An Autochthonous Acidithiobacillus ferrooxidans Metapopulation Exploited for Two-Step Pyrite Biooxidation Improves Au/Ag Particle Release from Mining Waste
Abstract
1. Introduction
2. Materials and Methods
2.1. Pyrite Concentrate
2.2. Origin, Acquisition, and Maintenance of Microorganisms
2.3. Identification of Bacteria
2.4. Two-Stage Oxidation of Pyrite
2.5. Analytical Methods
3. Results and Discussion
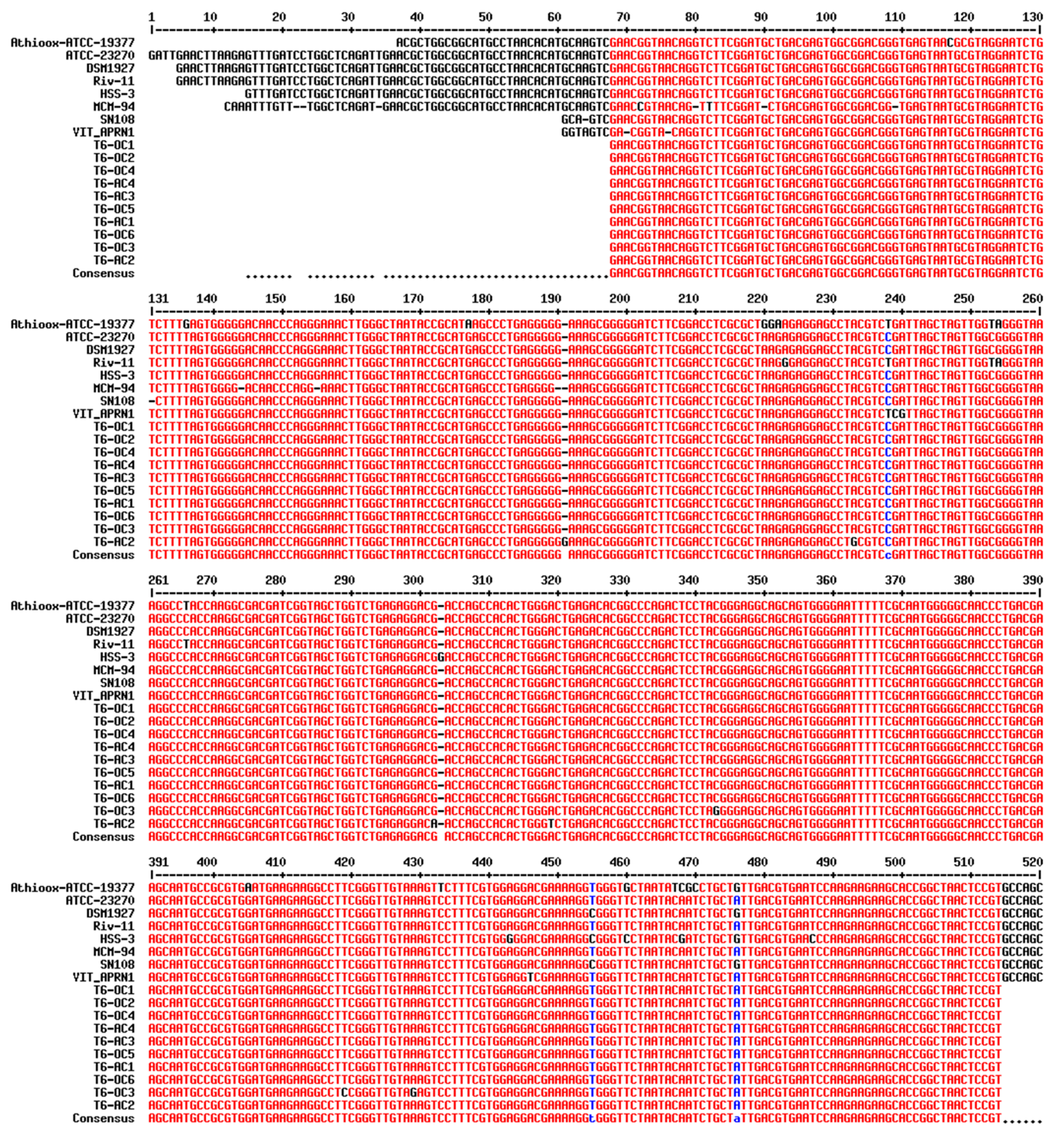
3.1. Strains of A. ferrooxidans Identified in the Remnant Solution T6
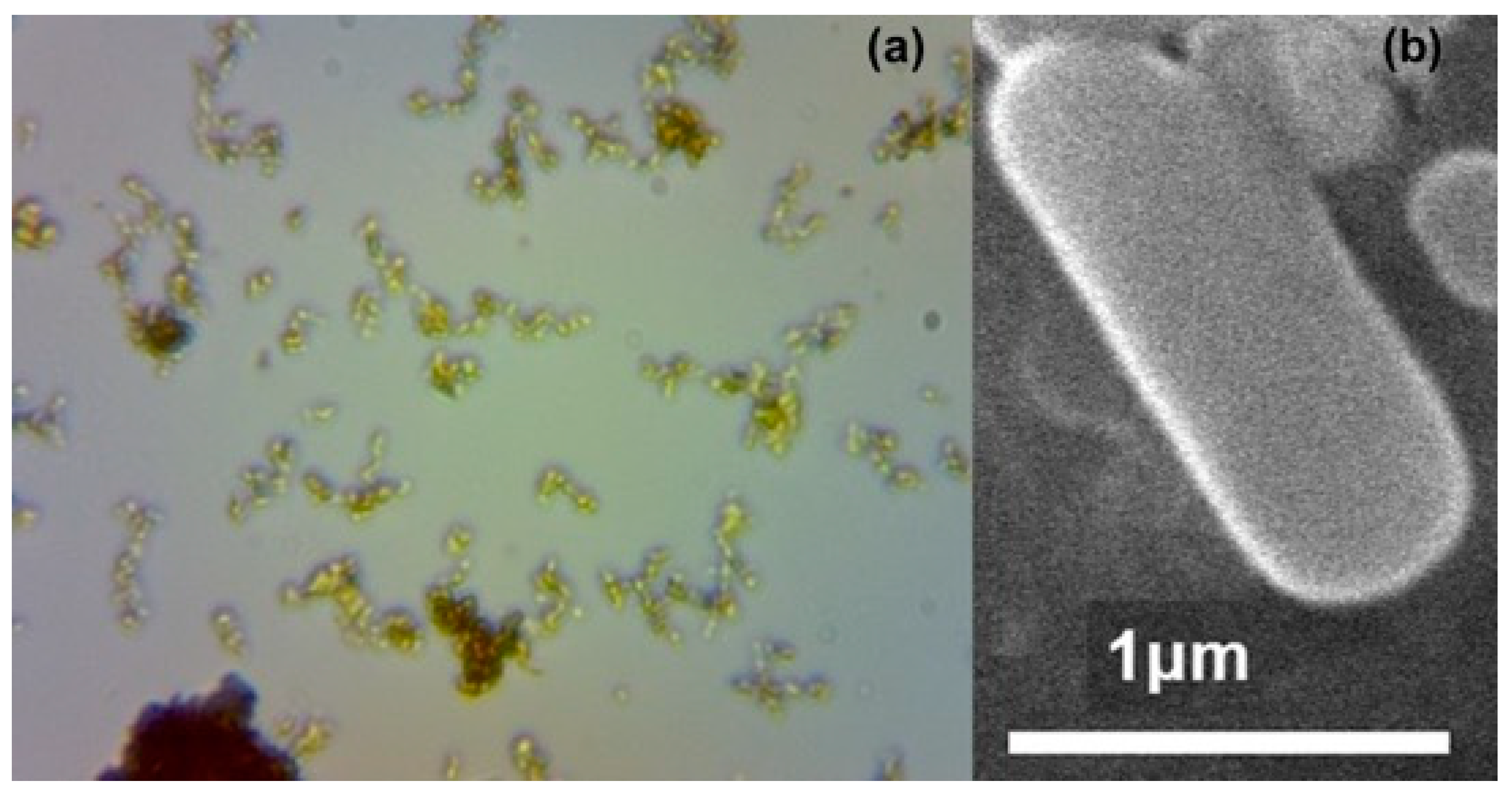
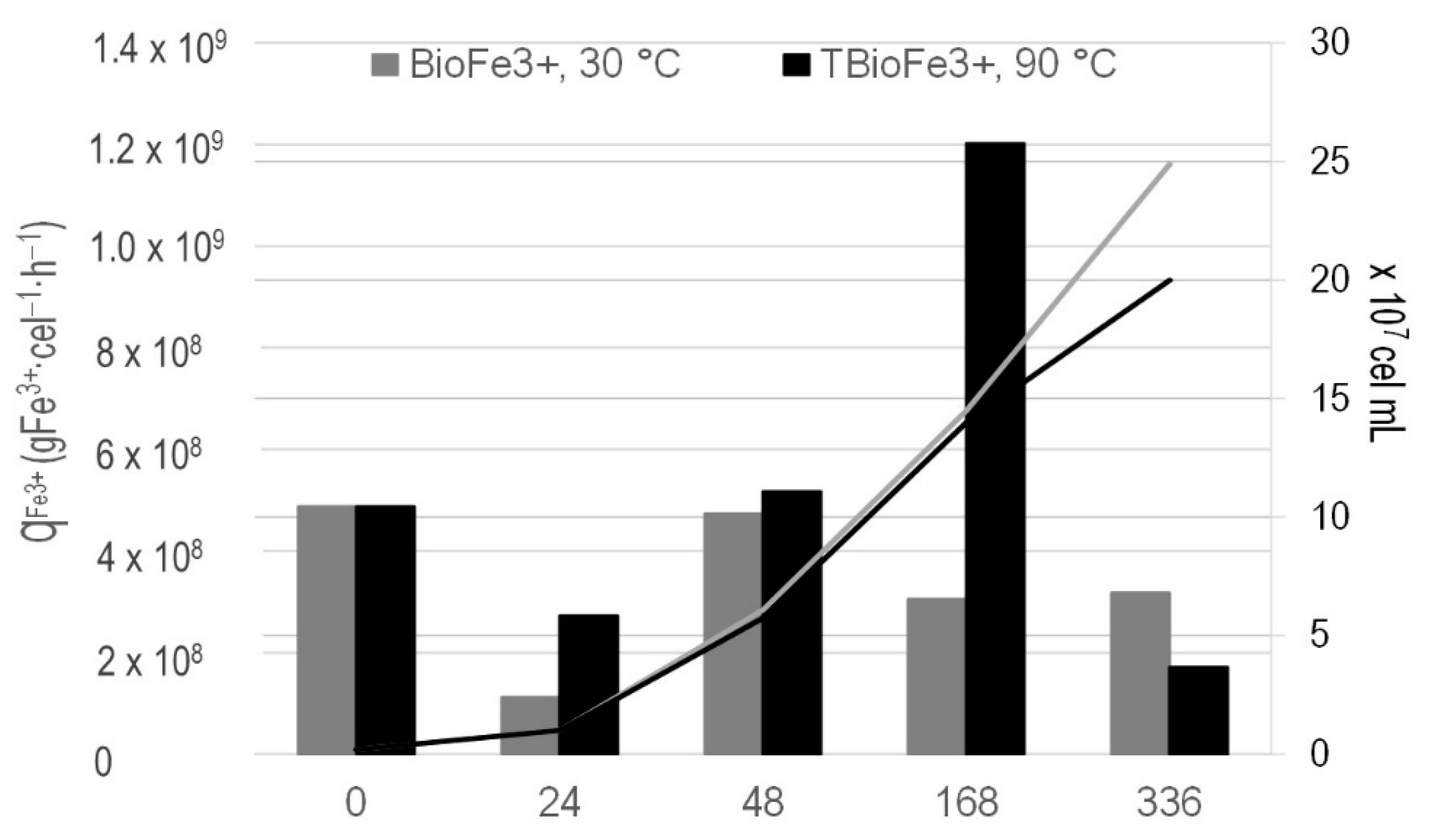
3.2. Bioferric Production by A. ferrooxidans Strains
3.3. Chemical Preoxidation Followed by Biological Oxidation of Pyrite
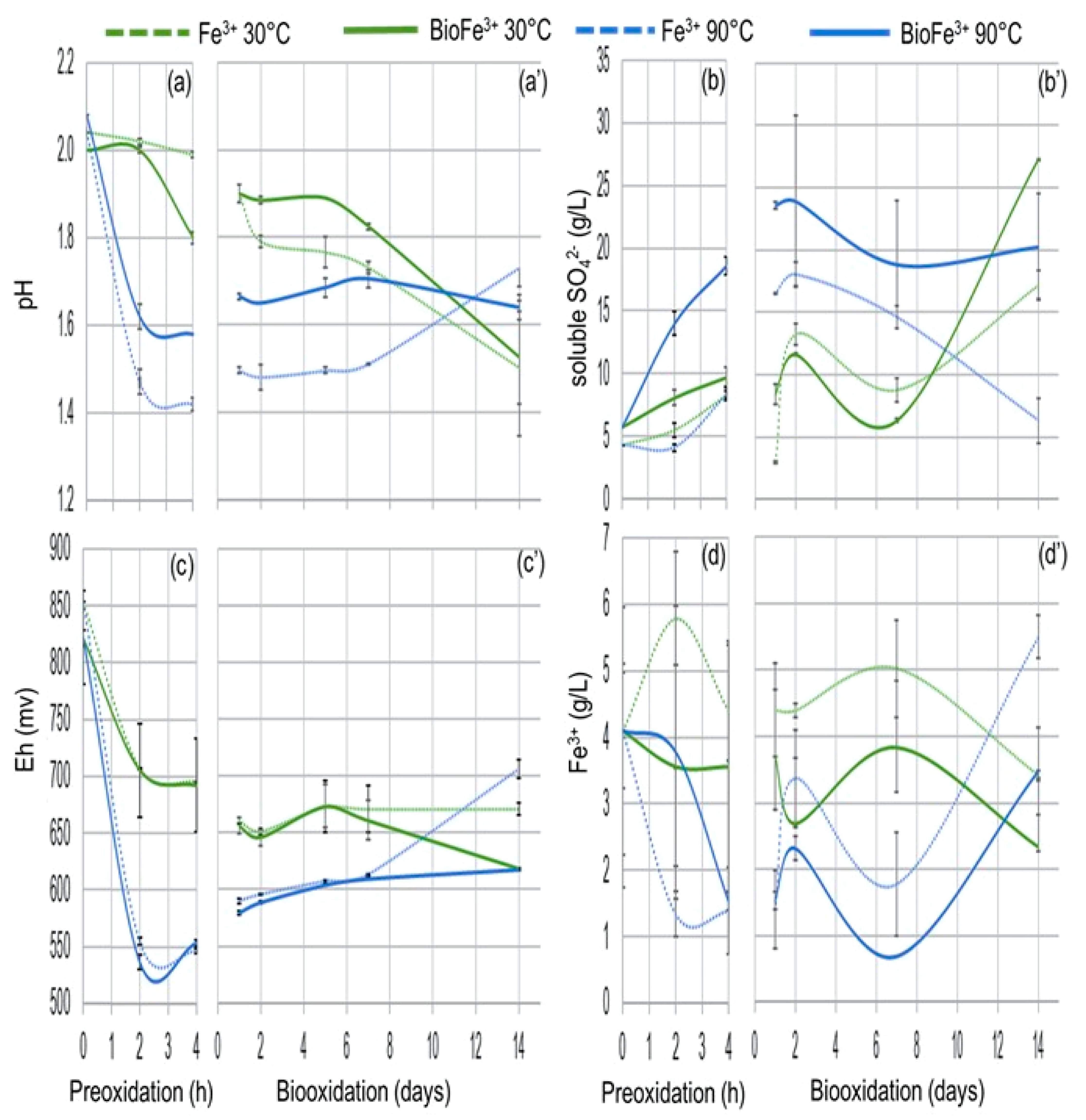
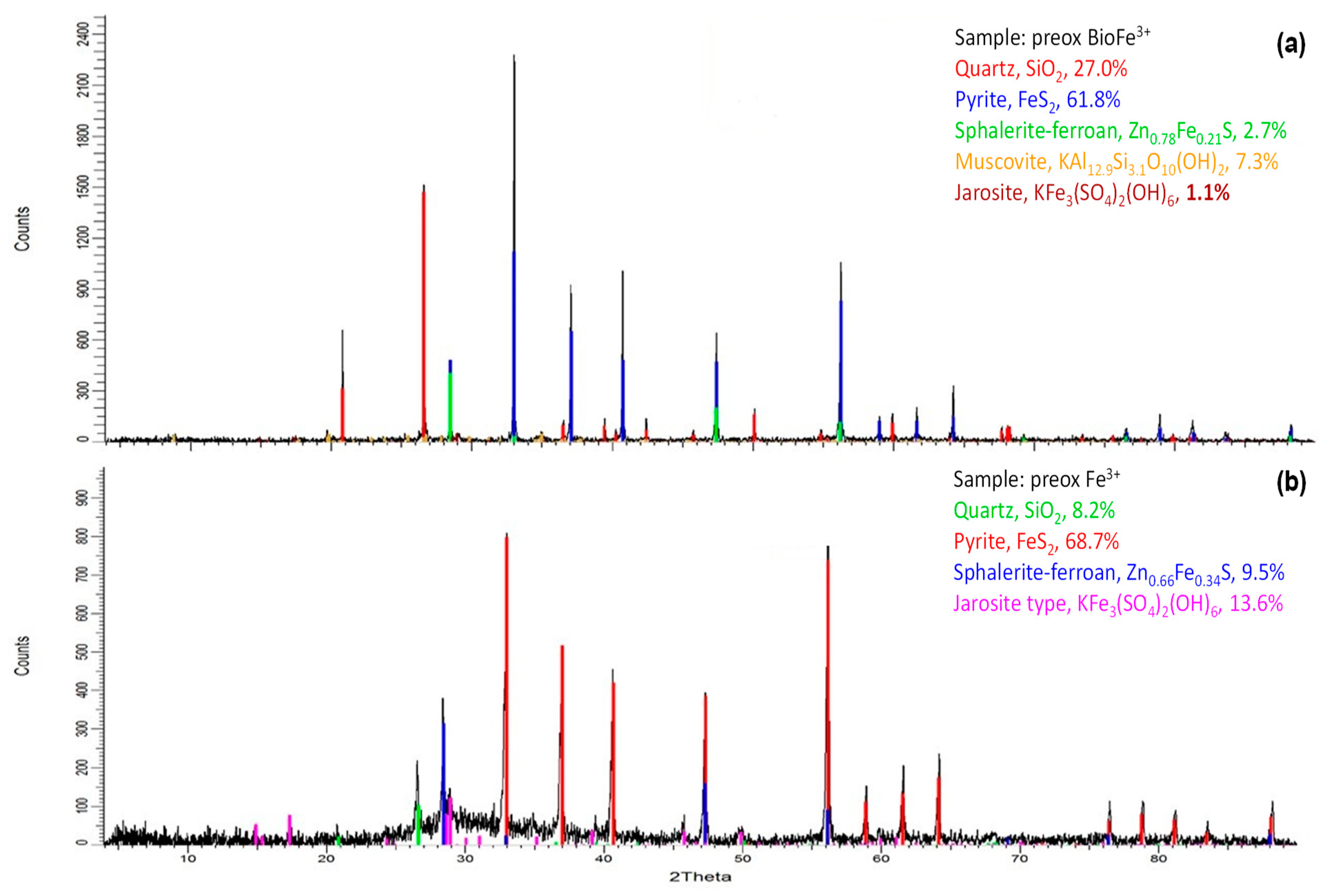
3.3.1. Chemical Preoxidation of Pyrite
3.3.2. Biological Oxidation of Pyrite Concentrate
3.3.3. Gold and Silver Extraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gorain, B.K.; Kondos, P.D.; Lakshmanan, V.I. Innovations in gold and silver processing. In Innovative Process Development in Metallurgical Industry; Lakshmanan, V., Roy, R., Ramachandran, V., Eds.; Springer: Cham, Switzerland, 2016; pp. 393–428. [Google Scholar] [CrossRef]
- Jafari, M.; Abdollahi, H.; Shafaei, S.Z.; Gharabaghi, M.; Jafari, H.; Akcil, A.; Panda, S. Acidophilic bioleaching: A review on the process and effect of organic-inorganic reagents and materials on its efficiency. Minrt. Proces. Extr. Metall. Rev. 2018, 40, 87. [Google Scholar] [CrossRef]
- Brune, K.; Bayer, T. Engineering microbial consortia to enhance biomining and bioremediation. Front. Microbiol. 2012, 3, 203. [Google Scholar] [CrossRef]
- He, J.; Kappler, A. Recovery of precious metals from waste streams. Microbial Biotechnol. 2017, 10, 1194. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.; Wilson, G. Bioleaching genomics. Microbial Biotechnol. 2009, 2, 297. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, V.; Zhang, Y. A review: Pyrite oxidation mechanisms and acid mine drainage prevention. Crit. Rev. Environ. Sci. Technol. 1995, 25, 141. [Google Scholar] [CrossRef]
- Tao, H.; Dongwei, L. Presentation on mechanisms and applications of chalcopyrite and pyrite bioleaching in biohydrometallurgy—A presentation. Biotechnol. Rep. 2014, 4, 107. [Google Scholar] [CrossRef]
- Sánchez-Andrea, I.; Rodríguez, N.; Amils, R.; Sanz, J. Microbial diversity in anaerobic sediments at Río Tinto, a naturally acidic environment with a high heavy metal content. App. Environ. Microbiol. 2011, 77, 6085. [Google Scholar] [CrossRef]
- Sun, H.; Chen, M.; Zou, L.; Shu, R.; Ruan, R. Study of kinetics of pyrite oxidation under controlled redox potential. Hydrometallurgy 2015, 155, 13. [Google Scholar] [CrossRef]
- Asamoah, R.K. Specific refractory gold flotation and bio-oxidation products: Research overview. Minerals 2021, 11, 93. [Google Scholar] [CrossRef]
- Ghassa, S.; Noaparast, M.; Shafaei, S.Z.; Abdollahi, H.; Gharabaghi, M.; Boruomand, Z. A study on the zinc sulfide dissolution kinetics with biological and chemical ferric reagents. Hydrometallurgy 2017, 171, 362. [Google Scholar] [CrossRef]
- Wang, G.; Xie, S.; Liu, X.; Wu, Y.; Liu, Y.; Zeng, T. Bio-oxidation of a high-sulfur and high-arsenic refractory gold concentrate using a two-stage process. Miner. Eng. 2018, 120, 94. [Google Scholar] [CrossRef]
- Fomchenko, N.Y.V.; Muravyov, M.I.; Kondrat’eva, T.F. Two-stage bacterial-chemical oxidation of refractory gold-bearing sulfidic concentrates. Hydrometallurgy 2010, 101, 28. [Google Scholar] [CrossRef]
- Fomchenko, N.; Muravyov, M. Sequential bioleaching of pyritic tailings and ferric leaching of nonferrous slags as a method for metal recovery from mining and metallurgical wastes. Minerals 2020, 10, 1097. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Mudunuru, B.M.; Hackl, R. The role of microorganisms in gold processing and recovery—A review. Hydrometallurgy 2014, 142, 70–83. [Google Scholar] [CrossRef]
- Li, Q.; Li, D.; Qian, F. Preoxidation of high-sulfur and high-arsenic refractory gold concentrate by ozone and ferric ions in acidic media. Hydrometallurgy 2009, 97, 61–66. [Google Scholar] [CrossRef]
- Muravyov, M.I.; Bulaev, A.G. Two-step oxidation of a refractory gold-bearing sulfidic concentrate and the effect of organic nutrients on its biooxidation. Miner. Eng. 2013, 45, 108–114. [Google Scholar] [CrossRef]
- Zheng, Y.; Wei, D.; Liu, W.; Duan, H.; Zhou, S. One novel two-step bio-oxidation pretreatment of arsenic-containing gold-bearing concentrate. Int. J. Electrochem. Sci. 2018, 13, 5983–5994. [Google Scholar] [CrossRef]
- Steijns, M.; Derks, F.; Verloop, A.; Mars, P. The mechanism of the catalytic oxidation of hydrogen sulfide: II. Kinetics and mechanism of hydrogen sulfide oxidation catalyzed by sulfur. J. Catal. 1976, 42, 87–95. [Google Scholar] [CrossRef]
- Fowler, T.A.; Holmes, P.R.; Crundwell, F. On the kinetics and mechanism of the dissolution of pyrite in the presence of Thiobacillus ferrooxidans. Hydrometallurgy 2001, 59, 257. [Google Scholar] [CrossRef]
- Rawlings, D. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 2005, 4, 13. [Google Scholar] [CrossRef]
- Rao, S.; Muyinda, N.; De Baets, B. Stability analysis of the coexistence equilibrium of a balanced metapopulation model. Nat. Sci. Rep. 2021, 11, 14084. [Google Scholar] [CrossRef]
- Kunin, V.; He, S.; Warnecke, F.; Peterson, S.B.; Garcia-Martin, H.; Haynes, M.; Ivanova, N.; Blackall, L.L.; Breitbart, M.; Rohwer, F.; et al. Bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008, 18, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Palacios, C.; Zettler, E.; Amils, R.; Amaral-Zettler, L. Contrasting microbial community assembly hypotheses: A reconciling tale from the Río Tinto. PLoS ONE 2008, 3, e3853. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, R.; Croucher, N.J.; Andam, C.P.; Corander, J.; Hanage, W.P.; Marttinen, P. Efficient inference of recent and ancestral recombination within bacterial populations. Mol. Biol. Evol. 2017, 34, 1167–1182. [Google Scholar] [CrossRef]
- Sipola, A.; Marttinen, P.; Corander, J.; Berger, B. Bacmeta: Simulator for genomic evolution in bacterial metapopulations. Bioinformatics 2018, 34, 2308–2310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, X.; Tao, J.; Ma, L.; Xiao, Y.; Liang, Y.; Liu, X.; Yin, H. Comparative genomics of the extreme acidophile Acidithiobacillus thiooxidans reveals intraspecific divergence and niche adaptation. Int. J. Mol. Sci. 2016, 17, 1355. [Google Scholar] [CrossRef]
- Karavaiko, G.I.; Turova, T.P.; Kondrat’eva, T.F.; Lysenko, A.M.; Kolganova, T.V.; Ageeva, S.N.; Muntyan, L.N.; Pivovarova, T.A. Phylogenetic heterogeneity of the species Acidithiobacillus ferrooxidans. Int. J. Syst. Evol. Microbiol. 2003, 53, 113. [Google Scholar] [CrossRef][Green Version]
- Saavedra, A.; Aguirre, P.; Gentina, J.C. Climbing the hill: The implications of ferrous iron pulse adaptation on biooxidation by Acidithiobacillus ferrooxidans. Hydrometallurgy 2020, 197, 105485. [Google Scholar] [CrossRef]
- Nemati, M.; Harrison, S.T.L. Effects of solid particles on thermophilic bioleaching of sulphide minerals. In Biohydrometallurgy and the Environment; Toward the Mining of the 21st Century-Part A; Amils, R., Ballester, A., Eds.; Elsevier: Amsterdam, The Netherland, 1999; pp. 473–482. [Google Scholar] [CrossRef]
- Callender, K.; Roy, S.; Khasa, D.; Whyte, L.; Greer, C. Actinorhizal alder phytostabilization alters microbial community dynamics in gold mine waste rock from Northern Quebec: A greenhouse study. PLoS ONE 2016, 11, e0150181. [Google Scholar] [CrossRef]
- Mesa, V.; Gallego, J.; González-Gil, R.; Lauga, B.; Sánchez, J.; Méndez-García, C.; Peláez, A. Bacterial, Archaeal, and Eukaryotic diversity across distinct microhabitats in an acid mine drainage. Front. Microbiol. 2017, 8, 1756. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.; Sato, T.; Weightman, A.; Martin, T.; Fry, J.; Hiom, S.; Wade, W. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 6, 795. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Madden, T.; Schäffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Allen, H.K.; Bayles, D.O.; Looft, T.; Trachsel, J.; Bass, B.E.; Alt, D.P.; Bearson, S.M.D.; Nicholson, T.; Casey, T.A. Pipeline for amplifying and analyzing amplicons of the V1-V3 region of the 16S rRNA gene. BMC Res. Notes 2016, 380. [Google Scholar] [CrossRef]
- Scholss, P.D. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 2010, 6, e1000844. [Google Scholar] [CrossRef]
- Coenye, T.; Vandamme, P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 2003, 228, 45. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Meth. 2003, 55, 541. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design ribosomal DNA amplicons in metagenomics studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R.; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genom. 2008, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, H.; Moya-Beltrán, A.; Covarrubias, P.C.; Issotta, F.; Cárdenas, J.P.; González, M.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Molecular systematics of the genus Acidithiobacillus: Insights into the phylogenetic structure and diversification of the taxon. Front. Microbiol. 2017, 8, 30. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; Van de Peer, Y.; Vandamme, P.; Thompson, F.L.; et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733. [Google Scholar] [CrossRef]
- Achtman, M.; Wagner, M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008, 6, 431–440. [Google Scholar] [CrossRef]
- Levins, R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull. Entomol. Soc. Am. 1969, 15, 237. [Google Scholar] [CrossRef]
- Keymer, J.E.; Galajda, P.; Muldoon, C.; Park, S.; Austin, R.H. Bacterial metapopulations in nanofabricated landscapes. Proc. Natl. Acad. Sci. USA 2006, 103, 17290–17295. [Google Scholar] [CrossRef] [PubMed]
- Mc Ginty, S.E.; Rankin, D.J.; Brown, S.P. Horizontal gene transfer and the evolution of bacterial cooperation. Evolution 2010, 65, 21. [Google Scholar] [CrossRef]
- Goethert, H.K.; Saviet, B.; Telford III, S.R. Metapopulation structure for perpetuation of Francisella tularensis tularensis. BMC Microbiol. 2009, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Pristas, P.; Kiskova, J.; Timkova, I.; Malinicova, L.; Luptakova, A.; Kusnierova, M.; Sedlakova-Kadukova, J. Genetic variability in Acidithiobacillus spp. -A working horse of environmental biotechnologies. Nova Biotechnol. Chim. 2018, 17, 125. [Google Scholar] [CrossRef]
- Nemati, M.; Lowenadler, J.; Harrison, S.T.L. Particle size effects in bioleaching of pyrite by acidophilic thermophile Sulfolobus metallicus (BC). Appl. Microbiol. Biotechnol. 2000, 53, 173. [Google Scholar] [CrossRef]
- Rawlings, D.; Tributsch, H.; Hansford, G. Reasons why “Leptospirillum”-like species rather than Thiobacillus ferrooxidans are the dominant iron-oxidizing bacteria in many commercial processes for the biooxidation of pyrite and related ores. Microbiol. 1999, 145, 5. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Yaghmaei, S.; Salimi, F.; Jafari, A. Influence of process variables on biooxidation of ferrous sulfate by an indigenous Acidithiobacillus ferrooxidans. Part I: Flask experiments. Fuel 2006, 85, 2555–2560. [Google Scholar] [CrossRef]
- Dopson, M. Growth in sulfidic mineral environments: Metal resistance mechanisms in acidophilic micro-organisms. Microbiology 2003, 149, 1959. [Google Scholar] [CrossRef] [PubMed]
- Pogliani, C.; Donati, E. The role of exopolymers in bioleaching of a non-ferrous metal sulphide. J. Ind. Microbiol. Biotechnol. 1999, 22, 88. [Google Scholar] [CrossRef]
- Dutrizac, J.E. Factors affecting alkali jarosite precipitation. Metall. Trans. B 1983, 14, 531–539. [Google Scholar] [CrossRef]
- Dutrizac, J.E.; Dinardo, O. The co-precipitation of copper and zinc with lead jarosite. Hydrometallurgy 1983, 11, 61–78. [Google Scholar] [CrossRef]
- Rohwerder, T.; Gehrke, T.; Kinzler, K.; Sand, W. Bioleaching review part A: Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulfide oxidation. App. Microbiol. Biotechnol. 2003, 63, 239. [Google Scholar] [CrossRef]

| Element | Fe | Cu | Zn | As | Pb | Ag | Au |
|---|---|---|---|---|---|---|---|
| (%) | (g/T) | ||||||
| Content | 36.2 | 0.04 | 0.30 | 0.46 | 0.23 | 78 | 22.4 |
| Solution | pH | Eh (mV) | Total Fe | Fe2+ | Cu | Zn | Pb | As | Biomass (Cells·mL−1) |
|---|---|---|---|---|---|---|---|---|---|
| T6 | 3.05 | 552 | 30.9 | 27 | 57 | 127 | 1.2 | 7.5 | 14 × 106 |
| Precipitate composition | Jarosite, KFe3(SO4)(OH)6 (83.7%); rozenite, FeSO4·4H2O (16.34%) | ||||||||
| Initial Fe2+ (g·L−1) | Final Fe2+ (g·L−1) | Final BioFe3+ (g·L−1) | Final %BioFe3+ | |
|---|---|---|---|---|
| 14 | 0.0 | 14.0 | 100 | 3.97 × 10−7 |
| 20 | 0.5 | 19.53 | 97.7 | 3.86 × 10−7 |
| Oxidation Duration, Day | Oxidation Residue 1 | Cyanidation Residue | Pregnant Solution | Extraction | ||||
|---|---|---|---|---|---|---|---|---|
| Au (g·T−1) | Ag (g·T−1) | Au (g·T−1) | Ag (g·T−1) | Au (mg·L−1) | Ag (mg·L−1) | Au (%) | Ag (%) | |
| 21 | 2.89 | 75 | 2.59 | 32 | 0.11 | 4.18 | 30.52 | 57.97 |
| 25 | 2.90 | 73 | 2.65 | 35 | 0.09 | 4.05 | 27.17 | 55.27 |
| 35 | 2.96 | 74 | 2.65 | 34 | 0.09 | 3.75 | 27.09 | 54.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Paredes, A.E.; Alfaro-Saldaña, E.F.; Hernández-Sánchez, A.; García-Meza, J.V. An Autochthonous Acidithiobacillus ferrooxidans Metapopulation Exploited for Two-Step Pyrite Biooxidation Improves Au/Ag Particle Release from Mining Waste. Mining 2021, 1, 335-350. https://doi.org/10.3390/mining1030021
Jiménez-Paredes AE, Alfaro-Saldaña EF, Hernández-Sánchez A, García-Meza JV. An Autochthonous Acidithiobacillus ferrooxidans Metapopulation Exploited for Two-Step Pyrite Biooxidation Improves Au/Ag Particle Release from Mining Waste. Mining. 2021; 1(3):335-350. https://doi.org/10.3390/mining1030021
Chicago/Turabian StyleJiménez-Paredes, Andrea E., Elvia F. Alfaro-Saldaña, Araceli Hernández-Sánchez, and J. Viridiana García-Meza. 2021. "An Autochthonous Acidithiobacillus ferrooxidans Metapopulation Exploited for Two-Step Pyrite Biooxidation Improves Au/Ag Particle Release from Mining Waste" Mining 1, no. 3: 335-350. https://doi.org/10.3390/mining1030021
APA StyleJiménez-Paredes, A. E., Alfaro-Saldaña, E. F., Hernández-Sánchez, A., & García-Meza, J. V. (2021). An Autochthonous Acidithiobacillus ferrooxidans Metapopulation Exploited for Two-Step Pyrite Biooxidation Improves Au/Ag Particle Release from Mining Waste. Mining, 1(3), 335-350. https://doi.org/10.3390/mining1030021





