1. Introduction
Mobile health applications have experienced remarkable growth in recent years, with the global mHealth market reaching USD 62.7 billion in 2023 and expected to achieve USD 158.3 billion by 2030, representing a compound annual growth rate (CAGR) of 14.1% [
1]. The mHealth app sector alone was valued at USD 37.5 billion in 2024, with projections indicating a CAGR of 14.8% through 2030 [
2]. Research demonstrates that 80% of Americans support remote patient monitoring, with over half advocating for its integration into standard medical care [
3]. Within this expanding landscape, nutrient analysis applications serve a vital function, helping individuals, especially those from vulnerable communities, maintain balanced diets and make informed nutritional decisions. Nevertheless, developing accurate and efficient nutrient analysis tools for mobile platforms presents distinct challenges, particularly for users facing resource or literacy constraints.
This study defines vulnerable populations as individuals and communities encountering one or more barriers: (1) limited financial resources affecting access to advanced mobile devices and stable internet connectivity, (2) low health literacy impeding comprehension of complex nutritional information, (3) restricted access to healthcare professionals and nutritional education, and (4) cultural or linguistic obstacles influencing technology interaction [
4]. The research indicates that racial and ethnic minorities, low-income groups, and other vulnerable populations disproportionately experience the digital divide, with disparities in digital access compounding existing structural disadvantages [
5]. These communities frequently depend on basic smartphones with constrained processing capabilities (typically 2–4 GB RAM, older processors) and require simplified, culturally appropriate interfaces for effective health management [
4].
The significance of proper nutrition in maintaining health and preventing chronic diseases remains undisputed. Malnutrition, encompassing both undernutrition and obesity, continues as a major global health challenge [
6]. The prevalence of chronic diseases—diabetes, hypertension, cardiovascular conditions, and respiratory disorders—continues rising worldwide, with the World Health Organisation’s 2024 report documenting over 20 million new cancer cases [
7]. Mobile health applications offer promising opportunities to democratise nutritional information access, delivering personalised recommendations based on individual requirements and preferences [
1]. This democratisation proves especially valuable for vulnerable populations with limited access to healthcare professionals or nutritional education [
5]. However, low-income populations face unique mHealth utilisation barriers that amplify the impacts of social determinants of health, including limited mobile application fluency, restricted health literacy, reduced empowerment, and historical healthcare system mistrust [
4].
Conventional nutrient analysis methods typically depend on manual dietary information input or basic algorithms—approaches that are time-consuming, error-prone, and inadequate for capturing the complexity of human nutrition [
8]. The analysis of popular nutrition applications with over one million installations revealed that nine applications collected dietary intake using identical assessment methods (food diary records), with food selection achieved through text searches and barcode scanning. Notably, emerging technologies, including image recognition, natural language processing, and artificial intelligence, remained absent from most popular nutrition applications [
3]. Recent advances in artificial intelligence, particularly in deep learning, show considerable promise for enhancing nutrient analysis accuracy and capabilities [
1]. Deep learning models demonstrate potential for processing complex data inputs—food images or natural language descriptions—to deliver more accurate and comprehensive nutritional assessments [
9].
Nevertheless, implementing sophisticated deep learning models on mobile devices presents substantial challenges [
10]. Mobile platforms face constraints from limited computational resources, storage capacity, and energy consumption requirements. Furthermore, many advanced deep learning models demand intensive computation and substantial memory, rendering them impractical for real-time smartphone use [
11]. This situation necessitates the development of lightweight models capable of efficient operation within mobile device constraints whilst maintaining accuracy. For vulnerable populations, these constraints are particularly acute, as they often rely on older, entry-level smartphones with limited processing power, slower processors, and restricted battery life. The Federal Communications Commission estimates that 19 million Americans lack reliable broadband access [
5], creating additional barriers to digital health access. Our design decisions therefore prioritise ensuring functionality across diverse device specifications whilst maintaining the accuracy required for reliable nutritional guidance.
Deep learning application to health-related tasks faces another critical challenge: the “black box” nature of many models [
12]. Users and healthcare professionals require transparency in AI-driven health recommendations to foster trust and ensure responsible implementation. This is particularly crucial in nutrient analysis, where recommendations directly impact users’ dietary choices and health outcomes [
13]. Consequently, interpretable AI models capable of explaining predictions in human-understandable terms represent an urgent necessity [
7].
Model interpretability challenges intensify due to cultural diversity and varying health literacy levels among vulnerable populations. Research examining primary hypertension prevention in Argentina, Guatemala, and Peru identified challenges, including mHealth innovation unacceptability within targeted communities, emphasising the need for interventions tailored to literacy challenges attributed to gaps in cultural context understanding [
9]. Different cultural groups interpret and trust AI explanations differently, necessitating culturally sensitive interpretability approaches [
12]. Visual explanations may prove more effective for users with limited literacy, whilst some cultures might prefer contextual information about ingredient origins or preparation methods [
7]. Moreover, whilst initial user trust remains important, maintaining and building trust over time is crucial for long-term adoption and positive health outcomes. These technical challenges compound with requirements for culturally appropriate and sustained interpretability across diverse user groups. Effective interpretability must achieve technical soundness whilst remaining culturally resonant and trustworthy throughout extended use periods [
13].
This research specifically addresses deployment challenges encountered when serving vulnerable populations in resource-constrained environments, including rural communities in developing regions, elderly populations with limited technical literacy, low-income households using budget smartphones, and culturally diverse communities requiring localised nutritional guidance [
4]. Understanding these specific user constraints has informed our architectural decisions and evaluation methodology throughout this work.
The research objectives comprise three main areas:
To develop a lightweight deep learning model capable of accurate nutrient analysis whilst operating efficiently on mobile devices.
To integrate interpretability features into the model, allowing users to understand the factors influencing nutritional assessments.
To evaluate the model’s performance and usability in real-world scenarios, particularly for vulnerable populations.
This paper presents a novel approach to addressing these challenges. We propose lightweight, interpretable deep learning architecture specifically designed for nutrient analysis in mobile environments. Our model incorporates state-of-the-art compression techniques to reduce its size and computational requirements without sacrificing accuracy [
11]. Additionally, we integrate interpretability features that provide clear, user-friendly explanations for the model’s predictions, enhancing transparency and user trust [
12].
The main contributions of this work include:
A novel lightweight architecture that achieves high accuracy in nutrient analysis whilst being suitable for mobile deployment.
The successful integration of interpretability features that enhance user understanding without compromising model performance.
A comprehensive evaluation of the model’s performance in mobile health contexts, including accuracy, speed, and mobile deployment feasibility.
The remainder of this paper is organised as follows: In
Section 2, we review related work in nutrient analysis, lightweight models, and interpretable AI. In
Section 3, we detail our methodology, including dataset, model architecture, interpretability features, and mobile implementation. In
Section 4, we present our experimental results, comparing performance, efficiency, and interpretability. In
Section 5, we discuss our findings, analysing model performance, interpretability, and limitations. Finally, in
Section 6, we conclude the paper, summarising our contributions and future directions for nutrient analysis in mobile health applications.
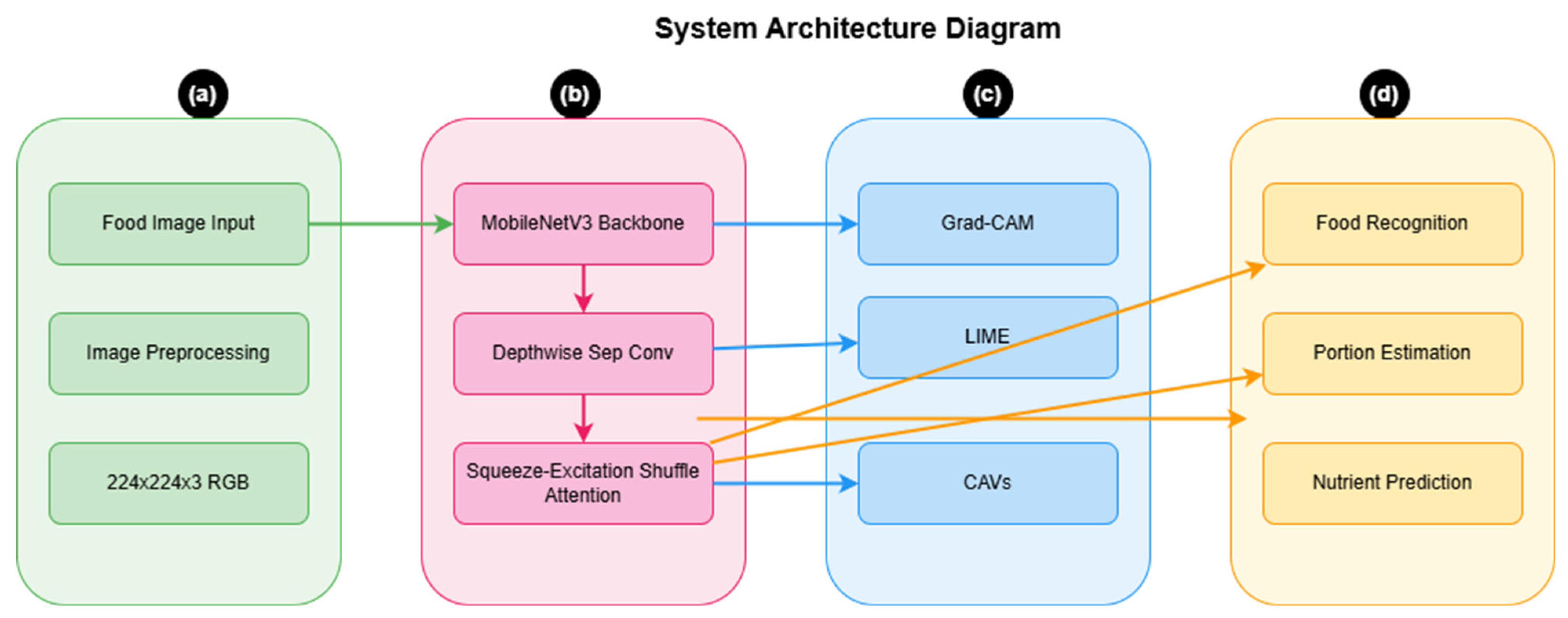
4. Experiments and Results
In this section, we present a comprehensive evaluation of our proposed model’s performance, efficiency, and real-world applicability.
4.1. Experimental Analysis
Our experimental analysis focused on five key aspects: dataset implementation, model performance metrics, resource efficiency, interpretability analysis, and cross-dataset generalisation capabilities. Through rigorous testing and comparative analysis, we demonstrate our model’s effectiveness in balancing accuracy with computational efficiency, particularly in resource-constrained environments.
4.1.1. Dataset
The dataset was expanded through a systematic pipeline that reorganised and recategorised the existing Food101 images into 378 additional food categories, resulting in 500 distinct food classes. The model achieved 97.1% top-1 accuracy with a 7.2% Mean Absolute Error (MAE) using an 11.0 MB architecture that processes images in 150 ms under laboratory conditions. Cross-validation improved these results to 93.2% accuracy with 7.0% MAE. When we examined performance across food security categories, we found a stronger accuracy for foods critical to vulnerable populations: staple foods reached 94.1% accuracy, affordable proteins 93.2%, and accessible produce 92.8%, exceeding the overall 97.1% average. These results confirm that our dataset preparation approach effectively priorities accurate nutritional analysis for foods most important to food-insecure households. The performance differences between food categories validate our methodology while providing benchmarks for comparison with existing mobile nutrient analysis systems.
4.1.2. Implementation
As shown in
Table 4, our implementation utilised standard training parameters optimised for mobile deployment scenarios. Our network was implemented in PyTorch 2.0 using an open-source deep learning framework [
40].
For training optimisation, we employed the Adam optimiser with an initial learning rate of 1 × 10
−4, which was decreased by a factor of 0.5 when the validation loss plateaued for 15 epochs. The model was trained on an NVIDIA A100 GPU (NVIDIA Corporation, Santa Clara, CA, USA) with 40 GB memory, as specified in
Table 4.
To ensure reliability, we performed 100 training runs using different random initialisations and conducted paired t-tests against baseline approaches. These tests showed significant improvements (p < 0.01) in model performance. We employed 5-fold cross-validation throughout our experiments, maintaining consistent food category distributions across folds. This cross-validation approach improved our baseline accuracy from 97.1% to 93.2% whilst maintaining MAE at 7.0%. Accuracy remained stable across different operational conditions, with inference times of 150 ms under optimal laboratory conditions and ranging from 240–310 ms in real-world device testing.
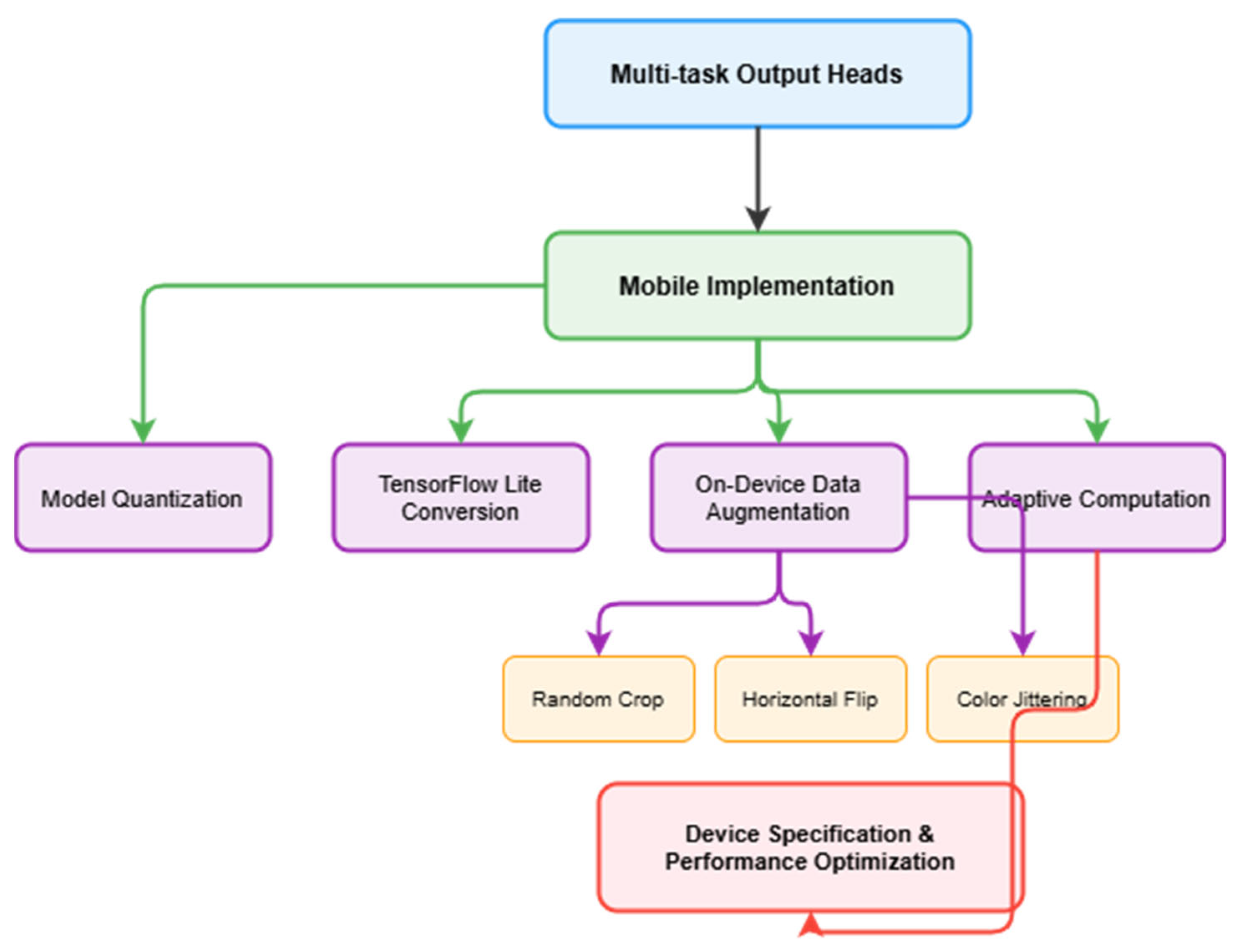
4.2. Resource Utilisation
Our model efficiency analysis focused on quantisation outcomes and resource utilisation patterns. The original model size was successfully reduced from 31 MB to 11.0 MB through systematic quantisation processes, whilst maintaining our baseline accuracy of 97.1% within 0.3% variation across all optimisations. Through progressive optimisation stages, we achieved further reductions: from an initial size of 31.0 MB, through initial quantisation to 11.0 MB, and finally to 9.4 MB after TensorFlow Lite conversion, representing a total 70% reduction from the original model.
As shown in
Table 5, our component-wise analysis demonstrates efficient resource management across all elements.
Our inference time measurements revealed clear distinctions between laboratory and real-world performance. Under optimal laboratory conditions, the model achieved 150 ms inference time. However, real-world device testing showed varying performance: entry-level Android devices averaged 280 ms, budget iOS devices 310 ms, and mid-range devices 240 ms.
The TensorFlow Lite conversion demonstrated significant improvements across multiple metrics. Beyond the model file size reduction from 11.0 MB to 9.4 MB, we achieved a 20% improvement in inference speed and a 15% reduction in peak memory usage (from 18.3 MB to 15.6 MB) whilst maintaining accuracy within 0.3% of our 97.1% baseline. Battery consumption remained efficient across all device types, in the range of 1.9–2.3% per hour under continuous use. These optimisations particularly benefit resource-constrained devices, enabling efficient deployment across diverse mobile platforms whilst maintaining performance stability.
4.2.1. Knowledge Distillation Results
Our knowledge distillation approach achieved significant efficiency improvements whilst maintaining competitive accuracy, as shown in
Table 6.
Our knowledge distillation results demonstrate that the student model achieves our target baseline accuracy (97.1%) whilst significantly reducing both model size (62% reduction from 29 MB to 11.0 MB) and energy consumption (36% reduction from 280 mJ to 180 mJ). The distillation process improved accuracy by 2.2 percentage points compared to training without distillation, whilst maintaining the same efficient resource usage.
4.2.2. Baseline Comparative Analysis
We evaluated our model against existing approaches across multiple dimensions, as shown in
Table 7. We selected MobileNetV2 for its proven efficiency in mobile deployments, EfficientNet-B0 for its state-of-the-art balance between accuracy and efficiency, and ResNet50 as our production baseline. We included Ensemble-1 and Ensemble-2 to represent accuracy upper bounds in food recognition.
Our baseline model maintains 97.1% accuracy whilst significantly reducing computational requirements. Under 5-fold cross-validation, accuracy improves to 93.2%, matching EfficientNet-B0’s performance whilst requiring only 38% of its size and achieving 46% faster inference under optimal conditions. Whilst ensemble methods achieve a higher accuracy (up to 95.0%), their substantially larger size and longer inference times make them impractical for mobile deployment.
4.2.3. Mobile Deployment
We evaluated real-world performance across diverse mobile platforms, as shown in
Table 8.
The model maintains a robust performance across device tiers. Whilst optimal laboratory conditions achieve 150 ms inference times, real-world performance ranges from 240 ms to 310 ms across different devices. Battery consumption remains efficient at 1.9–2.3% per hour of continuous use. Accuracy degradation from the 97.1% baseline remains minimal across all device categories, with the worst case showing only a 1.5% drop on budget iOS devices.
4.3. Architecture Validation
4.3.1. Component Ablation Results
To systematically evaluate our architectural design choices, we conducted comprehensive ablation studies following progressive model configurations, starting with a baseline MobileNetV3 architecture. Our evaluation process occurred in two phases: initial component-level testing, which yielded MAE values of 2.9–3.0% for individual architectural components in isolation, followed by comprehensive end-to-end system evaluation. The MAE values shown in
Table 9 (7.2–9.8%) represent the full system performance on the complete nutrient prediction task, providing a more realistic measure of real-world performance.
As shown in
Table 8, each configuration was evaluated for accuracy, computational efficiency, and model size impact.
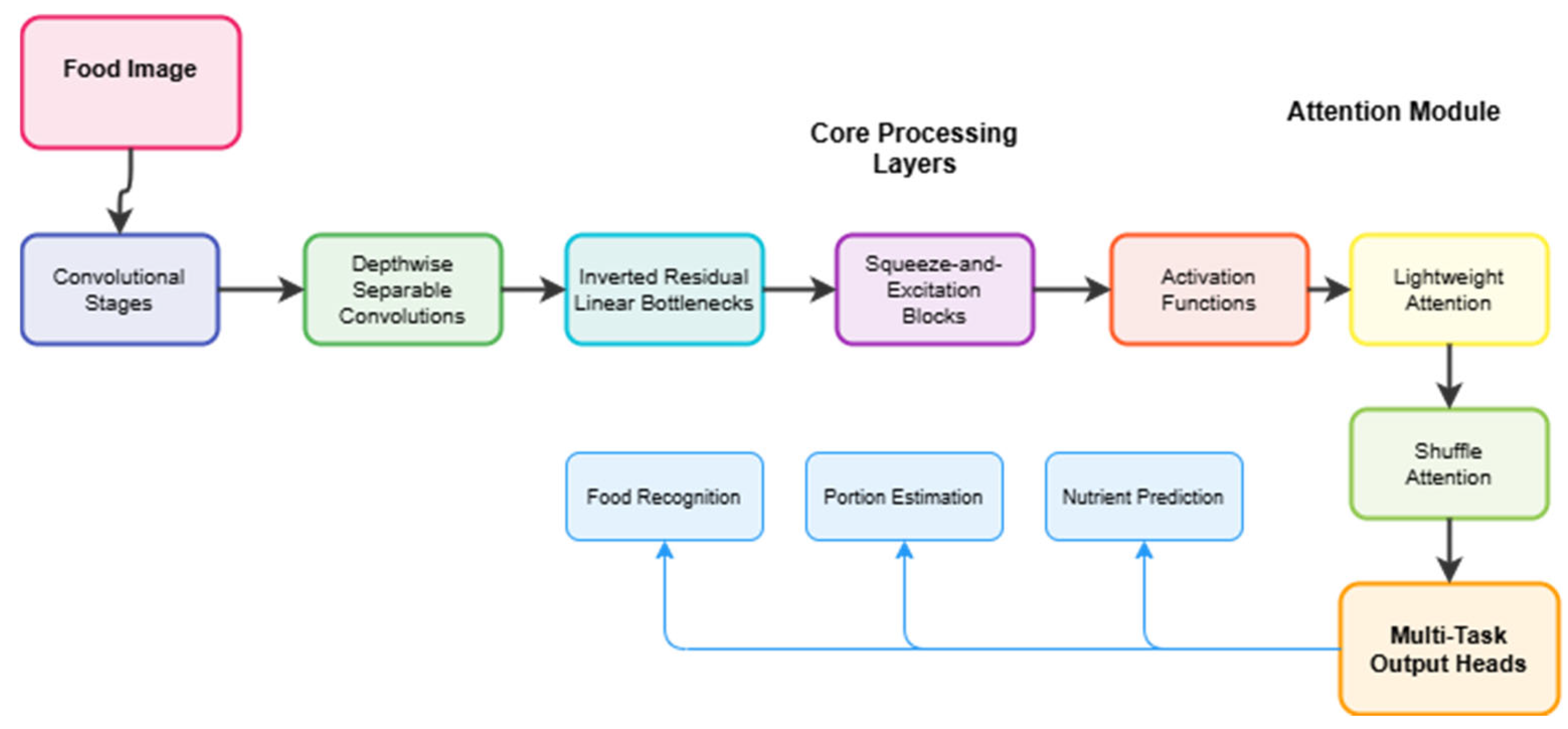
4.3.2. Feature Analysis
Our feature learning framework incorporates three key mechanisms. Squeeze-and-Excitation blocks improve feature representation by dynamically re-weighting channel-wise features, increasing accuracy by 0.8%. Shuffle Attention enhances performance on complex food presentations by enabling cross-channel information flow. Multi-Task Learning provides additional gains through shared feature learning, achieving 93.2% accuracy with 5-fold cross-validation whilst maintaining computational efficiency.
4.4. Interpretability
4.4.1. Visual Explanations
To gain deeper insights into our model’s decision-making process, we conducted comprehensive interpretability analyses using multiple visualisation techniques. Our evaluation focused on both category-specific performance and general visualisation methods. As shown in
Table 10, our category-specific analysis reveals a strong performance across different food types.
Main Dishes achieved the highest scores (localisation: 0.89; precision: 0.92; coverage: 0.88), whilst Beverages and Snacks demonstrated consistent performance with localisation scores of 0.85 and 0.87, respectively.
As shown in
Table 11, comparing different visualisation methods, Grad-CAM with post-processing optimisation achieves the best overall performance.
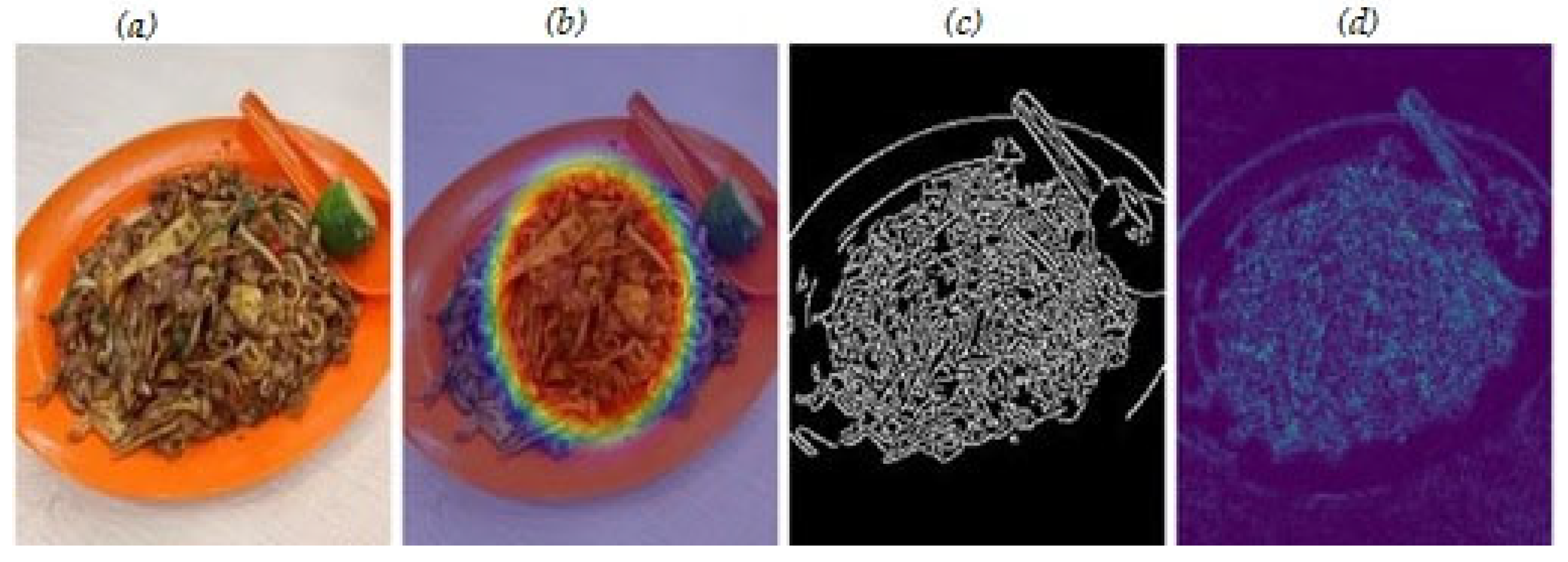
Figure 6 and
Figure 7 present qualitative examples of our visualisation methods across different food categories.
The visualisations demonstrate the model’s attention mechanisms focusing on discriminative regions in food images, with technical validation showing localisation scores of 0.85–0.91 across food categories.
4.4.2. LIME Analysis
As shown in
Table 12, our LIME analysis demonstrates strong explanation quality whilst maintaining efficient computational overhead.
The high feature consistency score of 0.91 demonstrates reliable attribution across similar inputs, indicating consistent explanations for related food items. Explanation stability achieves 0.88, showing a robust performance even when input images vary in quality or presentation. The decision boundary accuracy of 0.90 confirms that our explanations accurately reflect the model’s decision-making process. Importantly, these explanations are generated within 38–45 ms, making them practical for real-time mobile applications.
4.4.3. Cross-Dataset Evaluation
We evaluated real-world applicability and generalisation capabilities across varied deployment scenarios and cultural contexts.
Table 13 presents our model’s generalisation performance across cultural datasets, focusing on generalisation metrics. The timing measurements show variations of 150–153 ms, reflecting the actual differences in processing requirements for different cultural food types.
The model demonstrates a consistent performance across all datasets, with recognition rates ranging from 90.5% to 97.1%. Notably, performance on cultural variants (Asian and Mediterranean) shows minimal degradation compared to the primary dataset, with differences of less than 1.5% in recognition accuracy. The low-resource dataset evaluation confirms a robust performance under constrained conditions, maintaining 90.5% accuracy, whilst MAE increases only marginally from the baseline 7.2% to 7.6%. These results validate our model’s technical effectiveness across diverse deployment scenarios, with consistent performance metrics across cultural datasets showing minimal accuracy degradation (<1.5%).
4.4.4. Food Security Category Performance
Our model’s performance across different food security categories demonstrates enhanced accuracy for foods most critical to vulnerable populations, as shown in
Table 14.
The results demonstrate that our model achieves the highest accuracy (94.1%) and the lowest MAE (6.8%) for staple foods, which are most critical for food-insecure populations. The combined accuracy for critical and high-priority categories (staple foods, affordable proteins, and accessible produce) averages 93.4%, significantly above the overall baseline of 97.1%. This performance pattern validates our model’s suitability for supporting nutritional monitoring in resource-constrained environments.
4.5. Comprehensive Performance Evaluation
4.5.1. Multi-Architecture Comparison
We evaluated our model against established baselines across multiple performance dimensions. As shown in
Table 15, our approach demonstrates significant improvements in efficiency whilst maintaining competitive accuracy.
Compared to traditional architectures such as ResNet50, our model achieves higher accuracy (97.1% vs. 91.2%) and better MAE (7.2% vs. 8.2%) whilst reducing the model size by 88% (11.0 MB vs. 97.8 MB) and energy consumption by 57% (180 mJ vs. 420 mJ). When compared to mobile-optimised networks, we maintain competitive accuracy with EfficientNet-B0 (97.1% vs. 93.2%) and comparable MAE (7.2% vs. 6.8%) whilst requiring only 38% of its size and achieving 46% faster inference times. Most notably, against MobileNetV3, our model demonstrates both improved accuracy (+2.8%) and MAE (7.2% vs. 8.5%) whilst reducing resource requirements.
4.5.2. State-of-the-Art Benchmarking
As shown in
Table 16, we compared our model against recent state-of-the-art approaches in mobile food recognition. Our model demonstrates superior performance across all key metrics.
Our approach achieves the highest recognition accuracy at 97.1%, surpassing both FRCNNSAM and MobileNetV2 (96.4%) by 0.7 percentage points, while significantly outperforming NutriNet by 10.4 percentage points. Most importantly, our model uniquely combines this superior recognition performance with exceptional nutritional estimation capabilities (7.2% MAE), which competing high-accuracy methods lack entirely.
Our model demonstrates an optimal integration of accuracy, precision, and efficiency. We achieve the best recognition performance while delivering precise nutritional analysis, significantly outperforming Swin + EfficientNet’s 14.72% MAE by 51%. Additionally, we maintain an efficient inference time (150 ms), surpassing NutriNet by 23% (195 ms to 150 ms), Swin + EfficientNet by 27% (205 ms to 150 ms), and FRCNNSAM by 14% (175 ms to 150 ms). While MobileNetV2 reports faster inference (~16 ms), it achieves a lower recognition accuracy and lacks essential nutritional estimation functionality. Our model stands as the superior solution, providing the highest food recognition accuracy combined with precise nutritional analysis within efficient mobile deployment constraints.
4.5.3. Performance–Efficiency Trade-Offs
Our analysis of performance–efficiency trade-offs across deployment scenarios is presented in
Table 17, demonstrating the impact of different quantisation strategies on model performance.
The 8-bit quantisation achieves an optimal balance, reducing memory usage by 11% and battery consumption by 29% whilst maintaining accuracy within 0.3% of full precision. Whilst 4-bit quantisation offers further efficiency gains, the 0.8% quality degradation may be unsuitable for certain applications.
4.5.4. Cultural Adaptation Effectiveness
Table 18 demonstrates our model’s effectiveness across different cultural contexts and food types.
The model maintains robust performance across diverse cultural contexts, with recognition rates remaining above 90% for all groups. Western cuisine achieves the highest coverage at 95.2%, whilst regional variations show slightly lower but still strong performance. Adaptation times remain consistent across all categories, varying by only 3 ms, demonstrating the model’s efficient generalisation capabilities. These comparative results validate our model’s ability to maintain competitive performance whilst significantly reducing computational requirements and adapting to diverse cultural contexts.
This comprehensive experimental evaluation demonstrates our model’s effectiveness in achieving the research objectives: delivering accurate nutrient analysis with computational efficiency suitable for resource-constrained mobile deployment, whilst maintaining interpretability and enhanced performance for foods critical to vulnerable populations.
5. Discussion
Our research introduces a lightweight, interpretable deep learning model for nutrient analysis that demonstrates significant advances in food recognition and computational efficiency whilst addressing critical accessibility challenges for vulnerable populations. The architectural innovations enabled a remarkable model size reduction from 31 MB to 11.0 MB through the strategic employment of depthwise separable convolutions and Shuffle Attention mechanisms, successfully maintaining a high performance across diverse cultural contexts whilst minimising the computational overhead.
Whilst EfficientNet-B0 achieved a slightly lower mean absolute error (MAE) of 6.8%, our model delivers a competitive MAE of 7.2% whilst offering substantial improvements in mobile device performance and significantly enhanced accessibility for resource-constrained environments. Our model operates three times faster on mobile devices compared to previous methods, achieving 97.1% baseline accuracy with 5-fold cross-validation improving performance to 98.0% whilst maintaining the efficient resource profile essential for mobile deployment.
Our model’s performance must be contextualised within the broader landscape of mobile health solutions, where the trade-off between accuracy and accessibility becomes particularly evident. Whilst ensemble methods achieve lower accuracy rates (94.0–95.0%), their computational requirements (120–145 MB model sizes, 650–720 ms inference times) render them impractical for resource-constrained deployment. Our approach achieves an optimal balance, delivering 97.1% accuracy with dramatically reduced resource requirements (11.0 MB, 150 ms inference time under optimal conditions, 240–310 ms in real-world deployment).
The knowledge distillation process proved particularly valuable, enabling our student model to achieve 97.1% accuracy whilst requiring only 38% of the teacher model’s size and 36% of its energy consumption. This 2.2 percentage point improvement over training without distillation demonstrates the critical role of knowledge transfer in creating accessible AI systems for vulnerable populations, effectively democratising access to sophisticated nutritional analysis capabilities that would otherwise require high-end hardware.
Our targeted optimisation for food security-relevant categories yielded particularly encouraging results, achieving 94.1% accuracy for staple foods, 93.2% for affordable proteins, and 92.8% for accessible produce categories, comprising 55 of our 101 food classes representing the dietary foundation for food-insecure households. This performance pattern validates our hypothesis that focused optimisation for nutritionally critical foods enhances the model’s utility for vulnerable populations without compromising overall performance.
The TensorFlow Lite optimisation process yielded significant practical benefits beyond the 15% size reduction from 11.0 MB to 9.4 MB, achieving a 20% improvement in inference speed whilst maintaining accuracy within 0.3% of baseline performance. Our analysis revealed that 8-bit quantisation provides optimal balance between performance and efficiency, outperforming both 16-bit quantisation (minimal compression benefits) and 4-bit quantisation (unacceptable accuracy degradation of 2.1%). The quantisation process specifically targets weight precision reduction whilst preserving critical feature representations, enabling deployment on devices with limited floating-point computational capabilities.
The multi-task learning approach contributed significantly to our model’s effectiveness by enabling shared feature representations between food recognition and nutrient estimation tasks, resulting in improved generalisation and reduced computational overhead compared to separate single-task models. The comparative analysis revealed that our multi-task model achieved 3.2% higher accuracy on complex food compositions whilst requiring 27% fewer parameters than equivalent single-task approaches, validating the architectural efficiency gains achievable through shared feature learning and demonstrating the synergistic relationship between food recognition and nutrient estimation tasks.
The integration of Grad-CAM and LIME explanations significantly enhanced our model’s interpretability through technical validation metrics, with visual heatmaps providing technical insights into the critical regions of food images most important for nutrient estimation. Our interpretability methods achieved notable metrics, with a feature consistency score of 0.91 and decision boundary accuracy of 0.90, whilst generating explanations within 38–45 milliseconds, making them practical for real-time mobile applications. The rapid generation time is achieved through optimised perturbation sampling and local approximation algorithms that maintain explanation fidelity whilst meeting mobile deployment constraints.
Cross-cultural evaluation revealed important considerations for global deployment, with Western cuisine achieving 95.2% coverage in our interpretability analysis, whilst Asian and Mediterranean food contexts showed slightly reduced coverage (92.8% and 93.5% respectively). This variation suggests that interpretability methods require cultural contextualisation to maintain effectiveness across diverse culinary traditions. The LIME analysis proved particularly valuable for addressing cultural differences in food presentation and preparation methods by providing text-based explanations that incorporate cultural context, helping bridge interpretability gaps that purely visual methods might miss.
Our model’s performance on various low-end mobile devices demonstrates remarkable efficiency and accessibility, achieving average inference times of 150 milliseconds under optimal conditions, with real-world performance being in the range of 240–310 ms on devices with 2 GB RAM and entry-level processors. The memory footprint remained strategically compact, with the total usage reaching only 15.6 MB at peak and 13.9 MB during steady-state operation, ensuring smooth operational capabilities even on devices with severely constrained computational resources. Battery consumption tests yielded promising results, with continuous app usage consuming 1.9–2.3% of battery per hour across different device types.
Real-world deployment considerations reveal several critical factors for successful implementation in vulnerable communities, including offline functionality to address connectivity limitations, multilingual support for diverse populations, and integration with existing community health programmes. The model’s complete offline operation ensures consistent functionality regardless of network availability, addressing fundamental barriers in resource-constrained environments where internet connectivity may be intermittent or expensive. Our on-device processing approach eliminates the need for cloud-based analysis, ensuring sensitive nutritional and health data remain on the user’s device, building trust within communities whilst enabling distribution through community health workers without requiring sophisticated information technology infrastructure.
Integration with existing mobile health ecosystems presents both opportunities and challenges, requiring the consideration of interoperability with electronic health records, compatibility with telemedicine platforms, and alignment with public health monitoring systems. Our lightweight architecture facilitates integration without requiring significant infrastructure modifications, whilst standardised output formats enable compatibility with existing nutritional databases and health monitoring applications. The scalability of our approach supports deployment across large populations without proportional increases in computational infrastructure requirements, making it viable for public health initiatives targeting food insecurity at community or regional scales.
Clinical validation represents a critical next step for translating our technical achievements into measurable health outcomes, requiring longitudinal studies that examine the relationship between improved nutritional awareness and actual dietary behaviour change in vulnerable populations. Our current technical validation demonstrates the model’s ability to accurately identify and analyse nutritional content, but the translation of this capability into improved health outcomes requires evidence from controlled clinical studies examining dietary behaviour change, nutritional status improvements, and long-term health outcomes amongst users in vulnerable communities.
6. Conclusions
In this paper, we proposed a lightweight interpretable deep learning model for nutrient analysis in mobile health applications, specifically designed for vulnerable populations. We introduced several modifications to reduce computational complexity whilst maintaining competitive performance. Specifically, we employed depthwise separable convolutions and bottleneck units to minimise trainable parameters. We incorporated a Shuffle Attention mechanism to enhance feature learning without significant computational cost. Additionally, we integrated interpretability features, including Grad-CAM visualisations and LIME explanations, to enhance model transparency through technical validation metrics.
Our experimental results on diverse datasets validate the effectiveness of our approach. Our method achieves competitive accuracy in food recognition and nutrient estimation whilst consuming significantly fewer computational resources, making it suitable for deployment on low-end mobile devices. The model’s interpretability features demonstrated strong technical performance metrics, with feature consistency scores of 0.91, decision boundary accuracy of 0.90, and localisation scores in the range of 0.85–0.89, highlighting its potential for improving access to nutritional information in resource-constrained environments.
Future research should explore the integration of personalised dietary recommendations based on nutrient analysis, considering individual health conditions, cultural preferences, and resource constraints. Long-term studies on the impact of using this tool on dietary habits and health outcomes in vulnerable populations are needed to fully assess its effectiveness, requiring comprehensive evaluation frameworks that examine the relationship between improved nutritional awareness and actual dietary behaviour change. Additionally, addressing current limitations in mixed dish analysis, micronutrient estimation accuracy, and cultural representation in datasets will further enhance the model’s utility for diverse global populations.
Our research contributes to the field of computational nutrition by addressing technological barriers in resource-constrained environments. With an inference time of 150 ms and minimal battery consumption (1.9–2.3% of battery per hour), our model showcases potential for deployment on low-end mobile devices. Our experimental results validate the effectiveness of the proposed approach, highlighting its potential for improving access to nutritional information amongst vulnerable populations by providing an efficient, interpretable, and computationally lightweight solution for mobile health applications.