Genetic Evidence Confirms That the Porostomate Nudibranch Dendrodoris gunnamatta Allan, 1932 Is a Morphotype of Dendrodoris krusensternii (Gray, 1850) (Gastropoda: Nudibranchia)
Abstract
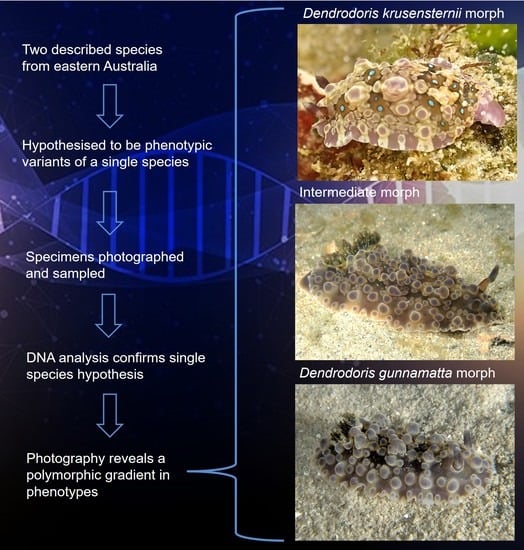
1. Introduction
2. Materials and Methods
2.1. Source of Material
2.2. Molecular Methods
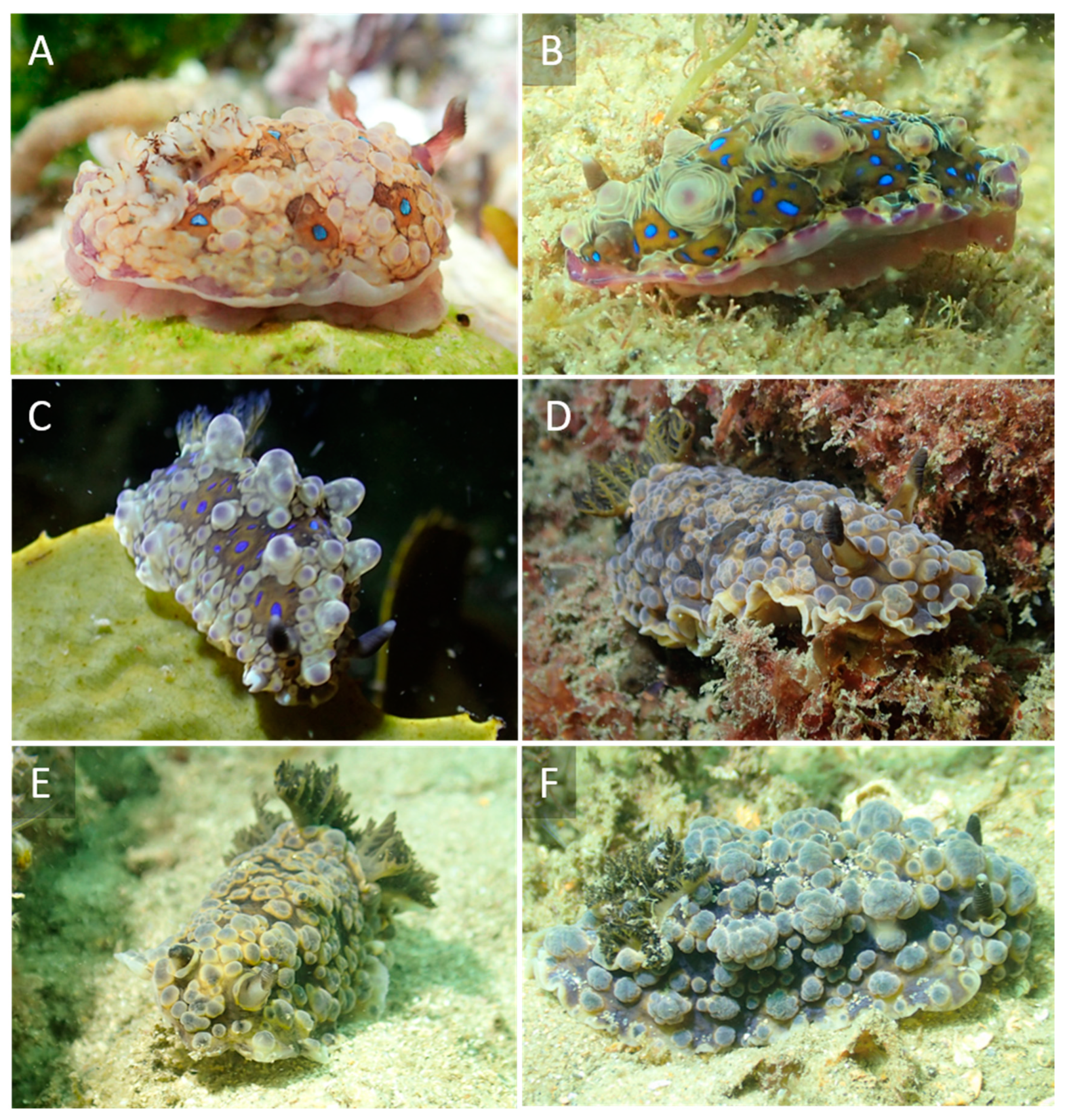
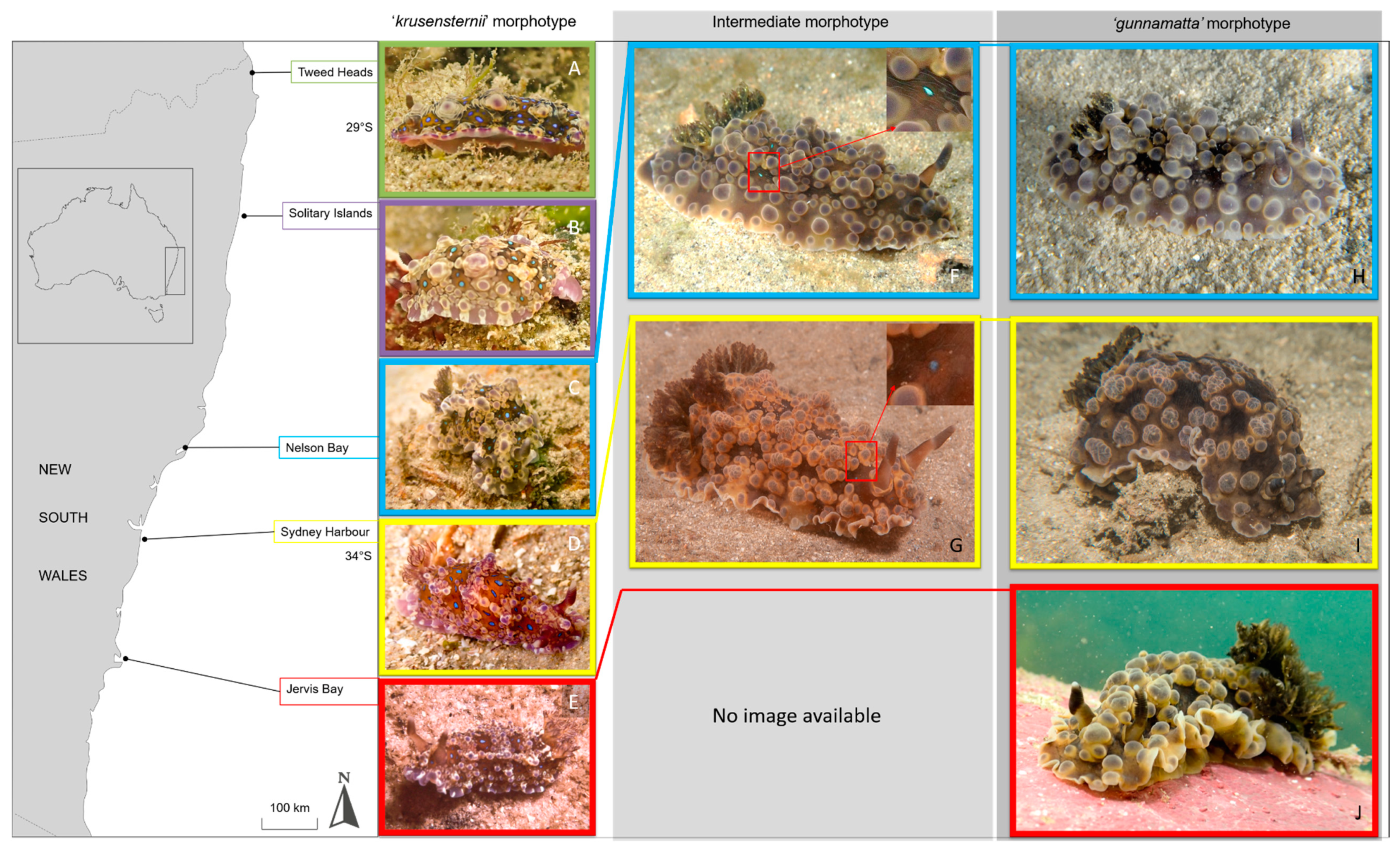
2.3. Photographic Methods
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MolluscaBase. Dendrodoris krusensternii (Gray, 1850). Available online: http://www.marinespecies.org/aphia.php?p=taxdetails&id=550448 (accessed on 5 April 2021).
- Hirose, M.; Hirose, E.; Kiyomoto, M. Identification of five species of Dendrodoris (Mollusca: Nudibranchia) from Japan, using DNA barcode and larval characters. Mar. Biol. 2015, 45, 769–780. [Google Scholar] [CrossRef]
- Rudman, W.B. Re: Dendrodoris denisoni or D. gunnamatta. Available online: http://www.seaslugforum.net/message/22773 (accessed on 5 April 2021).
- Thompson, T.E. Biology of Opisthobranch Molluscs; Ray Society: London, UK, 1976. [Google Scholar]
- Allan, J.K. A new genus and species of sea-slug, and two new species of sea-hares from Australia. Rec. Aust. Mus. 1932, 18, 314–320. [Google Scholar] [CrossRef][Green Version]
- Gosliner, T.M.; Valdés, Á.; Behrens, D.W. Nudibranch and Sea Slug Identification: Indo-Pacific; New World Publications: Jacksonville, FL, USA, 2015. [Google Scholar]
- Nimbs, M.J.; Hutton, I.; Davis, T.R.; Larkin, M.F.; Smith, S.D.A. The heterobranch sea slugs of Lord Howe Island, NSW, Australia (Mollusca: Gastropoda). Proc. Roy. Soc. Vic. 2020, 132, 12–41. [Google Scholar] [CrossRef]
- Nimbs, M.J.; Smith, S.D.A. An illustrated inventory of the sea slugs of New South Wales, Australia (Gastropoda: Heterobranchia). Proc. Roy. Soc. Vic. 2017, 128, 44–113. [Google Scholar] [CrossRef]
- Rudman, W.B. Dendrodoris denisoni (Angas, 1864). Available online: http://www.seaslugforum.net/find/denddeni (accessed on 5 April 2021).
- Valdés, Á.; Fahey, S.J. Dorid nudibranchs described by J. E. Gray in M. E. Gray, 1842-1857 (Mollusca: Opisthobranchia). Rec. West. Aust. Mus. Suppl. 2006, 69, 95–102. [Google Scholar] [CrossRef]
- Angas, G.F. Description D’espèces Nouvelles Appartenant À Plusieurs Genres de Mollusques Nudibranches des Environs de Port-Jackson (Nouvelle-Galles du Sud): Accompagnée de Dessins Faits D’après Nature. J. Conchyliol. 1864, 12, 43–70. [Google Scholar]
- Nimbs, M.J.; Smith, S.D.A. Beyond Capricornia: Tropical sea slugs (Gastropoda, Heterobranchia) extend their distributions into the Tasman Sea. Diversity 2018, 10, 99. [Google Scholar] [CrossRef]
- Smith, S.D.A.; Davis, T.R. Slugging it out for science: Volunteers provide valuable data on the diversity and distribution of heterobranch sea slugs. Moll. Res. 2019, 39, 214–223. [Google Scholar] [CrossRef]
- Dayrat, B.; Conrad, M.; Balayan, S.; White, T.R.; Albrecht, C.; Golding, R.; de Frias Martins, A.M. Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca): New insights from increased taxon sampling. Mol. Phylogenet. Evol. 2011, 59, 425–437. [Google Scholar] [CrossRef]
- Lindgren, A.R. Molecular inference of phylogenetic relationships among Decapodiformes (Mollusca: Cephalopoda) with special focus on the squid order Oegopsida. Mol. Phylogenet. Evol. 2010, 56, 77–90. [Google Scholar] [CrossRef]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 1991, 10, 506–513. [Google Scholar] [CrossRef]
- Donald, K.M.; Kennedy, M.; Spencer, H.G. Cladogenesis as the result of long-distance rafting events in South Pacific topshells (Gastropoda, Trochidae). Evolution 2005, 59, 1701–1711. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Bio. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Nishimura, D. Sequencher 3.1.1. Biotech Softw. Internet Rep. 2000, 1, 24–30. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Bio. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36 (Suppl. 2), W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Brodie, G.D.; Calado, G. Dendrodoris arborescens (Collingwood, 1881)(Mollusca: Nudibranchia): Larval characteristics reveal a masked porostome species. Rec. West. Aust. Mus. Suppl. 2006, 69, 119–126. [Google Scholar] [CrossRef][Green Version]
- Brodie, G.D.; Willan, R.C.; Collins, J.D. Taxonomy and occurrence of Dendrodoris nigra and Dendrodoris fumata (Nudibranchia: Dendrodorididae) in the Indo-West Pacific region. J. Molluscan Stud. 1997, 63, 407–423. [Google Scholar] [CrossRef]
- Padula, V.; Bahia, J.; Stöger, I.; Camacho-García, Y.; Malaquias, M.A.E.; Cervera, J.L.; Schrödl, M. A test of color-based taxonomy in nudibranchs: Molecular phylogeny and species delimitation of the Felimida clenchi (Mollusca: Chromodorididae) species complex. Mol. Phylogenet. Evol. 2016, 103, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Golestani, H.; Crocetta, F.; Padula, V.; Camacho-García, Y.; Langeneck, J.; Poursanidis, D.; Pola, M.; Yokeş, M.B.; Cervera, J.L.; Jung, D.-W.; et al. The little Aplysia coming of age: From one species to a complex of species complexes in Aplysia parvula (Mollusca: Gastropoda: Heterobranchia). Zool. J. Linn. Soc. 2019, 187, 279–330. [Google Scholar] [CrossRef]
- Nimbs, M.J.; Wilson, N.G. Saved by the shell: Molecular analysis detects the cryptic sea hare, Aplysia concava G. B. Sowerby I, 1833 (Mollusca: Heterobranchia: Aplysiidae), from Oceania, with a redescription. Taxonomy 2021, 1, 48–59. [Google Scholar] [CrossRef]
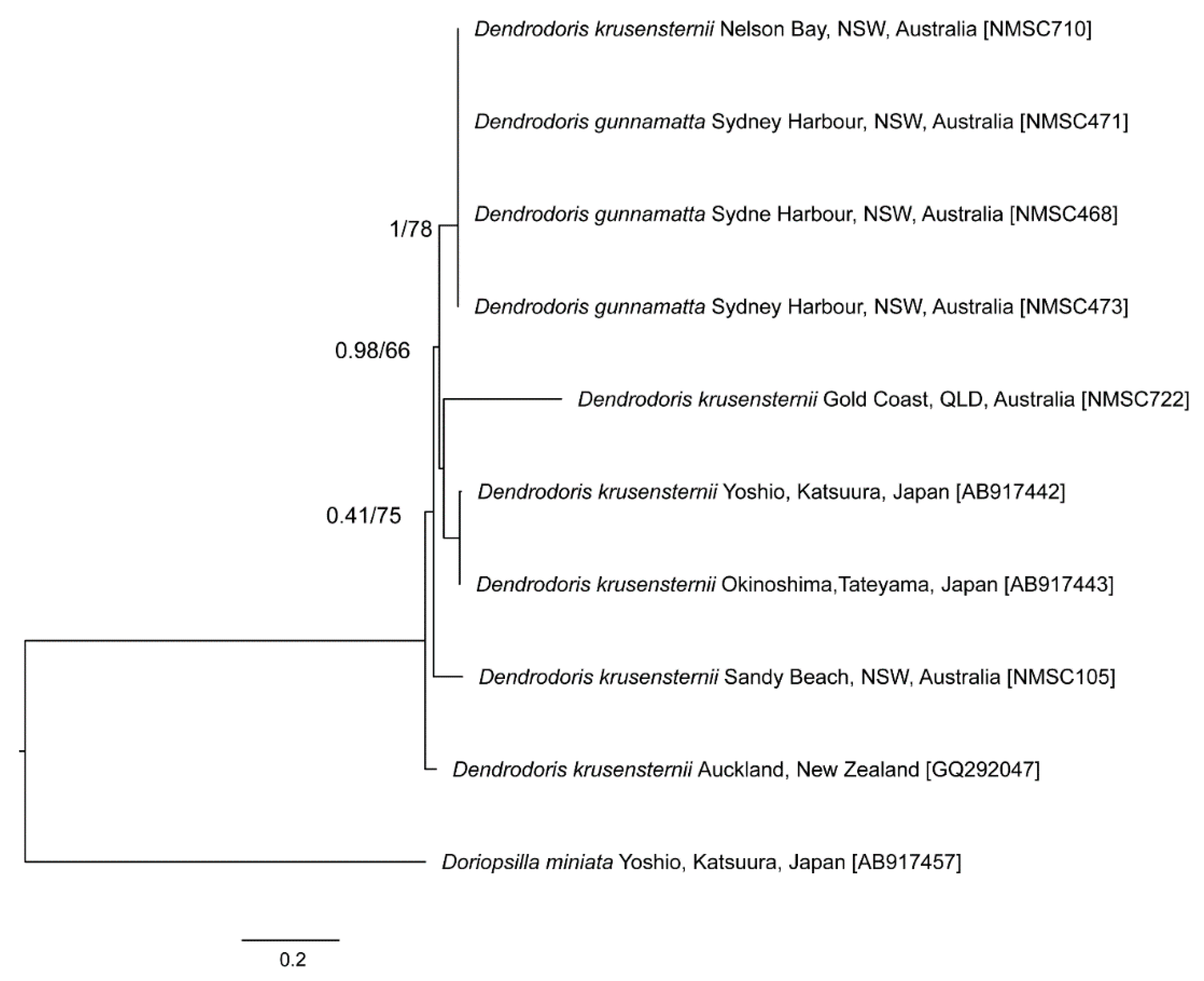

| Species | Locality | Voucher Number | GenBank Accession |
|---|---|---|---|
| D. krusensternii | Sawtell, NSW, Australia | NMSC105 | MZ373329 |
| Yoshio, Katsuura, Japan | TY64 | AB917442 | |
| Okinoshima, Tateyama, Japan | TY82 | AB917443 | |
| Auckland, New Zealand | GQ292047 | ||
| Tweed Heads, NSW, Australia | NMSC722 | MZ373328 | |
| Nelson Bay, NSW, Australia | NMSC710 | MZ373324 | |
| D. gunnamatta | Sydney Harbour, NSW, Australia | NMSC471 | MZ373326 |
| Sydney Harbour, NSW, Australia | NMSC468 | MZ373325 | |
| Sydney Harbour, NSW, Australia | NMSC473 | MZ373327 | |
| Do. miniata | Yoshio, Katsuura, Japan | TY65 | AB917457 |
| Distance between and within Species | ||
|---|---|---|
| D. krusensternii | D. gunnamatta | |
| D. krusensternii | 0.3613 (0.0783) | |
| D. gunnamatta | 0.0625 (0.0755) | 0.0000 (0.0000) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nimbs, M.J.; Smith, S.D.A. Genetic Evidence Confirms That the Porostomate Nudibranch Dendrodoris gunnamatta Allan, 1932 Is a Morphotype of Dendrodoris krusensternii (Gray, 1850) (Gastropoda: Nudibranchia). Taxonomy 2021, 1, 152-159. https://doi.org/10.3390/taxonomy1020012
Nimbs MJ, Smith SDA. Genetic Evidence Confirms That the Porostomate Nudibranch Dendrodoris gunnamatta Allan, 1932 Is a Morphotype of Dendrodoris krusensternii (Gray, 1850) (Gastropoda: Nudibranchia). Taxonomy. 2021; 1(2):152-159. https://doi.org/10.3390/taxonomy1020012
Chicago/Turabian StyleNimbs, Matt. J., and Stephen D. A. Smith. 2021. "Genetic Evidence Confirms That the Porostomate Nudibranch Dendrodoris gunnamatta Allan, 1932 Is a Morphotype of Dendrodoris krusensternii (Gray, 1850) (Gastropoda: Nudibranchia)" Taxonomy 1, no. 2: 152-159. https://doi.org/10.3390/taxonomy1020012
APA StyleNimbs, M. J., & Smith, S. D. A. (2021). Genetic Evidence Confirms That the Porostomate Nudibranch Dendrodoris gunnamatta Allan, 1932 Is a Morphotype of Dendrodoris krusensternii (Gray, 1850) (Gastropoda: Nudibranchia). Taxonomy, 1(2), 152-159. https://doi.org/10.3390/taxonomy1020012