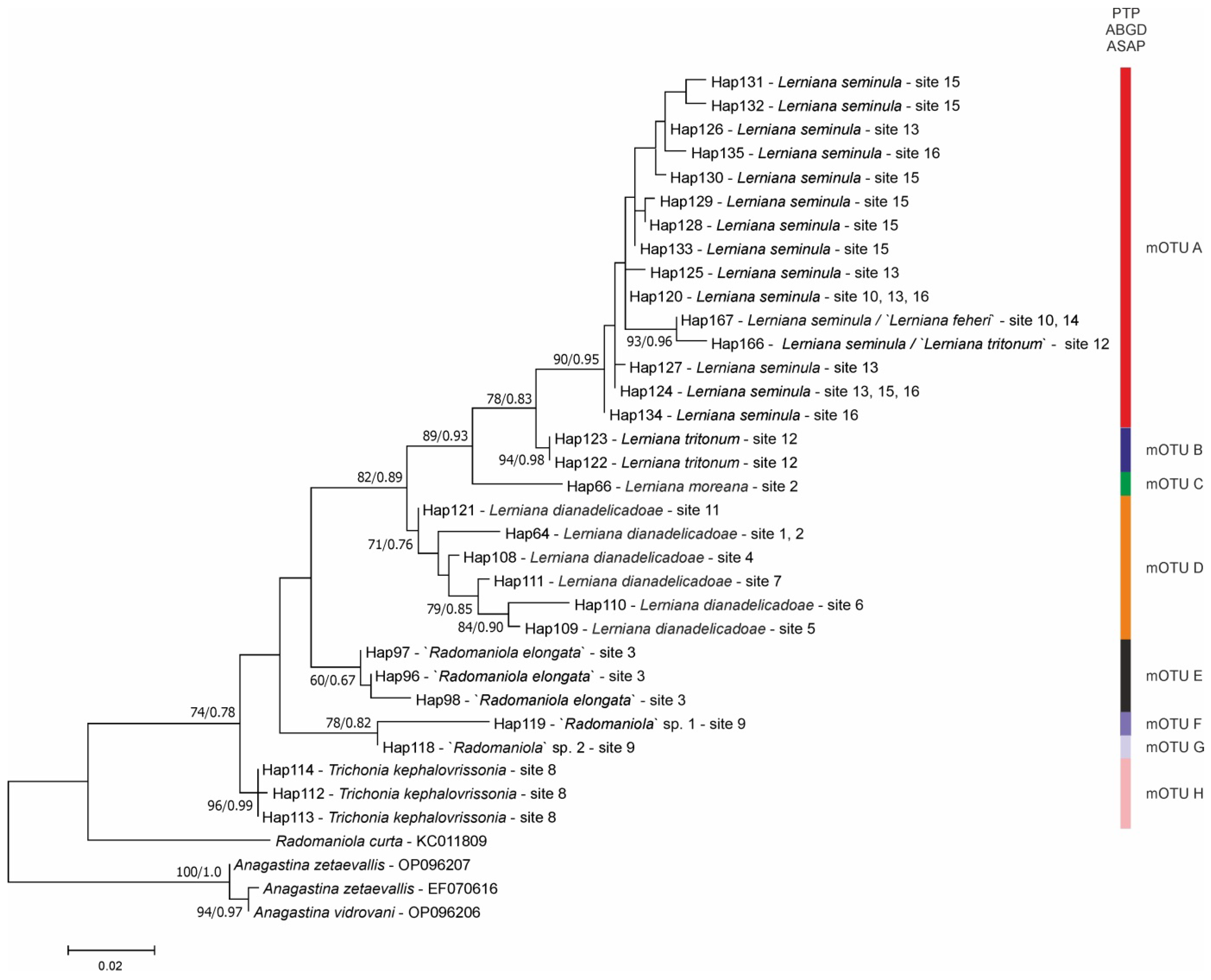
3.1. Molecular Part
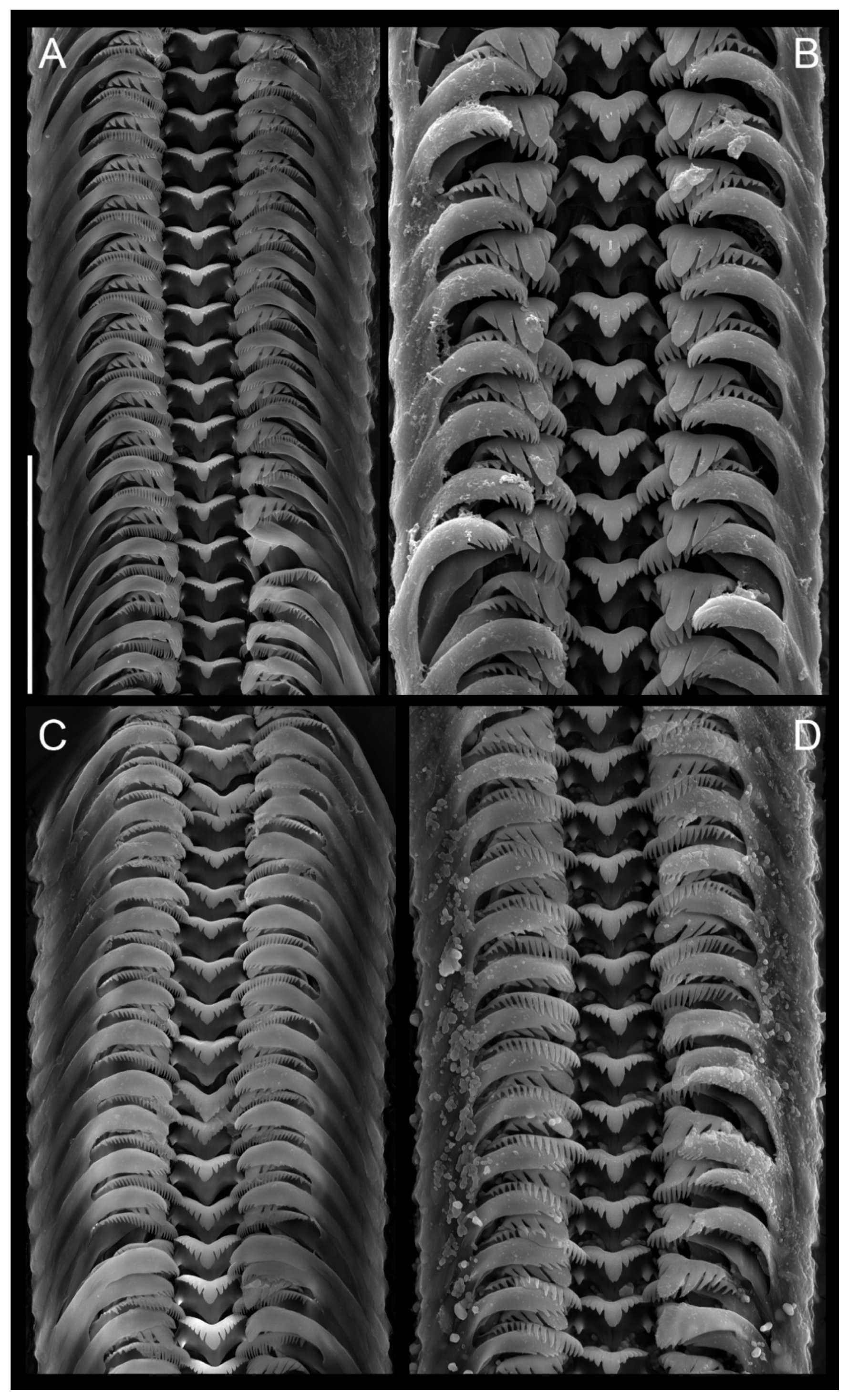
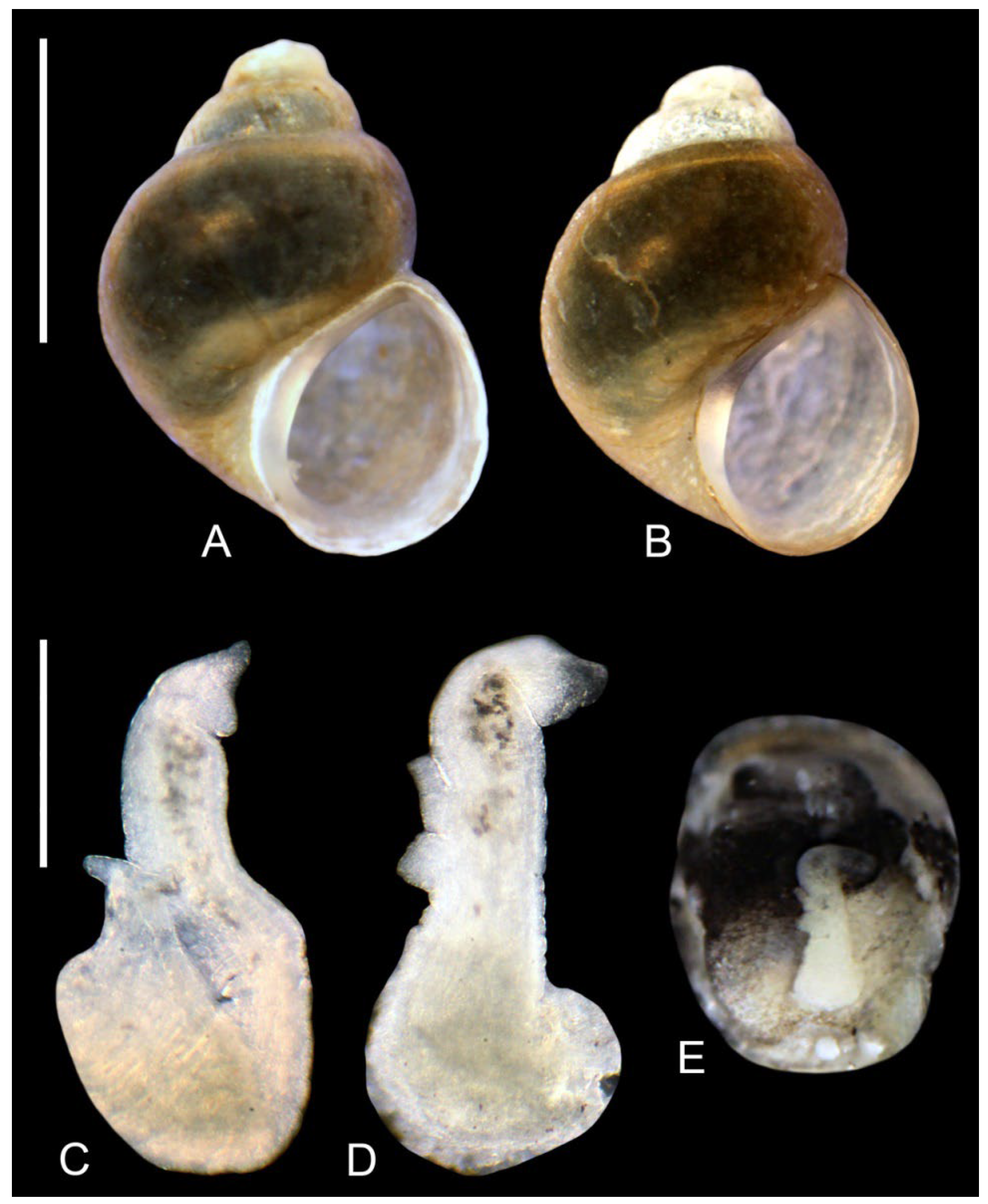
3.2. Systematic Part
- LSID urn:lsid:zoobank.org:act:FEF8E391-3B88-44D3-8C58-CB872E3EA076
- GenBank sequence numbers: PX096765 - PX096768, KC011734, KC011747, KC011762, KC011766, KC011772
- mOTU D
1–1 1–1
- LSID urn:lsid:zoobank.org:act:9C5EE953-14E9-4A50-B97A-CB35F72CA3E4
- GenBank sequence number: PX096769
- mOTU C
2 – 2 2 – 2
1 – 1
1 – 1 1 – 1 1 – 1
4. Discussion
- (1)
- “Radomaniola” elongata clearly belongs neither to the genus Radomaniola, nor to Lerniana, it is distinct both morphologically and molecularly. However, with only COI sequence, it is not possible to infer its phylogenetic relationships. Probably it represents a distinct genus, but some fresh material is badly needed, to sequence more loci. It is close to Lerniana phylogenetically, but not geographically, since Montenegro is far from southern Greece. Similarly. the phylogenetic relationships of another clade of “Radomaniola”, consisted of two rather molecularly distinct species, remain unclear.
- (2)
- Lerniana tritonum (mOTU B) is evidently distinct, both morphologically and molecularly from L. seminula (mOTU A). On the other hand, molecular identity and slight morphological differences between L. feheri and L. seminula legitimate, despite the doubts presented by Delicado & Hauffe [2], synonymizing of L. feheri as a younger synonym of L. seminula.
- (3)
- It is noteworthy that L. tritonum has been found only at its type locality so far, while L. seminula at six localities, all of them at Peloponnese. Newly described L. dianadelicadoae expands the geographic range of Lerniana—it can be found also in Attica (locality 7) and south Thessaly (Figure 1: locality 4–6). Another newly described species L. moreana has been found at only one locality (Figure 1: locality 2) at Peloponnese, in sympatry with L. dianadelicadoae.
- (4)
- “Radomaniola” elongata in our tree computed for all the 168 COI sequences of Radomaniola and allied taxa (also including the sequences of Delicado & Hauffe [2]), the tree not presented in this study, clustered outside of the clade grouping Radomaniola. As stated above, without freshly collected material, enabling to study other loci, its phylogenetic relationships remain enigmatic. Apart from its characteristically high-spired shell, all the other character states can be found also in Radomaniola.
- (5)
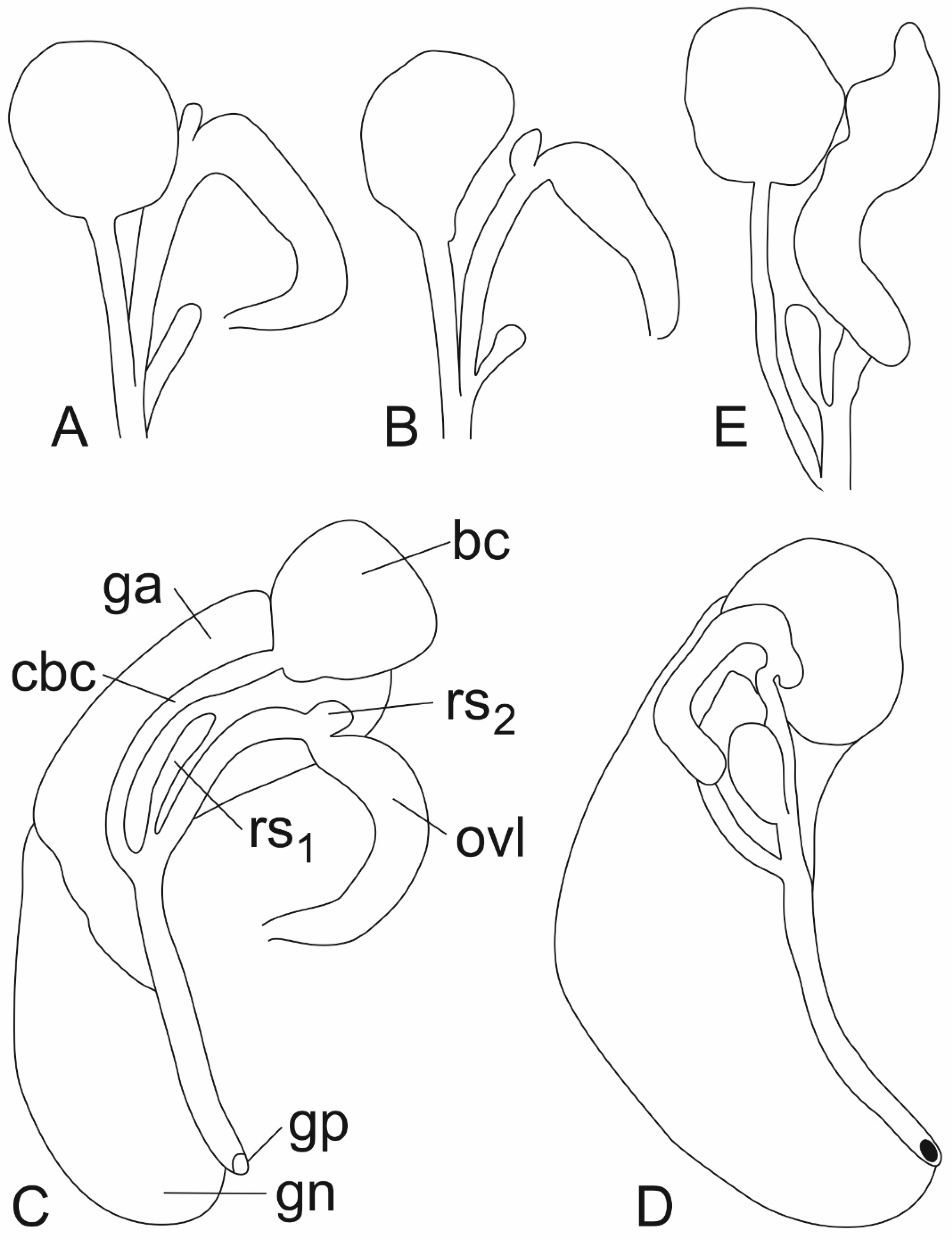
- Trichonia kephalovrissonia in the COI/18S tree [1] belonged to the clade grouping all the species of Radomaniola, also “R. elongata”, although the clade was formed by unresolved polytomy. This suggested the assignment of Trichonia kephalovrissonia to the genus Radomaniola, and this suggestion was followed by Radea et al. [18]. The assignment of T. kephanovrissonnia to the genus Radomaniola was later confirmed by the molecular data of Delicado & Hauffe [2], thus followed in WoRMS (Editorial Board) 2024 [47]. However, our COI tree places T. kephalovrissonia within this clade (bootstrap support 74%), not with Radomaniola. richonia kephalovrissonia falls close to the three “Radomaniola” species (“Radomaniola” elongata, “Radomaniola” sp. 1, “Radomaniola” sp. 2), and the phylogenetic position of all the four taxa remains unclear. It has to be noted that there are still no molecular data for Trichonia trichonica.
- (6)
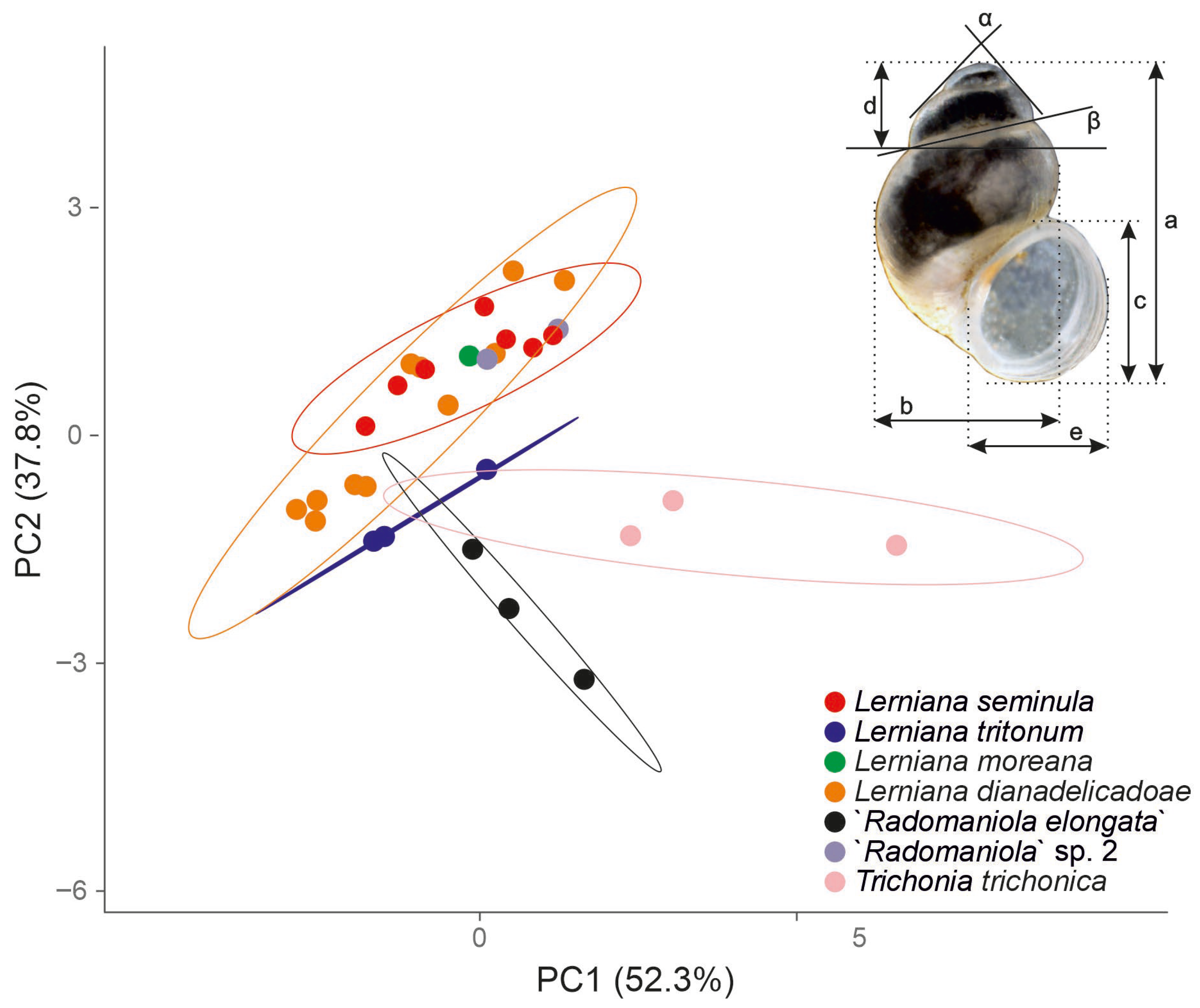
- In the principal component analysis (Figure 6) PC1 and PC2 explained cumulatively 90.1% of the total variation, the other PCs explained less than expected with a broken-stick model [48,49]. PC1 reflects mostly size differences, morphological polymorphism, and sexual dimorphism, PC2 differences in shape [50]. As could be seen in Figure 6, PC1 reflected small shells of Trichonia trichonica different from all the other taxa. The latter overlapped in the PC1 space, only the ranges of variation were different. Also, in PC2 space the most variable Lerniana dianadelicadoae overlapped with less variable L. seminula, L. moreana and “Radomaniola”sp. 1. Lerniana tritonum and especially “Radomaniola” elongata differed in PC2. Obviously, with such unnumerous specimens measured, and not representative samples, these conclusions should be interpreted with caution, as rather preliminary, but still PCA seems useful as a summary of the morphometric data.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falniowski, A.; Szarowska, M.; Glöer, P.; Pešić, V. Molecules vs morphology in the taxonomy of the Radomaniola/Grossuana group of Balkan Rissooidea (Mollusca: Caenogastropoda). J. Conchol. 2012, 41, 19–36. [Google Scholar]
- Delicado, D.; Hauffe, T. Shell features and anatomy of the springsnail genus Radomaniola (Caenogastropoda: Hydrobiidae) show a different pace and mode of evolution over five million years. Zool. J. Linn. Soc. 2022, 196, 393–441. [Google Scholar] [CrossRef]
- Bourguignat, J.-R. Testacea Novissima Quæ Cl. de Saulcy in Itinere per Orientem Annis 1850 et 1851, Collegit; Baillière: Paris, France, 1852; pp. 1, 5–31. Available online: https://gallica.bnf.fr/ark:/12148/bpt6k99037b/f1.image (accessed on 10 August 2025).
- Bourguignat, J.-R. Catalogue raisonné des mollusques terrestres et fluviatiles recueillis par M. F. de Saulcy pendant son voyage en Orient. In Voyage Autour de la Mer Morte et Dans les Terres Bibliques Exécuté de Décembre 1850 à Avril 1851, Livr. 15; de Saulcy, F., Ed.; Gide & J. Baudry: Paris, France, 1853; XXVI+96p, pp. 1–4, 64, Available online: http://gallica.bnf.fr/ark:/12148/bpt6k99038p (accessed on 24 August 2025).
- Roth, J.R. Spicilegium molluscorum orientalium annis 1852 et 1853 collectorum. Malakol. Blätter 1854, 2, 17–58. [Google Scholar]
- Von Frauenfeld, G. Vorläufige Aufzählung der Arten der Gattungen Hydrobia Htm. und Amnicola Gld. Hldm. in der kaiserlichen und in Cuming’s Sammlung. Verhandlungen Kais.-Königlichen Zool.-Bot. Ges. Wien Abh. 1863, 13, 17–1032. Available online: https://www.biodiversitylibrary.org/page/54758154 (accessed on 24 August 2025).
- Westerlund, C.A.; Blanc, H. Apercų sur la Faune Malacologique de la Grèce Inclus l’Epire et la Thessalie. Coquilles Extramarines; Imprimerie de Ferré & Tornese: Naples, Italy, 1879; Volume 19, pp. 1–61. Available online: https://www.molluscabase.org/aphia.php?p=sourcedetails&id=298691 (accessed on 10 August 2025).
- Martens, E. Griechische Mollusken. Gesammelt von Eberh. Von Örtzen. Arch. Für Naturgeschichten 1889, 1, 169–240. [Google Scholar]
- Radoman, P. New classification of fresh and brakish [sic!] water Prosobranchia from the Balkans and Asia Minor. Poseb. Izd. Prir. Muz. Beogr. 1973, 32, 1–30. [Google Scholar]
- Szarowska, M. Molecular phylogeny, systematics and morphological character evolution in the Balkan Rissooidea (Caenogastropoda). Folia Malacol. 2006, 14, 99–168. [Google Scholar] [CrossRef]
- Delicado, D.; Hauffe, T.; Wilke, T. Fifth mass extinction event triggered the diversification of the largest family of freshwater gastropods (Caenogastropoda: Truncatelloidea: Hydrobiidae). Cladistics 2024, 40, 82–96. [Google Scholar] [CrossRef]
- Haldeman, S.S. A Monograph of the Limniades and Other Fresh Water Univalve Shells of North America; J. Dobson: Philadelphia, PA, USA, 1840; Volume 1, pp. 1–16. [Google Scholar] [CrossRef]
- Kabat, A.R.; Hershler, R. The prosobranch snail family [Hydrobiidae (Gastropoda: Rissooidea): Review of classification and supraspecific taxa. Smithson. Contrib. Zool. 1993, 547, 1–94. [Google Scholar] [CrossRef]
- Schütt, H. Zur Kenntnis griechischer Hydrobiiden. Arch. Molluskenkd. 1980, 110, 115–149. [Google Scholar]
- Falniowski, A.; Jaszczyńska, A.; Osikowski, A.; Hofman, S. Litthabitellidae: A new family of the Truncatelloidea (Caenogastropoda). J. Nat. Hist. 2023, 57, 299–329. [Google Scholar] [CrossRef]
- Westerlund, C.A. Malakologiska bidrag. II. För Vetenskapen nya Land- och Sötvatten Mollusker. Öfversigt Af Kongliga Vetensk.-Akad. Förhandlingar 1881, 4, 50–70. [Google Scholar]
- Szarowska, M.; Falniowski, A. Destroyed and threatened localities of Rissooid snails (Gastropoda: Rissooidea) in Greece. Folia Malacol. 2011, 19, 35–39. [Google Scholar] [CrossRef]
- Radea, C.; Parmakelis, A.; Papadogiannis, V.; Charou, D.; Triantis, K.A. The hydrobioid freshwater gastropods (Caenogastropoda, Truncatelloidea) of Greece: New records, taxonomic re-assessments using DNA sequence data and update of the IUCN Red List Categories. ZooKeys 2013, 350, 1–20. [Google Scholar] [CrossRef]
- Radea, C.; Louvrou, I.; Bakolitsas, K.; Economou-Amilli, A. Local endemic and threatened freshwater hydrobiids of Western Greece: Elucidation of anatomy and new records. Folia Malacol. 2017, 25, 3–13. [Google Scholar] [CrossRef]
- Rueden, D.T.; Schindelin, J.; Hiner, M.C.; Dezonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, e529. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef]
- Falniowski, A. Anatomical characters and SEM structure of radula and shell in the species-level taxonomy of freshwater prosobranchs (Mollusca: Gastropoda: Prosobranchia): A comparative usefulness study. Folia Malacol. 1990, 4, 53–142. [Google Scholar] [CrossRef]
- Hershler, R.; Ponder, W.F. A review of morphological characters of hydrobioid snails. Smithson. Contrib. Zool. 1998, 600, 1–55. [Google Scholar] [CrossRef]
- Szarowska, M.; Osikowski, A.; Hofman, S.; Falniowski, A. Pseudamnicola Paulucci, 1878 (Caenogastropoda: Truncatelloidea) from the Aegean Islands: A long or short story? Org. Divers. Evol. 2016, 16, 121–139. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Xia, X. Data Analysis in Molecular Biology and Evolution; Kluwer Academic Publishers: Boston, MA, USA; Dordrecht, The Netherlands; London, UK, 2000; 296p. [Google Scholar]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenetics Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X. DAMBE: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; 8p. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v.2; Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohn, A.S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2010. [Google Scholar]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2011, 21, 1864–1877. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Merckelbach, L.; Borges, L. Make every species count: FastaChar software for rapid determination of molecular diagnostic characters to describe species. Mol. Ecol. Resour. 2020, 20, 1761–1768. [Google Scholar] [CrossRef] [PubMed]
- Hütter, T.; Ganser, M.H.; Kocher, M.; Halkic, M.; Agatha, S.; Augsten, N. DeSignate: Detecting signature characters in gene sequence alignments for taxon diagnoses. BMC Bioinform. 2020, 21, 151. [Google Scholar] [CrossRef] [PubMed]
- Radoman, P. Hydrobioidea a superfamily of Prosobranchia (Gastropoda). I. Systematics. Monogr. Serbian Acad. Sci. Arts DXLVII Dep. Sci. 1983, 57, 1–256. [Google Scholar]
- Frogley, M.; Preece, R. A review of the aquatic Mollusca from Lake pamvotis, Ioannina, an ancient lake in NW Greece. J. Conchol. 2007, 39, 271–296. [Google Scholar]
- Georgiev, D. A new species of Radomaniola Szarowska, 2006 (Gastropoda: Hydrobiidae) from Peloponnese, Greece. Acta Zool. Bulg. 2013, 65, 293–295. [Google Scholar]
- Glöer, P.; Hirschfelder, H.-J. A new hydrobiid genus and species from Peloponnese (Greece), with a note on Achaiohydrobia (Gastropoda: Hydrobiidae). Basteria 2023, 87, 127–129. [Google Scholar]
- Falniowski, A. Species Distinction and Speciation in Hydrobioid gastropods (Mollusca: Caenogastropoda: Truncatelloidea). Arch. Zool. Stud. 2018, 1, 3. [Google Scholar] [CrossRef]
- WoRMS (Editorial Board). World Register of Marine Species. 2024. Available online: https://www.marinespecies.org (accessed on 29 July 2025).
- Rohlf, F.J. NTSYSpc, Numerical Taxonomy and Multivariate Analysis System; Version 2.0. [Computer Software and Manual]; Exeter Software: Setauket, NY, USA, 1998. [Google Scholar]
- Peres-Neto, P.R.; Jackson, D.A.; Somers, K.M. How many principal components? Stopping rules for determining the number of non-trivial axes revisited. Comput. Stat. Data Anal. 2005, 49, 974–997. [Google Scholar] [CrossRef]
- Falniowski, A. Metody Numeryczne w Taksonomii [Numerical Techniques in Taxonomy]; Wydawnictwo Uniwersytetu Jagiellońskiego: Kraków, Poland, 2003. [Google Scholar]
- Cuttelod, A.; Seddon, M.; Neubert, E. European Red List of Non-marine Molluscs; Publications Office of the European Union: Luxembourg, 2011. [Google Scholar]
- Szarowska, M.; Hofman, S.; Falniowski, A. Vinodolia fiumana Radoman, 1973 (Caenogastropoda: Rissooidea): Rediscovery and relationships of a species presumed extinct. Folia Malacol. 2013, 21, 135–142. [Google Scholar] [CrossRef]
- Boeters, H.D.; Glöer, P.; Slavevska-Stamenković, V. The Radomaniola/Grossuana group from the Balkan Peninsula, with a description of Grossuana maceradica n. sp. and the designation of a neotype of Paludina hohenackeri Küster, 1853 (Caenogastropoda: Truncatelloidea: Hydrobiidae). Arch. Molluskenkd. 2017, 146, 187–202. [Google Scholar] [CrossRef]
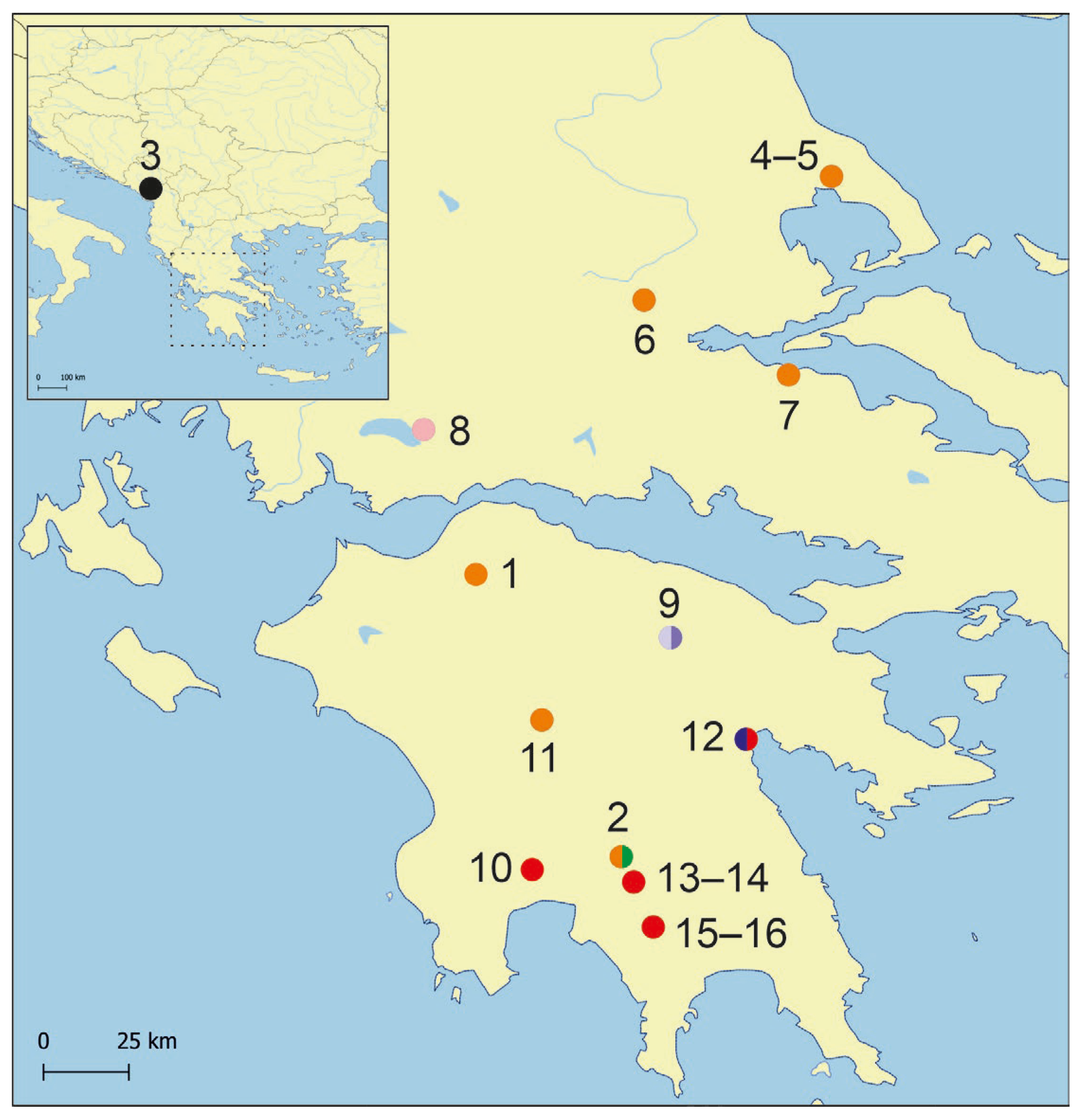









| Id | Locality | Coordinates | GB Numbers | Haplotypes | |
|---|---|---|---|---|---|
| 1 | Katarraktis centre, spring, and limestone cliffs, Achaia, Peloponnese, Greece | 38°05′55″ N | 21°50′08″ E | PX096765 PX096766 PX096767 | Hap_64 Hap_64 Hap_64 |
| 2 | Peloponnese—Laconia regional unit, Kastorio, spring 3.3 km N of village at Aqueduct source in Virtaliotiko firangi | 37°10′01″ N | 22°18′25″ E | PX096768 PX096769 | Hap_64 Hap_66 |
| 3 | spring on island Vranjina, Skadar Lake area—M17 | 42°16′21″ N | 19°28′09″ E | KC011811 KC011812 KC011813 | Hap_96 Hap_97 Hap_98 |
| 4 | spring at Makrinitsa/Koukourava, Oros Pilion, E of Volos—G18 | 39°23′34″ N | 22°59′55″ E | KC011734 | Hap_108 |
| 5 | spring NW of Dhrakia, Oros Pilion, E of Volos—G15 | 39°23′03″ N | 23°00′04″ E | KC011772 | Hap_109 |
| 6 | spring of Achilles, ESE of Kalamakion, NW of Lamia—G14 | 38°59′12″ N | 22°22′43″ E | KC011747 | Hap_110 |
| 7 | spring S of Agios Konstantinos, N of Mt. Spartias, Attica—G13 | 38°45′05″ N | 22°51′11″ E | KC011766 | Hap_111 |
| 8 | spring in city centre of Thérmon, NE of Trichonida Lake—G11 | 38°34′22″ N | 21°39′58″ E | KC011728 KC011729 KC011730 | Hap_112 Hap_113 Hap_114 |
| 9 | spring near Stimfalia Lake, between Bouzion and Kalanoi, Peloponnese—G7 | 37°53′38″ N | 22°28′27″ E | KC011763 KC011764 | Hap_118 Hap_119 |
| 10 | springs Piges Pamisou, Peloponnese—G4, D | 37°10′04″ N | 22°01′35″ E | KC011733 | Hap_120 |
| 11 | spring between Palaiochori and Karkalou, WNW of Tripolis, Peloponnese—G6 | 37°37′21″ N | 22°02′58″ E | KC011762 | Hap_121 |
| 12 | spring at Mili (Lérni), Peloponnese—G5, D | 37°33′06″ N | 22°43′03″ E | KC011731 KC011732 MZ539670 MZ539671 | Hap_122 Hap_123 Hap_166 Hap_166 |
| 13 | spring at Tripi, N. Taigetos Mts., Peloponnese—G3 | 37°05′37″ N | 22°20′49″ E | KC011735 KC011736 KC011737 KC011738 KC011739 KC011740 KC011741 KC011742 | Hap_124 Hap_120 Hap_125 Hap_126 Hap_124 Hap_127 Hap_124 Hap_127 |
| 14 | 19a: Greece, spring at Tripi west of Sparta—D | 37°05′31″ N | 22°21′07″ E | MZ539644 | Hap_167 |
| 15 | spring S of Koumousta, Taigetos Mts., Peloponnese—G1 | 36°56′44″ N | 22°24′33″ E | KC011751 KC011752 KC011753 KC011754 KC011755 KC011756 KC011757 | Hap_128 Hap_129 Hap_130 Hap_131 Hap_132 Hap_124 Hap_133 |
| 16 | spring S of Koumousta, Taigetos Mts., Peloponnese—G2 | 36°56′30″ N | 22°24′23″ E | KC011758 KC011759 KC011760 KC011761 | Hap_120 Hap_124 Hap_134 Hap_135 |
| A | B | C | D | E | F | G | H | Rad. | Ana. | |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.013 | |||||||||
| B | 0.027 | 0.000 | ||||||||
| C | 0.043 | 0.039 | - | |||||||
| D | 0.069 | 0.043 | 0.047 | 0.021 | ||||||
| E | 0.081 | 0.055 | 0.067 | 0.044 | 0.008 | |||||
| F | 0.106 | 0.095 | 0.101 | 0.079 | 0.066 | - | ||||
| G | 0.093 | 0.072 | 0.077 | 0.050 | 0.038 | 0.026 | - | |||
| H | 0.079 | 0.052 | 0.057 | 0.046 | 0.037 | 0.063 | 0.037 | 0.002 | ||
| Radomaniola | 0.123 | 0.111 | 0.115 | 0.104 | 0.064 | 0.079 | 0.082 | 0.082 | - | |
| Anagastina | 0.139 | 0.136 | 0.120 | 0.128 | 0.132 | 0.112 | 0.123 | 0.109 | 0.116 | 0.005 |
| a | b | c | d | e | α | β | |
|---|---|---|---|---|---|---|---|
| Lerniana dianadelicadoae | |||||||
| holotype | 1.58 | 0.91 | 0.79 | 0.42 | 0.70 | 93 | 11 |
| 2M25 | 1.81 | 1.15 | 0.99 | 0.35 | 0.83 | 100 | 7 |
| 2M26 | 1.54 | 1.04 | 0.90 | 0.33 | 0.76 | 97 | 8 |
| 2M27 | 1.70 | 1.08 | 0.92 | 0.39 | 0.73 | 92 | 8 |
| 2N15 | 1.75 | 1.08 | 0.93 | 0.40 | 0.79 | 103 | 9 |
| LD1 | 1.64 | 1.10 | 0.98 | 0.29 | 0.82 | 96 | 8 |
| LD2 | 1.54 | 1.00 | 0.92 | 0.32 | 0.75 | 97 | 9 |
| LD3 | 1.58 | 0.90 | 0.81 | 0.44 | 0.67 | 92 | 13 |
| LD4 | 1.66 | 0.92 | 0.83 | 0.47 | 0.71 | 91 | 14 |
| LD5 | 1.56 | 0.89 | 0.82 | 0.41 | 0.68 | 92 | 13 |
| LD6 | 1.65 | 0.93 | 0.83 | 0.41 | 0.70 | 91 | 12 |
| LD7 | 1.50 | 0.91 | 0.77 | 0.38 | 0.69 | 92 | 11 |
| mean | 1.63 | 0.99 | 0.87 | 0.38 | 0.74 | 94.67 | 10.25 |
| SD | 0.09 | 0.09 | 0.07 | 0.05 | 0.06 | 3.927 | 2.38 |
| min | 1.50 | 0.89 | 0.77 | 0.29 | 0.67 | 91.00 | 7.00 |
| max | 1.81 | 1.15 | 0.99 | 0.47 | 0.83 | 103.00 | 14.00 |
| Lerniana moreana | |||||||
| holotype | 1.67 | 1.04 | 0.96 | 0.40 | 0.76 | 96 | 12 |
| Lerniana seminula | |||||||
| LS1 | 1.87 | 1.13 | 0.99 | 0.44 | 0.79 | 89 | 11 |
| LS2 | 1.77 | 1.09 | 0.98 | 0.37 | 0.77 | 90 | 10 |
| LS3 | 1.65 | 1.05 | 0.96 | 0.32 | 0.80 | 86 | 10 |
| LS4 | 1.85 | 1.12 | 0.96 | 0.42 | 0.79 | 89 | 13 |
| LS5 | 1.55 | 0.98 | 0.92 | 0.32 | 0.68 | 92 | 13 |
| LS6 | 1.59 | 0.96 | 0.91 | 0.33 | 0.74 | 89 | 8 |
| LS7 | 1.60 | 1.01 | 0.91 | 0.34 | 0.76 | 92 | 9 |
| mean | 1.70 | 1.05 | 0.95 | 0.36 | 0.76 | 89.57 | 10.57 |
| SD | 0.13 | 0.07 | 0.03 | 0.05 | 0.04 | 2.07 | 1.90 |
| min | 1.55 | 0.96 | 0.91 | 0.32 | 0.68 | 86.00 | 8.00 |
| max | 1.87 | 1.13 | 0.99 | 0.44 | 0.80 | 92.00 | 13.00 |
| Lerniana tritonum | |||||||
| LT1 | 1.77 | 0.98 | 0.81 | 0.55 | 0.69 | 86 | 14 |
| LT2 | 1.90 | 1.03 | 0.87 | 0.66 | 0.76 | 82 | 15 |
| LT3 | 1.75 | 0.95 | 0.79 | 0.57 | 0.70 | 84 | 15 |
| mean | 1.81 | 0.99 | 0.82 | 0.59 | 0.72 | 84.00 | 14.67 |
| SD | 0.08 | 0.04 | 0.04 | 0.06 | 0.04 | 2.00 | 0.58 |
| min | 1.75 | 0.95 | 0.79 | 0.55 | 0.69 | 82.00 | 14.00 |
| max | 1.90 | 1.03 | 0.87 | 0.66 | 0.76 | 86.00 | 15.00 |
| “Radomaniola elongata” | |||||||
| RE1 | 2.48 | 1.08 | 0.80 | 1.19 | 0.72 | 68 | 18 |
| RE2 | 2.20 | 0.99 | 0.85 | 0.96 | 0.70 | 68 | 17 |
| RE3 | 2.02 | 0.99 | 0.87 | 0.76 | 0.70 | 68 | 18 |
| mean | 2.23 | 1.02 | 0.84 | 0.97 | 0.71 | 68.00 | 17.67 |
| SD | 0.23 | 0.05 | 0.04 | 0.22 | 0.01 | 0.00 | 0.58 |
| min | 2.02 | 0.99 | 0.80 | 0.76 | 0.70 | 68.00 | 17.00 |
| max | 2.48 | 1.08 | 0.87 | 1.19 | 0.72 | 68.00 | 18.00 |
| “Radomaniola” sp. 2 | |||||||
| R1 | 1.73 | 1.08 | 0.99 | 0.39 | 0.74 | 89 | 9 |
| R2 | 1.83 | 1.16 | 0.95 | 0.45 | 0.82 | 90 | 13 |
| mean | 1.78 | 1.12 | 0.97 | 0.42 | 0.78 | 89.50 | 11.00 |
| SD | 0.07 | 0.06 | 0.03 | 0.04 | 0.06 | 0.71 | 2.83 |
| min | 1.73 | 1.08 | 0.95 | 0.39 | 0.74 | 89.00 | 9.00 |
| max | 1.83 | 1.16 | 0.99 | 0.45 | 0.82 | 90.00 | 13.00 |
| Trichonia trichonica | |||||||
| TT1 | 2.43 | 1.16 | 0.95 | 0.97 | 0.78 | 71 | 17 |
| TT2 | 2.43 | 1.05 | 0.95 | 1.01 | 0.75 | 68 | 18 |
| TT3 | 2.70 | 1.78 | 0.98 | 1.19 | 0.76 | 67 | 16 |
| mean | 2.52 | 1.33 | 0.96 | 1.06 | 0.76 | 68.67 | 17.00 |
| SD | 0.16 | 0.39 | 0.02 | 0.12 | 0.02 | 2.08 | 1.00 |
| min | 2.43 | 1.05 | 0.95 | 0.97 | 0.75 | 67.00 | 16.00 |
| max | 2.70 | 1.78 | 0.98 | 1.19 | 0.78 | 71.00 | 18.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaszczyńska, A.; Grego, J.; Hofman, S.; Osikowski, A.; Falniowski, A. Taxonomic Notes on Lerniana Delicado et Hauffe, 2022, Trichonia Schütt, 1980 (Truncatelloidea: Hydrobiidae: Horatiinae) and Allied Taxa. Taxonomy 2025, 5, 45. https://doi.org/10.3390/taxonomy5030045
Jaszczyńska A, Grego J, Hofman S, Osikowski A, Falniowski A. Taxonomic Notes on Lerniana Delicado et Hauffe, 2022, Trichonia Schütt, 1980 (Truncatelloidea: Hydrobiidae: Horatiinae) and Allied Taxa. Taxonomy. 2025; 5(3):45. https://doi.org/10.3390/taxonomy5030045
Chicago/Turabian StyleJaszczyńska, Aleksandra, Jozef Grego, Sebastian Hofman, Artur Osikowski, and Andrzej Falniowski. 2025. "Taxonomic Notes on Lerniana Delicado et Hauffe, 2022, Trichonia Schütt, 1980 (Truncatelloidea: Hydrobiidae: Horatiinae) and Allied Taxa" Taxonomy 5, no. 3: 45. https://doi.org/10.3390/taxonomy5030045
APA StyleJaszczyńska, A., Grego, J., Hofman, S., Osikowski, A., & Falniowski, A. (2025). Taxonomic Notes on Lerniana Delicado et Hauffe, 2022, Trichonia Schütt, 1980 (Truncatelloidea: Hydrobiidae: Horatiinae) and Allied Taxa. Taxonomy, 5(3), 45. https://doi.org/10.3390/taxonomy5030045






