Abstract
In the race against COVID-19 for timely therapeutic developments, mass spectrometry-based high-throughput methods have been valuable. COVID-19 manifests an extremely diverse spectrum of phenotypes from asymptomatic to life-threatening, drastic elevations in immune response or cytokine storm, multiple organ failure and death. These observations warrant a detailed understanding of associated molecular mechanisms to develop therapies. In this direction, high-throughput methods that generate large datasets focusing on changes in protein interactions, lipid metabolism, transcription, and epigenetic regulation of gene expression are extremely beneficial sources of information. Hence, mass spectrometry-based methods have been employed in several studies to detect changes in interactions among host proteins, and between host and viral proteins in COVID-19 patients. The methods have also been used to characterize host and viral proteins, and analyze lipid metabolism in COVID-19 patients. Information obtained using the above methods are complemented by high-throughput analysis of transcriptomic and epigenomic changes associated with COVID-19, coupled with next-generation sequencing. Hence, this review discusses the most recent studies focusing on the methods described above. The results establish the importance of mass spectrometry-based studies towards understanding the infection process, immune imbalance, disease mechanism, and indicate the potential of the methods’ therapeutic developments and biomarker screening against COVID-19 and future outbreaks.
1. Introduction
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has resulted in over 4.6 million deaths worldwide in a span of less than 2 years. COVID-19 is caused by a coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronaviruses are a family of enveloped viruses with a large single-stranded positive-sense RNA genome. Although several human coronaviruses have been linked to the common cold, more notorious human coronaviruses have emerged. According to the National Institute of Allergy and Infectious Diseases, much higher case fatality rates have been observed with these coronaviruses, namely SARS-associated coronavirus (SARS-CoV; case fatality rate ~ 10%) and Middle East respiratory syndrome coronavirus (MERS-CoV; case fatality rate ~ 34%).
Despite higher case fatality rates, coronaviruses such as SARS-CoV and MERS-CoV are self-limiting and, therefore, have been well contained. SARS-CoV-2 is a novel coronavirus with a much lower case fatality rate than SARS-CoV or MERS-CoV. Yet, SARS-CoV-2 has proven harder to contain. The difficulty in containing SARS-CoV-2 has underscored the need for techniques such as mass spectrometry in the diagnosis and treatment of COVID-19 [1,2,3].
1.1. Mass Spectrometry
Mass spectrometry (MS) is a technique used to analyze the composition of biological samples. Using a mass spectrometer, the molecular masses of ions can be determined through the evaluation of an ion’s mass-to-charge ratio (m/z). Common methodologies in large molecule liquid chromatography-mass spectrometry (LC-MS) include collision-induced dissociation (CID). Collision induced dissociation (CID) leads to the breakage of bonds for precursor ion fragmentation [4]. Other fragmentation methods include higher-energy C-trap dissociation (HCD) and electron-transfer dissociation (ETD) [5]. Subsequent tandem mass spectrometry (MS-MS) generates large datasets that facilitate elucidation of structure for proteins (proteomics) and lipids (lipidomics).
Different types of mass spectrometry method are available which include data-dependent and data-independent, label and label-free methods. Based on their features, methods are chosen to detect membrane-bound proteins and soluble proteins. Conventional data-dependent acquisition (DDA) methods involve the digestion of a proteomic sample into peptides, followed by ionization and mass spectrometry analysis [6]. In a full-scan mass spectrum, peptide signals which appear greater than the noise are selected for fragmentation leading to the production of tandem (MS/MS) mass spectra [6]. The results are subjected to matching with spectra in a database. MS assays are used for high-sensitivity detection of peptides with high accuracy in targeted proteomics approaches like selected reaction monitoring (SRM) [6]. Data-independent acquisition (DIA) utilizes the advantages of DDA and SRM, where all peptides in a specified mass-to-charge (m/z) window undergo fragmentation. Repetition of the analysis is performed with the mass spectrometer exploring the full range of m/z window [6]. Hence, peptides are quantified accurately bypassing the limitations associated with profiling only peptides that were previously determined to be of interest [6].
MS-based isobaric labeling is used for quantitative proteomics where chemical groups which are isobaric, i.e., identical masses, are used to label peptides or proteins. The tags differ in terms of heavy isotopes distribution around their structure and are called tandem mass tags. In MS-based label-free protein quantification, relative changes in two or more biological samples are identified and quantified, instead of labeling proteins using a stable isotope-containing compound. This method is good for high-throughput clinical- and biomarker-related studies because it does not limit sample numbers or types. Various non-labeled techniques are used in combination with statistical analysis for identification of differential expression and absolute quantification of proteins [7].
1.2. Mass Spectrometry and Proteomics
Proteomic MS studies often utilize two approaches: bottom-up and top-down methods. These methods differ in both the size of the protein that is analyzed by the mass spectrometer and the complexity of the sample that can be analyzed. In bottom-up methods, protein samples are digested into fragments prior to mass spectrometric analysis. In top-down methods, complete proteins are fragmented as the sample is nebulized within a mass spectrometer. While LC-MS/MS bottom-up methods are more appropriate for complex biological samples (e.g., blood or tissue homogenates), the top-down approach is often adequate for purified proteins [8].
MS proteomics has identified specific interactions between proteins in the SARS-CoV-2 and human proteomes. Gordon et al. (2020) utilized affinity purification mass spectrometry (AP-MS) to identify 332 protein interactions between proteins in the SARS-CoV-2 and human proteomes [1]. As described by the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI), AP-MS is a technique specifically used for identifying interactions between proteins. It works by coupling affinity purification, a technique which segregates molecules based upon binding ability, with mass spectrometry. The interactions between the human and SARS-CoV-2 proteins were identified after observing human cells exhibiting 26 SARS-CoV-2 proteins. While the success of this study was achieved without the use of a lung tissue derived cell line, as lung tissue is the main location of viral infection, the increased presence of the examined “prey proteins” in lung tissue proves the efficacy of the experiment [1].
MS proteomics has also identified case-severity related proteins and protein interactions. Notably, LC-MS/MS proteomics has identified specific proteins and protein–protein interactions linked to mild and severe COVID-19 cases. Upregulation of heparin cofactor II and downregulation of cubilin are associated with COVID-19 severity [9]. Differential regulation of 27 proteins discovered through LC-MS/MS was likewise linked to COVID-19 severity [10]. Another study discovered peptides specific to SARS-CoV-2 that may be of potential diagnostic relevance [11]. The assessment of protein–protein interactions can also be accomplished by in situ cross-linking and mass spectrometry, which has been recently used to structurally probe Nsp1, Nsp2, and nucleocapsid (N) proteins of SARS-CoV-2 [12]. The method includes integrative modeling to computationally combine cross-linking data with domain structures for determination of full-length atomic models. The study was able to generate a single consistent all-atom model of the full-length Nsp2 whose cross-links reported a complex topology with long-range interactions. The model presents three putative metal binding sites indicative of Nsp2’s role in zinc regulation within the replication-transcription complex [12]. Multiple intra- and interdomain cross-links were detected for the N protein. Hence, the technique provides significant stereochemical and electrostatic information on proteins, and their structural probing to highlight their importance in the cellular context [12].
1.3. Mass Spectrometry and Lipidomics
MS is also a powerful technique for the interrogation of lipid metabolism and membrane biophysics. Lipidomic implications of COVID-19 are less understood than changes to the proteome or transcriptome. Lipidomic profiles of COVID-19 patients could prove useful for disease diagnosis and treatment. Altered circulating lipoprotein levels are associated with COVID-19 pathophysiology [13]. In a comparative analysis of mild, severe, and critical COVID-19 patient status, serum levels of LDL-cholesterol (LDL-C) and total cholesterol (TC) progressively declined from mild to critical [13]. Despite some limitations in this study as mentioned by the authors, the observations suggests a diminished hepatic function with greater severity of respiratory distress. Diminished hepatic lipoprotein production and mobilization is likely downstream of organ failure induced by lower respiratory capacity (and thus bulk oxygen transport) in COVID-19 patients.
Another important element of lipid metabolic response to COVID-19 is intracellular signaling through long chain polyunsaturated fatty acid oxygenation. Twenty carbon and 22 carbon fatty acids (eicosanoic and docosanoic acids) are commonly linked to glycerol and sphingosine phospholipids in the plasma-membrane fraction of mammalian cells, including those of COVID-19 infected lung and serum leukocytes and lymphocytes [14]. Phospholipase A2 and fatty acid hydrolases can cleave these lipids to release free fatty acids, whereupon intracellular acyl-CoA synthetase re-esterifies them as coenzyme A-linked thioesters. C20/C22-CoA esters are substrates for cyclooxygenases, lipoxygenases, and P450 oxygenases to produce eicosanoids and docosanoids (local stress hormones often during active COVID-19 infection) [15].
In a study examining the bronchoalveolar lavage (BAL) specimens from COVID-19 patients, cyclooxygenase (COX) generated eicosanoids (e.g., vasoconstrictive thromboxanes and leukotrienes) were elevated above healthy controls [16]. Lipoxygenase (LOX) products (e.g., hydroxy and oxy-eicosa- and docosa (C22) tri- and tetra-, penta- and hexaenoic acids) were also elevated in these patients [16].
COVID-19 infection associated pro-inflammatory cytokines like IL-1β, are known to increase expression of COX2, which synthesizes TXA2 from arachidonic acid (20:4 ω-6) [17]. TXA2 promotes bronchoconstriction while acting as an activator of both leucocytes and platelets. Other COX products such as the prostaglandins can activate lymphocytes. Both leucocytes and lymphocytes are integral in the hyper-inflammatory respiratory distress syndrome (RDS) implicated in COVID-19 [18,19].
Membrane raft components (e.g., ceramide and cholesterol) are also likely critical for SARS-CoV-2 intracellular entry. While ceramide promoted uptake of SARS-CoV-2 virion into epithelial cells, sphingosine-1-phosphate (S1P) receptor activation inhibited entry [20]. Neutral and acid sphingomyelinase regulate distribution of voltage gated channels, glucose and lipid uptake [21,22] and may, therefore, also regulate SARS-CoV-2 spike protein binding and proteolytic processing. Multiple biochemical pathways produce ceramide, with S1P and ceramide interconversion potentially altering the balance of viral transmission. Taken together, these findings suggest that the COVID-19 lipidome is a biomedically relevant target that deserves further inquiry.
1.4. Mass Spectrometry in Disease Research
With the improvement of MS-based proteomics, over 5000 new plasma protein biomarkers have been discovered [23]. In the context of COVID-19, proteomic studies revealed COVID-19 pathology [24,25,26,27,28] and associated characteristics of plasma protein [29], serum [30], and prognostic markers [31]. MS-based lipidomics has shed significant insights into biomarkers, inflammation, infection, and biological profiles associated with COVID-19 [32,33,34,35,36,37,38], gut microbiome alterations [39], and drug response [40].
MS-based studies on proteomics and lipidomics complement transcriptomic and epigenomic studies well. This is because transcriptomics and epigenomics analyses can reveal further upstream biological processes, and help to connect them with resulting proteomics and lipidomics profiles. Hence, there is a growing popularity of all the above high-throughput multiomics methods involving next-generation sequencing for a well-rounded understanding of COVID-19. MS-based methods have been employed in several clinical studies worldwide, some of which are summarized in Table 1, and have also been indicated to be a superior method for COVID-19 screening [41,42]. This review will, therefore, discuss both mass spectrometry and next generation sequencing-based approaches to interrogate COVID-19.

Table 1.
Some clinical trials against coronavirus disease 2019 (COVID-19) using mass spectrometry.
2. Proteomics and Mass Spectrometry in Disease Research
Proteomics is a discipline that investigates the full complement of proteins expressed by a cell, tissue, or organism. Proteins play a significant role in cellular function, structure, and regulation. Therefore, the ability to identify the location and function of proteins is an integral component of biological sciences, especially when determining the differential expression and turnover of polypeptides in an active disease state such as an infection by COVID-19.
Although proteomics is a relatively recent discipline, with its own methodology, proteomics has quickly become a major biochemical technique. When attempting to analyze biological networks or patterns, proteomics is often the most applicable discipline, when compared to disciplines such as genomics or transcriptomics, because proteins are a final product of gene expression.
Proteomics has proven to be a principal method in diagnosing and treating disease through the identification of the proteins present in a healthy sample compared to a diseased sample [44]. The discovery of these proteins, known as biological markers or biomarkers, has been especially important to the improvement of biomedical sciences.
While current proteomic workflows can differ, one of the most efficient workflows includes the use of MS-based approaches. The generic structure of a proteomic workflow using MS involves the simplification of the sample through fractionation or depletion, digestion of the sample, purification of the sample, and finally MS analysis [23,45]. The development of MS-based proteomics has greatly improved the rate at which protein biomarkers are discovered, enabling a much more efficient system for disease diagnosis and treatment.
The discovery of the first disease-related protein biomarkers provided a foundation for modern day biomarker research. One of the earliest protein biomarkers to be discovered was C-reactive protein, which was identified in 1930 and has implications in inflammatory diseases [46]. Because modern technologies were not readily available while early biomarker discoveries were being made, the comprehensive list of protein biomarkers grew at a slow rate for many years.
Even as recently as 2010, approximately only two protein biomarkers were being discovered annually due to the limitations of the methods used to analyze protein samples. These methods included the more often used immunoassay-based approaches, which were limited by specificity and multiplexing, and less advanced MS-based proteomics approaches, which were limited by the large number of proteins within samples. In recent years, the limitations that hindered MS-based proteomics have been addressed.
Proteomic studies have since identified numerous disease-specific protein biomarkers. Many of these disease-related protein biomarkers exhibit differential expression. One disease-specific protein biomarker that exhibits differential expression is pancreatic cancer-related apolipoprotein E, for which increased expression has been observed in pancreatic cancer patients [47]. In addition to apolipoprotein E, raised levels of lipoprotein (a) in plasma have been implicated in atherosclerosis [48].
Several other disease-specific protein biomarkers exhibit dysregulation. A disease-specific protein biomarker that exhibits dysregulation is galectin-3, for which dysregulation has been connected to diseases of the kidney, diseases of the heart, and cancers [49]. Another disease-specific protein biomarker that exhibits dysregulation is Mucin 1, for which upregulation has been observed in breast cancer samples [50].
4. Lipidomics in Disease Research
Much like proteomics, lipidomics has become a central feature for disease diagnostics and progression including therapeutic recovery. Mass spectrometry coupled with gas and liquid chromatography have become essential for determining lipid profiles in healthy vs. diseased states. To understand the plenum of lipids in biochemistry, it is necessary to provide a primer on nomenclature and biosynthesis.
Viral infection and the host immune response are directed via multiple lipid signaling circuits including those associated with obesity, fatty acid oxygenation, phospholipase induction and membrane lipid raft dynamics. Therefore, the mass-spectral investigation of these parameters is necessary to apprehend their role in COVID-19 transmission and severity [66,67,68,69].
4.1. General Features of Fatty acid Metabolism and Nomenclature
Polyunsaturated fatty acids (PUFAs) are synthesized from two sources: dietary and endogenous de novo fatty acid synthesis (FAS). PUFAs can be characterized by the number, geometry and position of double bonds. These double bonds are introduced into preformed saturated fatty acids (long-chain aliphatic monocarboxylic acids) most commonly of C16, C18, C20, and C22 chain length. Some of these double bonds can be introduced prior to consumption and two in particular of these polyunsaturated fatty acids are essential in the diet. These are linoleic acid (18:2 Δ9, 12) and α linolenic acid (18:3 Δ9, 12, 15). The position of the double bonds as counted from the CH3 terminus of the fatty acid provides the omega (ω, last C atom) classification system. Hence linoleic acid is an ω-6 fatty acid and α-linolenic acid is an ω-3.
The position of these double bonds is biologically significant as this chemical structure is responsible for differential function subsequent to elongation and further desaturation of these essential fatty acids to arachidonic acid (AA: 20:4 Δ5, 8, 11, 14), eicosapentaenoic acid (EPA: 20:5 Δ 5, 8, 11, 14, 17) and docosahexaenoic acid 22:6 Δ(DHA: Δ4, 7, 10, 13, 16, 19). There are also intermediates in these pathways Some dietary and endogenously derived saturated, monoenoic and polyenoic acids of different omega series classification are also produced. There are also geometric isomers such as the naturally occurring ω-7, trans fatty acid found in sphingolipids called palmitoleic acid (16:1 (t) Δ7).
Once the fatty acids have been fully elongated and desaturated, they become substrates for complex lipid synthesis involving sphingosine and glycerol biochemical pathways involving CoA thioester metabolism. Fatty acids are rarely free in the cell except in severely dyslipidaemic patients. In this case, serum albumin can non-covalently transport free fatty acids throughout the body where they may confer lipotoxicity due in part to their ability to disrupt membrane physiology [70]. The study of lipids causing bioenergetic, signaling and membrane structural damage has been extensively studied using MS and much emphasis has been placed on saturated fatty acids such as palmitate (16:0) and stearate (18:0) as major contributors to microsomal and mitochondrial events in lipotoxicity [71]. Free fatty acid-induced lipotoxicity is typically a component of illnesses associated with the obesity epidemic [72,73].
To what extent free fatty acid-induced lipotoxicity plays a role in COVID-19 pathophysiology has not yet been fully characterized. However, MS studies involving viral protein palmitoylation and ER-associated lipotoxicity have been implicated in COVID-19 disease progression and transmittance although the roles of TAG and membrane lipid turnover require a more complete lipid metabolic analysis. Viral protein acylation plays a role in transmission and this includes several viral protein products in all tissues where the virus is replicating [74].
The complex lipid-associated omega-3 and omega-6 PUFA’s will be removed from their oxygen or amide linked ester bonding in response to cellular stress including infection and inflammation only to be re-esterified to CoA as a thioester, where they may be oxygenated by three types of enzyme that reside in multiple subcellular compartments. These include cyclooxygenases, lipoxygenases and P450 oxygenases thus producing eicosanoids and docosanoids which act as potent local hormones-autocoids and paracoids. These oxygenation pathways are common in lung epithelial cells and contribute directly to the immune responses including the trafficking of neutrophils, the activation of macrophages plus activation of T-helper cell acuity to APC’s [75].
These oxygenated fatty acids are then linked to regulating the guanylate cyclase mediated synthesis of cGMP which is a component of the control molecular switch for K+ and Ca++ conductance in muscle-including the myocardia. In severe pathological states, some of these eicosanoids (especially those derived from AA) may result in vasoconstriction which can cause systemic hypertension and with that decrease in blood flow; this may cause ischemic heart attack or MI. Cardiomyocytes, lung and cardiovascular endothelial cells plus circulating lymphocytes are implicated in the pathogenesis of viral transmission and immune response [76].
4.2. Inflammatory Response Linked to Fatty Acid Oxygenation
In general, local eicosanoids serve to initialize the stress preconditions that lead to an immune response and the potential for inflammation. This has been well described in both the innate (macrophage, neutrophil, dendritic cells, NK cells) and acquired (T and B cell lineages [77,78].
Tandem mass spectrometric analyses of C20 eicosanoids and C22 docosanoids have been implicated in the inflammatory rise observed in lung epithelia during active COVID-19 infection [16]. This evidence confirms decades long biomedical research establishing oxygenated fatty acid association with endothelial membrane-linked signaling (via phospholipase removal of VLCPUFA’s and subsequent in situ oxygenation) This response leads to a pro-inflammatory induction via innate immune cell signaling [79]. These oxygenated fatty acids thus serve as an initiator of the inflammatory response by pathogens including COVID-19.
Once oxygenated fatty acid metabolism is initiated, a cascade of events may occur that involve the neuroendocrine system and a stimulation of the corticosteroid axis in the CNS along with the induction of transcription, translation, glycosylation and secretion of pro-inflammatory cytokine, chemokines and growth factors which all serve to further the potential for a system -wide hyperimmune response. This response includes vasoconstriction, diapedesis of leucocytes and lymphocytes, and a cascade of REDOX mediated ROS production that can lead to cellular degeneration via apoptosis, necroptosis and ferroptosis in local tissue beds such as that associated with the pulmonary system. These activities occur in various endothelial and epithelial cell lineages especially those in the infected lung [77].
5. Lipidomics and Mass Spectrometry in COVID-19
5.1. The Immunomodulatory Axis of COVID-19 Disease Burden
During an active COVID-19 infection with lung presentation, a burst of hyperinflammation involving the production of pro-inflammatory secreted glycoproteins (cytokines) exacerbates the immune reaction and leads to loss of cellular integrity leading to a profound enhancement in immune-mediated responses. This cytokine eruption in the infected lung epithelia and endothelia is the product of cellular membrane VLCPUFAs transformation to eicosanoids which promote the migration and activation of macrophages and granulocytes including basophils and neutrophils [80]. High levels of the growth factor, macrophage-stimulating colony factor (MSCF) will promote myeloid differentiation to macrophages along with costimulatory IL-3 and IL-6. The granulocyte lineage are differentiated from the myeloid precursor population via GSCF along with the same pro-inflammatory cytokines [81].
Type 1 macrophages (M1) are pro-inflammatory and induce virally infected cellular death while type 2 macrophages (M2) inhibit this process while secondarily functioning to remove cellular debris via the phagocytic functions of this subgroup. PPARα transcriptionally regulates the production of fatty acid beta oxidation genes. M1 pro-inflammatory macrophages work through the TLR 2 receptor to promote glycolysis over fatty acid β-oxidation and thus remain polarized as potent pro-inflammatory macrophages [82].
When the transcription factor, PPAR α is expressed and shuttled to the nucleus, this results in the polarization to the M2 macrophages which produce anti-inflammatory cytokines. Underlying this transcription pattern is a transition toward fatty acid vs. glucose oxidation for bioenergetics. Fatty acid oxidation is necessary for M2 polarization and thus decreases the pro-inflammatory status encountered in diabetic and obese patients. This is a significant clinical consideration as these patients are high risk COVID-19 patients [82].
When apoptosis is induced in infected cells, macrophages will clear the cellular debris and little signaling will occur thus avoiding a massive recruitment of pro-inflammatory cells and mediators. However, if the programmed cell death is directed toward necroptosis of ferroptosis, the cellular debris is released from the dying cell and this will trigger a neutrophil -granulocyte response that will enhance the production of pro-inflammatory cytokines such as IL1β, IL6 and TNFα plus the pro-trafficking chemokine, MCP1. COVID-19 replication and advancing infection will thus enhance the endoplasmic reticulum (ER) stress response which involves the induction of COX, LOX and the epoxide synthases and hydrolases which form multiple lineages of AA based eicosnaoids.
While PGE 2 (a COX product) enhances the proinflammatoiry response, epoxide fatty acids from the soluble epoxide hydrolase will shut down further inflammation. The ER stress response specifically activates the COX lineage enzymes microsomal prostaglandin E synthase-1 and prostaglandin-endoperoxide synthase 2 [82]. The products of this pathway include a constellation of pro-inflammatory eicosanoids including prostaglandins, thromboxanes and the peptido-leukotrienes.
Conversely, the P450 epoxide synthases generate epoxyeicosatrienoic acids which are anti-inflammatory and vaso-dilating but they can be metabolized by the soluble epoxide hydrolase. Thus, there is feed-forward and feedback regulation of the immediate proximal eicosanoid autocoid response regulating a balance between pro and anti-inflammatory eicosanoid biosynthesis. These enzymatic modifications are found in pulmonary vascular beds especially those associated with active infection and viral replication [83].
Once eicosatrienoic fatty acids are produced they can also be shuttled into the production of a class of eicosanoids known as resolvins. These eicosnoids mediate the resolution of the inflammatory response which started with the granulocyte induced necroptosis or ferroptosis cellular destruction that launched the hyperinflammation initially [84].
The induction of eicosanoid biosynthesis during active infection or comorbid respiratory disease is under neuroendocrine control. Specifically, corticosteroids perform a suppressive function during cellular and vascular inflammation in the lung and cardiac muscle [85].
COVID-19 infection can occur during pregnancy so the immediate concern is related to the use of pharmacotherapy for pregnant women and their unborn child. There are American Obstetrics and Gynecology Association (AOGA) recommendations for this infection that have included the use of immune-suppressive corticosteroids either in lieu of or with a currently available vaccine. Corticosteroids are cholesterol-derived lipid mediators that are synthesized via the hypothalamo–pituitary–adrenocortical (HPA) axis [86].
Since endogenous corticosteroids suppress the respiratory disease inflammatory response (at least in part) through the down-regulation of lung eicosanoid metabolism via interferon mediated responses, the COVID-19 pandemic and its symptomatic association with ARDS has been evaluated. During the SARS and MERS viral outbreaks, which were far more virulent and, therefore, self contained, the timing of lung epithelia and endothelia corticosteroid suppression was highly correlated to the valence of disease pathophysiology [87].
COVID-19 transmission and infection kinetics appear to map onto a biphasic progression wherein there is an asymptomatic phase that may end the disease progression or, if virion titre is sufficient and co-morbidities obtain, there can be a second phase where frank respiratory disease presents with a sizable increase in cytokine and chemokine expression and secretion followed by a hyperimmune response [88].
Depending on the progression curve of the disease presentation, the inappropriate introduction of pharmacotherapeutic corticosteroids to COVID-19 patients, prior to massive cytokine and chemokine expression, can actually increase disease severity and enhance the possibility of poor prognosis and the ultimate outcome axis [86]. It has been shown that classical epithelial immune initiation starts with the production of pro-inflammatory mediators including the eicosanoids, LTB4 and PG2 which enhance inflammation until the “eicosanoid switch”, which marks the end of initiation; this is when the relative cellular concentrations of PGE2 plus PGD2 are equal to LTB4 [52].
At this point, resolution is apprehended by the production of anti-inflammatory eicosanoids, the resolvins and PGF2a. Pathogenic induction of endocrine stress hormones such as norepinephrine trigger subsequent cortisol resistance such that the “eicosanoid switch” is invoked when the molar concentration of norepinephrine balances out the concentrations of glucose-sensing serum insulin plus that of cortisol, after which, corticosteroid efficacy is restored. When norepinephrine levels drop to the non-stimulated nadir, steady-state corticosteroid tonicity is restored [52]. Therefore, the alteration of cortisol efficacy with the inflammatory signaling axis recommends that the pharmacological introduction of corticosteroids during an active infection, of COVID-19, could corrupt this balance leading to a hyper-inflammatory response and poor disease outcome.
Given previous evidence on IFN responses in SARS, a delayed corticosteroid therapy would clearly be indicated as more beneficial. The study examined above describes sequential change in eicosanoids, catecholamines and cortocsteroids as emblematic of these in vivo interactions during disease progression. During the early stages of infection, the induction of the stress response involves the expression and activation of COX and LOX activities which cause an increase in AA derived pro-inflammatory eicosanoids including PGE2 and LTB4. The stress hormones produced by the HPA and other neuroendocrine axes are also intimately involved in the immune response.
Subsequent to a pathogen-associated infection, the norepinephrine (NE) systemic stress response links both obesity and type II diabetes induced insulin and cortisol resistance to an alteration in eicosanoid molecular species accumulation thus ending the initial phase of the stress response. This is represented by the change in the ratio of COX activity linked PGE2 plus PGD2 concentration resting at the same levels of LOX generated LTB4 in lung tissue [89].
During this initiation phase of disease progression (e.g., COVID-19), the serum ratio of NE/cortisol goes from exceedingly high to roughly 1:1 at the same time LTB levels increase hyperbolically while PGE2 increases in a curvilinear pattern ultimately reaching that ratio of PGE2 + PGD2/LTB4 = 1. This constitutes the peak of the proinflammatory stage and it is followed by a steady decline in inflammation thus entering the resolution phase of the stress response curve. This inflammatory phase transition maps on to the COVID-19 disease presentation in the interstitia of the lung of infected patients [90].
Once the resolution phase of the systemic inflammatory response is invoked, the production of anti-inflammatory eicosanoids such as the resolvin D and E series which are derived from ω-3 VLCPUFAs are produced. The resolvins represent the antagonism between ω-3 and ω-6 fatty acid endothelial autocoid agency [91].
Resolvins abrogate the inflammatory response and limit its potential for causing massive tissue damage that can result in cellular degeneration, hyperinflammation and collapse of the endothelial support of lung capacity. Resolvins thus resolve the inflammatory response by promoting neutrophil apoptosis while promoting the anti-inflammatory type 2 macrophage lineage which serves to clean up cellular debris and thus close the infection court from further inflammatory signaling [92]. The resolvin, 14S,21R-dihydroxy docosahexaenoic acid (14S,21R-diHDHA) acts as an autocrine to stimulate the M2 mediated repair of vascular endothelia [93,94].
The corpus of this research effort was entirely supported by host endothelial and serum associated membrane fractions from circulating leucocytes and lymphocytes analyzed via LC-MS-MS [95]. This method is the only reliable and precise means for the determination of a multitude of lipid molecular species including positional and geometrical isomeric structures from biological specimens where the molar concentration can reach the very limits of accurate detection and quantification.
5.2. Focus on the Innate Immune Response
Much of what we have been focusing on with regard to COVID-19 lipidomics has been associated with the innate immune response. While lymphocytic based immunity including the Th1 responses and the cytotoxic T lymphocytes from the CD8+ lineage are critical for a robust acquired-memory based assault against COVID-19, B cells and antibody producing Plasma cells are also of critical importance depending on the progression of the disease. These lymphocytes play the essential role in generating natural multivalent long term immunity when memory cells become paramount [96,97]. Vaccines are also used to target these responses [98,99].
This review will continue to focus on the plenum of innate immune responses for two reasons:
- 1.
- extensive literature exists on omics of the innate response;
- 2.
- the emphasis on reducing transmission necessarily places the innate immune response to the front of clinical and diagnostic agenda.
Toll-like receptors (TLRs) play a major role with the innate immune response. Recently it has been demonstrated that TLR2 and MYD88 may be a key component of the inflammatory response linked to COVID-19 infection [100].
There are two functionally different major classes of pattern-recognition receptors (PRRs): endocytic pattern-recognition receptors and signaling pattern-recognition receptors. Endocytic PRRs are found on the surface of phagocytes and promote the attachment of microorganisms to them, leading to their subsequent engulfment and destruction. These PRRs include:
- 1.
- Mannose receptors on the surface of phagocytes bind mannose-rich glycans, the short carbohydrate chains with the sugar mannose or fructose as the terminal sugar that are commonly found in microbial glycoproteins and glycolipids but are rare in those of humans. Human glycoproteins and glycolipids typically have terminal N-acetylglucosamine and sialic acid groups. C-type lectins found on the surface of phagocytes are mannose receptors
- 2.
- Scavenger receptors found on the surface of phagocytic cells bind to bacterial cell wall components such as LPS, peptidoglycan and lipoteichoic acids. There are also scavenger receptors for certain components of other types of microorganisms, as well as for stressed, infected, or injured cells. Scavenger receptors include CD-36, CD-68, and SRB-1.
- 3.
- Opsonin receptors. Opsonins are soluble molecules produced as a part of the body’s immune defenses that bind microbes to phagocytes. One portion of the opsonin binds to a pathogen-associated pattern receptor (PAMP) on the microbial surface and another portion binds to a specific receptor on the phagocytic cell.
5.3. Mass Spectrometry-Based Studies on Spike Protein Helps in Vaccine Development
Techniques based on mass spectrometry have been critical for understanding the glycosylation of SARS-CoV-2 spike protein [27,101,102,103,104,105,106]. Glycosylation of the spike protein of coronavirus and other viral proteins are crucial to the infection process because the modification shields immunogenic epitopes, and impact immunological pressure across the protein surface [107,108]. In this regard, the entire signature of carbohydrates in a cell or organism is called glycome, which is under immense focus in SARS-CoV-2 research [109,110,111,112]. The studies have contributed towards the design and development of some of the vaccines against COVID-19 [113,114,115,116].
5.4. Signaling Pattern-Recognition Receptors (PRRs) Are Found on Multiple Host-Cell Surfaces
A series of signaling pattern-recognition receptors known as toll-like receptors (TLRs) are found on the surface of a variety of defense cells and other cells. These TLRs play a major role in the induction of innate immunity and contribute to the induction of adaptive immunity.
The binding of a microbial PAMP to its intracellular immune or epithelial cell TLR (or other PRR) transmits a signal to the host cell’s nucleus inducing the expression of genes coding for the synthesis of intracellular regulatory molecules including cytokines [117]. The cytokines, in turn, bind to cytokine receptors on other defense cells. Unique combinations of TLRs appear in different cell types and may occur in pairs. Different TLRs directly or indirectly bind different microbial surface molecules. These signaling PRRs, which are found in the membranes of the endosomes (phagolysosomes) are used to enhance the intracellular degredatioin of the pathogens.
It has been established that TLR-3 binds to the replicative (sub-genomic) double-stranded viral RNA, while TLR-7—binds uracil-rich single-stranded viral RNA.TLR-8—also binds single-stranded viral RNA and TLR-9—binds unmethylated cytosine-guanine dinucleotide sequences (CpG DNA) found in either bacterial and viral genomes. Most of the TLRs that bind to viral components trigger the synthesis of cytokines called interferons that block viral replication within infected host cells. TLR’s work through a set of transcription factors and adaptor molecules including Myd88 [118].
It has been reported that dysregulated type I interferon and inflammatory monocyte-macrophages (IMM) are directly linked to the lethal pneumonia obtained in SARS-CoV-infected mice. While SARS is far more virulent that COVID-19, it is a virus with very comparable aetiology as there is a spectrum of respiratory-illness related coronaviruses, some of which can cause an acute lethal disease which presents with innate immune associated lung inflammation [119].
Coronavirus genomic replication is controlled by the delayed type I interferon (IFN-I) signaling pathway which initiates the inflammatory phase of the infection ultimately associated with severe immunopathology, morbidity and death. In this study, which predates the COVID-19 pandemic, evidence show that endogenous lung associated IFN-I is measurable even until viral titers peak. However, early pharmacotherapeutic administration of IFN-I is associated with a blockade of lung immunopathology. This strongly supports a role for IFN-I promoting the observed enhanced accumulation of disease severity-linked inflammatory monocyte-macrophages (IMMs). The appearance and activity of IMMs results in an increase in pro-inflammatory cytokine and chemokine accumulation coupled with endo-vascular damage and a nadir of T lymphocyte immune-based responses that would otherwise control disease severity. Indeed, knockout of the IFN-αβ receptor (IFNAR) or the removal of IMM’s abrogates lethality but has no immediate effect on viral titre [119].
To demonstrate the link between the IFN responses and associated macrophage hyperinflammation and coronavirus carriage, it was learned that CNS-resident microglia and astrocytes also produce these type 1 interferons (T1 IFNs) signaling through the heterodimeric IFN-α/β receptor (IFNAR). Binding of T1 IFNs with the receptor stimulates JAK/STAT-signaling leading to the transcription of IFN-stimulated genes (ISGs) which mediate either pro- or anti-inflammatory sequelae depending on down-stream signaling including lipid metabolism. Fatty acids and complex membrane and signaling lipids play major roles in bioenergetics and cell fate (e.g., apoptosis, senescence, or autophagy [120].
This interferon system also impacts sphingolipid metabolic pathways expressed in various cellular environments including those of lung epithelia and both leucocytes and lymphocyte lineages contributing to viral respiratory lung pathology and disease severity [20].
Indeed, the T1 IFN response in the CNS may arise from viral infections and traumatic brain injury (TBI) and peripheral or neurodegeneration will activate this response systemically. Therefore, T1 IFNs can be protective or deleterious as in multiple sclerosis (MS), where T1 IFNs are thought to exert and perform an anti-inflammatory response via production of anti-inflammatory cytokine IL10 with simultaneous suppression of pro-inflammatory cytokine IL-1. IFN-β pharmacotherapy is common in MS patients since it has been shown to inhibit lymphocyte infiltration into the brain where this event is associated with a decline in relapse rate. Paradoxically, overexpression of the IFN-α gene has been observed in the murine CNS with links to neuroinflammation and neurodegeneration. Obviously IFN subtype, concentration, titration and cellular residency are all critical to this pathology [120].
Hence, it is important to emphasize that innate immunity as afforded by neutrophils, macrophages, dendritic cells and accompanying intracellular TLR’s and signaling through classical lipid pathways are the initial and most formidable means to thwart viral infection. Virions of appropriate host recognition epitope synapse formation will trigger IFN mediated gene expression via ISGs which are controlled via the IFN-regulatory factor (IRF) family and signal transducer and activator of transcription (STAT) family transcription factors. The canonical IRF and STAT signal transduction cascade involves post-translational covalent modifications, protein aggregate and subunit complex formation, leading to nuclear translocation of these nascent transcription factors [121].
5.5. Sphingolipids at the Center of COVID-19 Infection Dynamics
As shown in animal model muscle tissue, sphingomyelinase (SMase) -mediated ceramide accumulation will generate membrane microdomains. These membrane fragments will merge spontaneously into mobilized raft platforms that can aggregate protease (e.g., ACE2) receptors plus associated signaling lipids and proteins thus functioning to transduce a signal first generated via eicosanoid/pro-inflammatory cytokine response to viral transmission [122].
Several sphingosine-based bioactive lipids that are found within membranes or trafficking through them, have been implicated in normal and pathophysiological states as regulatory signaling molecules. Besides sphingomyelin (SM), the major lipid found in the myelin sheath of critical neuronal tracts in the CNS are the associated metabolites, ceramide, ceramide-1-phosphate (C1P), sphingosine, and sphingosine-1-phosphate (S1P), which are necessary bioactive sphingolipids in lung epithelia, various circulating leucocytes and lymphocytes during active infection [123].
The role of cerebrosides (galactosyl, glucosyl, sialyl and sulfonyl sphingolipids) are very well described in the biomedical literature as associated with inborn errors of metabolism, neurotransmission, immune responses and basic developmental biology. These cellular and extracellular-trafficking SM sphingolipids are interconvertible via enzymatic pathways that are not always expressed constitutively in any cell or tissue type, across time [124]. It has been shown that cerebroside turnover is associated with HIV infection so future MS-based omics research on COVID-19 needs to examine these complex lipids in infected individuals [125].
Sphingolipids and glycerol phosophlipids are implicated in various physiological phenomena including inflammation, stress resistance, proliferation, differentiation, and in the plastic and elastic remodeling of neuronal and glial cell fates in the CNS. The dynamic and potentiating sphingolipids are implicated in cell division, growth, apoptosis, autophagy, senescence, adhesion, migration, inflammation, angiogenesis, and endosomal plus organellar intracellular trafficking.
Opposing or reinforcing modes of action have been ascribed to individual sphingolipid classes and molecular specific subclasses; (e.g., CER as harbinger of PCD, while C1P and S1P sustain cell vitality and transformational viability). Mere addition or removal of ATP-derived anhydride phosphate as an ester to preformed polypeptides can reverse the direction of signaling without any change in transcription or translation of the participating molecules thus proving a molecular circuitry network to mediate cell fate. It is therefore not a surprise that bioactive sphingolipids will be involved in the initial and closing arguments toward viral pathogenesis, aging, and the morbidity of obesity.
Unlike biochemical reactions requiring the synthesis of loss of covalent, ionic and H-bonds, additional activation energy needed for specific annular-held proteins to translocate into specific membrane rafts is of a distinct character that is typically disassociated with transcription and translation. Thus, lipid signaling in health and disease has much more to do with biophysical phenomena such as hydrophobic interactions wherein the more solvation-associated chemistry of a fully mature and folded polypeptide 3-D state is the more likely it will diffuse into a lipid raft or membrane upon its synthesis when its chemical structure formation allows for a propensity toward a lower energy state.
Hence, lipid interactions are in the realm of a hydrophobic effect, where protein and lipid raft inclusion will titrate according to delocalized distributional processing, where there is a meta-stabilized lower energy potential. Besides these dominant hydrophobic interactions, there are processive activation energy potentials for non-homeostatic transition state free energy obtaining polypeptide inclusion. Whether a protein such as the COVID-19 receptor ACE 2 is included in a CER/cholesterol rich membrane raft requires Van der Walls limiting co-segregation of the protein/annular lipid during co-biosynthesis, processing and turnover.
Whether these processes are the result of non-specific chaperonin flux or unique monomer-polymer associations depending on membrane fluidity, cellular mixing and regular turnover of oxidized or oxygenated membrane lipids according to age or stress is largely unexplored. This research requires new techniques in mass spectrometry and solvent chemistry manipulation combined with in situ SMase and phospholipase bioorganic reaction kinetics and dynamics.
There is good reason to assume the protein transmembrane domains, typically composed of hydrophobic amino acid side chains, induce their insertion into the membrane via higher order lipid domain transformations from helix type II to bilayer transitions as coordinated by surface tension molecular regulation [126].
In a recent review of sphingolipids and SARS-CoV-2, it was established that lung cell ceramide facilitated the infection process, while sphingosine-1P inhibited it [20].
This implicates the sphingomyelinase biosynthetic route since the neutral SMAse isoform of the enzyme is located in the plasma membrane and upon activation would generate in situ CER from membrane sphingomyelin thus promoting this metabolic route for viral entry instead of de novo synthesis from palmitoyl CoA plus L-serine or the salvage routes through sphingosine phosphate or glycosphingolipid pathways.
However, acid SMase (aSMase) inhibitors such as amitriptyline and pharmacotherapeutics such as hydroxychloroquine that raise the pH of the phagolysosome (thus inhibiting aSMase) may suggest intracellular CER synthesis and thus membrane lipid raft assembly is also involved in the decrease of COVID-19 transmission and disease severity. Figure 1 provides a general outline for the proteomic/lipidomic axis in COVID-19 transmittance and pathology.
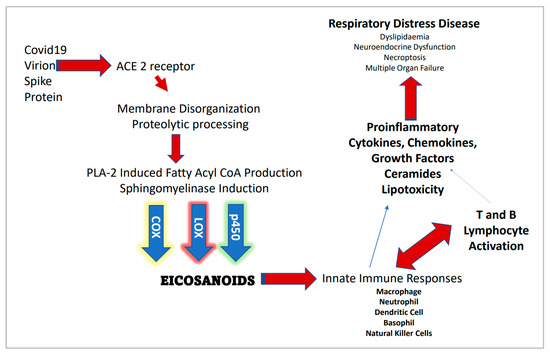
Figure 1.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and associated proteomic/lipidomic axis.
5.6. Contributions of Lipidomics in Understanding and Treating COVID-19
Overall, lipidomics studies reveal several important phenomenon associated with COVID-19 as outlined below. The viral replication and life-cycle is critically impacted by host lipids serving as double-membrane vesicles and other roles. Viral entry and propagation are regulated by lipid biogenesis [33]. Lipidomic analysis of COVID-19 patients in conjunction with quantification of proinflammatory cytokines and alarmins show a correlation between patient lipid profiles and factors like IL-26, TSLP (thymic stromal lymphopoietin) and adiponectin [33].
Lipidomic and metabolomic profiles showed common signatures despite heterogeneous clinical symptoms of COVID-19 patients, where treatment with the immunosuppressive drug tocilizumab partially reversed the metabolic changes caused by COVID-19 [40]. Dislipidemia, i.e., lipid imbalance with pathological consequences, is reported in a study which observed changes in levels of ceramides [35], increased triglycerides and decreased cholesteryl esters [34]. A study on COVID-19 acute phase patients reported severe dyslipidemia which impacts size and distribution of lipoprotein particles, dysregulation of central metabolism, accumulation of ketone bodies, and upregulation of succinate similar to the oncogenic pseudohypoxic environment [38].
Another study showed dyslipidemia in COVID-19 patients, and alterations in malic acid and carbamoyl phosphate leading to disruption in energy metabolism and hepatic dysfunction, respectively [32]. Patients who did not survive exhibited drastic downregulation of carbamoyl phosphate. Another biomolecule, guanosine monophosphate, also significantly varied between healthy controls versus COVID-19 patients, and between mild and fatal cases [32]. A study investigating oral and gut microbiome correlations with lipidomics of COVID-19 patients showed that in comparison to confirmed patients, 47 lipid molecules, including sphingomyelin and monoglyceride were depleted, and 122 lipid molecules, including phosphatidylcholine, phosphatidylethanolamine, and diglyceride, were enriched in confirmed patients recovery [39].
The contributions of lipidomics studies to COVID-19 research has been extensively reviewed in previous publications [36,37]. Collectively, the above lipidomics studies can potentially contribute to disease classification and course, monitor treatment outcomes, biomarker and therapeutic developments. Highlights of findings on COVID-19 from proteomics and lipodomics studies are outlined in Figure 2.

Figure 2.
Information obtained from proteomics and lipidomics analyses of COVID-19 patients. ↔ interactions between viral and host proteins; colored rectangles—elevated proteins and lipids.
6. Multiomics-Based Approaches to Understand COVID-19: Transcriptomics
Studies against COVID-19 have integrated mass spectrometry-based methods with other multiomics approaches. Multiomics methods have proven to be extremely beneficial in COVID-19 research ranging from discovery to clinical levels [127,128,129,130,131,132,133,134]. In addition to proteomics and lipidomics, other “omics” approaches that have been implemented in COVID-19 research are transcriptomics and epigenomics. Collectively, the high-throughput methods provide well-rounded understanding of molecular mechanisms associated with COVID-19.
6.1. Bulk and Single-Cell RNA-Sequencing
Transcriptomic profiling or RNA-sequencing (RNA-seq) helps to quantify and sequence all RNA transcripts [135]. Briefly, RNA is extracted from cells followed by reverse transcription to generate cDNA libraries which are sequenced and analyzed bioinformatically. RNA-seq can be performed on bulk or single cells. Bulk RNA-seq reveals the collective transcriptome of all kinds of cells in a sample, i.e., it reveals overall expression levels of each gene as a collective output from a given population of cells.
On the other hand, single-cell RNA-seq distinguishes between transcriptomes of individual cells in a sample, which requires that cells be bar-coded prior to sequencing. Processing single cells through a channel where individual cells are tagged enables a unique RNA library to be obtained from each cell. Raw library sequencing data (reads) undergo alignment and quantification against reference transcriptomes using bioinformatic software to enable transcript identification and quantitation. For human studies, the current human reference assembly is Genome Reference Consortium Human Build 38 (GRCh38).
6.2. RNA-Sequencing and COVID-19
To date, most COVID-19 transcriptomic studies are based on RNA-seq of peripheral blood mononuclear cells (PBMCs) obtained from COVID-19 patients with varying degrees of disease severity. A recent study by Xianwen Ren et al., employed a single-cell RNA-seq on 1.46 million cells using 284 samples from 196 COVID-19 cases and healthy controls [136]. The authors assessed PBMCs (obtained by standard density gradient centrifugation of whole blood), and lymphocytes from bronchoalveolar lavage fluid (BALF), and sputum from oropharyngeal swabs. In addition to transcriptomic alterations of the human host, the study also investigated viral RNA in the single cells. Sequencing reads were aligned against a customized reference genome where the SARS-CoV-2 genome (NC_045512, NCBI Refseq) was integrated as an additional chromosome. Parameters were set to screen for single cells having desired quantitation of viral RNA reads.
The large number of samples and multi-tissue comparison in this study provided an integrated analysis to understand how characteristics like age, gender, COVID-19 severity, and stage associate with molecular changes in immune cells, which may explain some of the clinical implications. Transcriptomic changes in precursor cells of B and T cells, and diversity of T and B cell receptor repertoires were found to be impacted by gender and age, while plasma B and proliferative T cells correlated with disease severity. The study discusses how potential cell–cell interactions are likely affected by the presence of SARS-CoV-2 mRNA in epithelial cells. Subsets of monocytes and megakaryocytes may be critical peripheral causes of cytokine storms due to interactions between hyper-inflammatory cell subtypes belonging in peripheral blood and pulmonary tissue. Accordingly, data from this study could inform targeted therapies against COVID-19 that specifically focus on infected cells.
Single-cell omics analysis was utilized by a large cohort study for combined analysis of transcriptome and surface proteome of PBMCs obtained from COVID-19 patients classified as asymptomatic, mild, moderate, severe and critical across different geographical locations [127]. One of the “omics” methods used in the study is cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), in which antibodies are labeled with oligonucleotides to integrate the analysis of transcriptome and cellular protein, in a format compatible with single-cell sequencing technology [137].
Among 1.1 million cells from 143 samples, ~800,000 cells satisfied quality control criteria. Cells were grouped into clusters using a computational tool called the Leiden algorithm (an improved method for grouping of related cells into communities) [138]. Cells were then subjected to manual annotation according to proteins expressed on their surfaces corresponding to RNA expression of known marker genes, with the help of flow cytometry. This process led to the classification of distinct subpopulations.
The study found relative expansion of proliferating immune cells like lymphocytes and monocytes, platelets, mobilized hematopoietic stem and progenitor cells (HSPCs) as COVID-19 severity worsened. Severe and critical COVID-19 showed expansion of plasmablasts and B cells. Disease severity was found to impact immune landscape alterations when cells were compartmentalized into phenotypic hyperspheres according to surface protein expression. A hypersphere is a four-dimensional analog of a sphere. Cell number of individual severity groups in the hyperspheres was quantified to reveal that 608 hyperspheres have significant differences in abundance with rising severity. Every major immune compartment contained hyperspheres of differential abundance. Elevation was observed for B cells, plasma cells, HSPCs, and myeloid compartment remodeling.
Ren et al. [136], and Stephenson et al. [127] employed transcriptomics-heavy pipeline, sorting cell populations via single-cell sequencing, and characterizing protein activity based on transcriptomic data [136], or with sequencer-compatible oligo-tags [127]. However, correlational studies, such as that conducted by Overmyer et al. [129] demonstrate how direct assays of patient proteomes can be analyzed alongside transcriptomic data to paint a more holistic picture of COVID-19 disease phenotypes. Researchers conducted high-throughput sequencing along with MS of 128 clinically validated patient samples, identified 17,000 unique biomolecules (either sequencer-identified transcripts or mass-spec-identified protein markers). Researchers then cross-analyzed the clinical data with the transcriptomics-and proteomics-characterized biomolecules to find 219 molecules that were correlated with COVID-19 disease phenotype [129]. Not only do broad-scope correlation studies illuminate new targets of study, but they also begin to provide clues about the cellular mechanisms of COVID infection, disease progression, and immune response. More clues are found the more clinical variables are combined with more samples for transcriptomic and proteomic studies employing high-throughput methods such as RNA-seq and MS.
Other examples which used RNA-seq is a study which implemented single-cell RNA-seq and metagenomic next generation sequencing of RNA from tracheal aspirate of COVID-19 patients [139]. Transcriptomic and proteomic methods have been implemented in understanding host responses during fatal cases of COVID-19 [140]. Studies have also focused on only bioinformatics analysis of available datasets of bulk and single-cell RNA-seq obtained from COVID-19 patients to understand how viral transcripts impact the pathogenesis of moderate and severe cases [141].
7. Epigenomics
A major application of omics-based methods is focused on the epigenome or chromatin landscape. Chromatin is composed of nucleosomes which are constituted by ~146 bp of DNA wrapped around an octamer of histone proteins for condense packaging inside nuclei of eukaryotic cells [142,143]. In this direction, popular omics-based methods are Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) [144] and chromatin immunoprecipitation followed by sequencing (ChIP-seq) [145]. ATAC-seq maps genome-wide regions of transcriptionally accessible chromatin versus closed/repressed chromatin. ChIP-seq maps genome-wide localizations of proteins of interest. COVID-19 studies have performed multi-omic profiling using a combination of single-cell ATAC-seq and RNA-seq [146,147] and the importance of epigenetic processes associated with COVID-19 has been established [148,149,150,151,152,153].
In one study, single-cell ATAC-seq was performed for epigenomic profiling of peripheral immune cells of 10 convalescent individuals at 4–12 weeks following recovery from moderate or severe COVID-19 [154]. Furthermore, T-cells were analyzed using a combinatorial method of single-cell T cell-receptor (TCR) sequencing (scTCR-seq), fluorescence-activated cell sorting (FACS) with index sorting and ATAC-seq, called TCR-FACS-index-ATAC sequencing (Ti-ATAC-seq). Data were obtained for cell-surface markers expression, paired TCR sequences and chromatin accessibility profiles at single cell resolution. Key revelations from this study include the identification and annotation of 13 subclusters of monocytes, a facilitated program for B-cell development, roles of effector CD8+ T cells in initial viral control, and formation of memory CD8+ T cells.
Epigenomic technology like ChIP-seq was employed in a study on COVID-19 using topotecan (TPT), an FDA-approved inhibitor of topoisomerase 1 (TOP1) [155]. ChIP-seq was performed to map the genome-wide localization of the epigenetic mark of H3K27ac which occupies regions of active transcription like promoters and enhancers. The results reveal a strong association between dynamic reorganization of genome compartmentalization during SARS-CoV-2 infection and transcriptional activity.
Overall, the study showed that lethal inflammation response resulting from SARS-CoV-2 infection is suppressed by TOP1 inhibition, and it implemented multidimensional epigenomic and transcriptomic analyses. The results indicate that TOP1 inhibitors likely can contribute to precise host-directed therapy to combat severe COVID-19. The study also implemented techniques like high-throughput chromosome conformation capture (Hi-C) and promoter capture Hi-C (PCHi-C). Hi-C is a technique that measures changes in chromatin architecture [156,157,158], while PCHi-C analyzes promoter interactions which was combined with Hi-C library generation [159,160].
Other factors that have previously been associated with epigenetic regulations of gene expression, disease, and recently implicated in COVID-19 are microRNA (miRNA) [161,162,163,164,165] and competitive endogenous RNA (ceRNA) [166,167,168,169]. miRNA has been associated with active COVID-19 infection with several molecular species upregulated (e.g., miR-31-5p) and down regulated [164]. However, the molecular targets for these miRNAs have not been extensively examined so it remains unclear if these are associated with specific clades of mRNA or, rather, function as an associated consortium of miRNAs as in the ceRNA mechanism where they function as a regulatory network to control the level of protein in the cell at the RNA molecular level [170].
8. Conclusions
Overall, the various studies on mass spectrometry-based approaches and multiomics technology have shown promise in providing significant new insights towards our understanding of COVID-19 (Figure 3). Collectively, the studies have provided molecular details for how COVID-19 impacts cellular content across stages of disease severity. Large datasets generated by the high-throughput multiomics methods contain a wealth of information that will surely inform therapeutic developments. With more research being focused in this direction, it is likely that the understanding and prediction of such outbreaks will improve, leading to better clinical outcomes in the future.

Figure 3.
Epigenomics and transcriptomics methods reveal molecular perturbations caused by SARS-CoV-2 infection. COVID-19 alters epigenomic and transcriptomic events in host cells which are detected by high-throughput technology involving next-generation sequencing (NGS) like RNA-seq, ChIP-seq, ATAC-seq, Hi-C, etc. Contributions of results include therapeutic developments and biomarker identification.
Author Contributions
Conceptualization, R.S., D.A. and D.J.G.; writing—original draft preparation, R.S., D.A., K.T., D.D.G., S.J. and D.J.G.; writing—review and editing, R.S., D.A., K.T., D.D.G., S.J. and D.J.G.; supervision, R.S. and D.J.G.; All authors have read and agreed to the published version of the manuscript.
Funding
This manuscript received no external funding.
Acknowledgments
We apologize to the authors who could not be cited due to space constraints.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, R.; Preianò, M.; Fregola, A.; Pelaia, C.; Montalcini, T.; Savino, R. Mapping the SARS-CoV-2-Host Protein-Protein Interactome by Affinity Purification Mass Spectrometry and Proximity-Dependent Biotin Labeling: A Rational and Straightforward Route to Discover Host-Directed Anti-SARS-CoV-2 Therapeutics. Int. J. Mol. Sci. 2021, 22, 532. [Google Scholar] [CrossRef]
- Rardin, M.J.; Schilling, B.; Cheng, L.-Y.; MacLean, B.X.; Sorensen, D.J.; Sahu, A.K.; MacCoss, M.J.; Vitek, O.; Gibson, B.W. MS1 Peptide Ion Intensity Chromatograms in MS2 (SWATH) Data Independent Acquisitions. Improving Post Acquisition Analysis of Proteomic Experiments. Mol. Cell. Proteom. 2015, 14, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Tolić, N.; Xie, F.; Zhao, R.; Purvine, S.O.; Schepmoes, A.A.; Ronald, J.M.; Anderson, G.A.; Smith, R.D. Effectiveness of CID, HCD, and ETD with FT MS/MS for Degradomic-Peptidomic Analysis: Comparison of Peptide Identification Methods. J. Proteome Res. 2011, 10, 3929–3943. [Google Scholar] [CrossRef]
- Doerr, A. DIA mass spectrometry. Nat. Methods 2014, 12, 35. [Google Scholar] [CrossRef]
- Patel, V.J.; Thalassinos, K.; Slade, S.E.; Connolly, J.B.; Crombie, A.; Murrell, J.C.; Scrivens, J.H. A Comparison of Labeling and Label-Free Mass Spectrometry-Based Proteomics Approaches. J. Proteome Res. 2009, 8, 3752–3759. [Google Scholar] [CrossRef]
- Girolamo, F.D.; Lante, I.; Muraca, M.; Putignani, L. The Role of Mass Spectrometry in the “Omics” Era. Curr. Org. Chem. 2013, 17, 2891–2905. [Google Scholar] [CrossRef]
- Bezstarosti, K.; Lamers, M.M.; Doff, W.A.; Wever, P.C.; Thai, K.T.; van Kampen, J.J.; Haagmans, B.L.; Demmers, J.A. Targeted proteomics as a tool to detect SARS-CoV-2 proteins in clinical specimens. bioRxiv 2021, 2020–2024. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef]
- Mahmud, I.; Garrett, T.J. Mass Spectrometry Techniques in Emerging Pathogens Studies: COVID-19 Perspectives. J. Am. Soc. Mass Spectrom. 2020, 31, 2013–2024. [Google Scholar] [CrossRef]
- Slavin, M.; Zamel, J.; Zohar, K.; Eliyahu, S.; Braitbard, M.; Brielle, E.; Baraz, L.; Stolovich-Rain, M.; Friedman, A.; Wolf, D.G.; et al. Targeted in situ cross-linking mass spectrometry and integrative modeling reveal the architectures of three proteins from SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2103554118. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zeng, W.; Su, J.; Wan, H.; Yu, X.; Cao, X.; Tan, W.; Wang, H. Hypolipidemia is associated with the severity of COVID-19. J. Clin. Lipidol. 2020, 14, 297–304. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef]
- Casari, I.; Manfredi, M.; Metharom, P.; Falasca, M. Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 2021, 82, 101092. [Google Scholar] [CrossRef] [PubMed]
- Archambault, A.; Zaid, Y.; Rakotoarivelo, V.; Turcotte, C.; Doré, É.; Dubuc, I.; Martin, C.; Flamand, O.; Amar, Y.; Cheikh, A.; et al. High levels of eicosanoids and docosanoids in the lungs of intubated COVID-19 patients. FASEB J. 2021, 35, e21666. [Google Scholar] [CrossRef]
- Conti, P.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Ronconi, G.; Toniato, E. IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: Inhibitory effect of the IL-1 receptor antagonist (IL-1Ra). J. Biol. Regul. Homeost. Agents 2020, 34, 1623–1627. [Google Scholar]
- Bos, L.D.J. COVID-19–related Acute Respiratory Distress Syndrome: Not So Atypical. Am. J. Respir. Crit. Care Med. 2020, 202, 622–624. [Google Scholar] [CrossRef]
- Navas-Blanco, J.R.; Dudaryk, R. Management of Respiratory Distress Syndrome due to COVID-19 infection. BMC Anesthesiol. 2020, 20, 177. [Google Scholar] [CrossRef] [PubMed]
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854. [Google Scholar] [CrossRef] [PubMed]
- Osawa, Y.; Seki, E.; Kodama, Y.; Suetsugu, A.; Miura, K.; Adachi, M.; Ito, H.; Shiratori, Y.; Banno, Y.; Olefsky, J.M.; et al. Acid sphingomyelinase regulates glucose and lipid metabolism in hepatocytes through AKT activation and AMP-activated protein kinase suppression. FASEB J. 2010, 25, 1133–1144. [Google Scholar] [CrossRef]
- De Lira, M.N.; Raman, S.J.; Schulze, A.; Schneider-Schaulies, S.; Avota, E. Neutral Sphingomyelinase-2 (NSM 2) Controls T Cell Metabolic Homeostasis and Reprogramming During Activation. Front. Mol. Biosci. 2020, 7, 217. [Google Scholar] [CrossRef]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef]
- Haas, P.; Muralidharan, M.; Krogan, N.J.; Kaake, R.M.; Hüttenhain, R. Proteomic Approaches to Study SARS-CoV-2 Biology and COVID-19 Pathology. J. Proteome Res. 2021, 20, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Lachén-Montes, M.; Corrales, F.J.; Fernández-Irigoyen, J.; Santamaría, E. Proteomics Insights Into the Molecular Basis of SARS-CoV-2 Infection: What We Can Learn From the Human Olfactory Axis. Front. Microbiol. 2020, 11, 2101. [Google Scholar] [CrossRef] [PubMed]
- McArdle, A.; Washington, K.E.; Orgel, B.C.; Binek, A.; Manalo, D.-M.; Rivas, A.; Ayres, M.; Pandey, R.; Phebus, C.; Raedschelders, K.; et al. Discovery Proteomics for COVID-19: Where We Are Now. J. Proteome Res. 2021, 20, 4627–4639. [Google Scholar] [CrossRef]
- Praissman, J.; Wells, L. Proteomics-Based Insights Into the SARS-CoV-2-Mediated COVID-19 Pandemic: A Review of the First Year of Research. Mol. Cell. Proteom. 2021, 20, 100103. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Armengaud, J. Proteomics in the COVID-19 Battlefield: First Semester Check-Up. Proteomics 2021, 21, e2000198. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, H.; Kim, S.Y.; Kim, Y.; Lee, J.-S.; Dan, K.; Seong, M.-W.; Han, D. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020, 10, 22418. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Suvarna, K.; Biswas, D.; Pai, M.G.J.; Acharjee, A.; Bankar, R.; Palanivel, V.; Salkar, A.; Verma, A.; Mukherjee, A.; Choudhury, M.; et al. Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity With Drug Repurposing Potential. Front. Physiol. 2021, 12, 652799. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Caterino, M.; Gelzo, M.; Sol, S.; Fedele, R.; Annunziata, A.; Calabrese, C.; Fiorentino, G.; D’Abbraccio, M.; Dell’Isola, C.; Fusco, F.M.; et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 2021, 11, 2941. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Huang, W.; Li, Y.; Lai, C.; Huang, S.; Wang, G.; He, Y.; Hu, L.; Chen, C. Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci. Rep. 2021, 41, BSR20204305. [Google Scholar] [CrossRef]
- Spick, M.; Longman, K.; Frampas, C.; Lewis, H.; Costa, C.; Walters, D.D.; Stewart, A.; Wilde, M.; Greener, D.; Evetts, G.; et al. Changes to the sebum lipidome upon COVID-19 infection observed via rapid sampling from the skin. EClinicalMedicine 2021, 33, 100786. [Google Scholar] [CrossRef] [PubMed]
- Mussap, M.; Fanos, V. Could metabolomics drive the fate of COVID-19 pandemic? A narrative review on lights and shadows. Clin. Chem. Lab. Med. 2021, 59. [Google Scholar] [CrossRef]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020, 21, 3544. [Google Scholar] [CrossRef]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; de Vicuña, A.G.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, H.; Cui, G.; Lu, H.; Wang, L.; Luo, H.; Chen, X.; Ren, H.; Sun, R.; Liu, W.; et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021, 70, 1253–1265. [Google Scholar] [CrossRef]
- Meoni, G.; Ghini, V.; Maggi, L.; Vignoli, A.; Mazzoni, A.; Salvati, L.; Capone, M.; Vanni, A.; Tenori, L.; Fontanari, P.; et al. Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 2021, 17, e1009243. [Google Scholar] [CrossRef]
- Rybicka, M.; Milosz, E.; Bielawski, K.P. Superiority of MALDI-TOF Mass Spectrometry over Real-Time PCR for SARS-CoV-2 RNA Detection. Viruses 2021, 13, 730. [Google Scholar] [CrossRef]
- Tran, N.K.; Howard, T.; Walsh, R.; Pepper, J.; Loegering, J.; Phinney, B.; Salemi, M.R.; Rashidi, H.H. Novel application of automated machine learning with MALDI-TOF-MS for rapid high-throughput screening of COVID-19: A proof of concept. Sci. Rep. 2021, 11, 8219. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, D.; Miotello, G.; Gallais, F.; Gaillard, J.C.; Debroas, S.; Bellanger, L.; Lavigne, J.P.; Sotto, A.; Grenga, L.; Pible, O.; et al. Proteotyping SARS-CoV-2 Virus from Nasopharyngeal Swabs: A Proof-of-Concept Focused on a 3 Min Mass Spectrometry Window. J. Proteome Res. 2020, 19, 4407–4416. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2016, 55, 182–196. [Google Scholar] [CrossRef]
- Darie, C.C. Mass Spectrometry and Proteomics: Principle, Workflow, Challenges and Perspectives. Mod. Chem. Appl. 2013, 1, e105. [Google Scholar] [CrossRef]
- Ridker, P.M. C-Reactive Protein: Eighty Years from Discovery to Emergence as a Major Risk Marker for Cardiovascular Disease. Clin. Chem. 2009, 55, 209–215. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, W.; Wang, W.; Shen, H.; Liu, L.; Lou, W.; Wang, X.; Yang, P. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br. J. Cancer 2017, 117, 1846–1854. [Google Scholar] [CrossRef]
- Jensen, M.K.; Bertoia, M.L.; Cahill, L.; Agarwal, I.; Rimm, E.B.; Mukamal, K.J. Novel metabolic biomarkers of cardiovascular disease. Nat. Rev. Endocrinol. 2014, 10, 659–672. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, M.; Hu, Q.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy (Review). Int. J. Mol. Med. 2017, 41, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Gam, L.-H. Breast cancer and protein biomarkers. World J. Exp. Med. 2012, 2, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Jannetto, P.J.; Danso, D. Mass spectrometry. Clin. Biochem. 2020, 82, 1. [Google Scholar] [CrossRef]
- Beccaria, M.; Cabooter, D. Current developments in LC-MS for pharmaceutical analysis. Analyst 2020, 145, 1129–1157. [Google Scholar] [CrossRef] [PubMed]
- Seger, C.; Salzmann, L. After another decade: LC–MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef]
- Nikolaev, E.N.; Indeykina, M.I.; Brzhozovskiy, A.G.; Bugrova, A.E.; Kononikhin, A.S.; Starodubtseva, N.L.; Petrotchenko, E.V.; Kovalev, G.I.; Borchers, C.H.; Sukhikh, G.T. Mass-Spectrometric Detection of SARS-CoV-2 Virus in Scrapings of the Epithelium of the Nasopharynx of Infected Patients via Nucleocapsid N Protein. J. Proteome Res. 2020, 19, 4393–4397. [Google Scholar] [CrossRef]
- Hober, A.; Hua, T.M.K.; Foley, D.; McDonald, T.; Vissers, J.P.; Pattison, R.; Ferries, S.; Hermansson, S.; Betner, I.; Uhlen, M.; et al. Rapid and Sensitive Detection of SARS-CoV-2 Infection Using Quantitative Peptide Enrichment LC-MS/MS Analysis. medRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Q.; Huang, G.; Lin, B.-Y.; Lin, D.-Z.; Ma, Y.; Zhang, Z.; Chen, T.; Zhou, J. Tandem Mass Tag-Based Proteomic Analysis of Potential Biomarkers for Hepatocellular Carcinoma Differentiation. OncoTargets Ther. 2021, 14, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Georgila, K.; Vyrla, D.; Drakos, E. Apolipoprotein A-I (ApoA-I), Immunity, Inflammation and Cancer. Cancers 2019, 11, 1097. [Google Scholar] [CrossRef]
- Cai, X.; Ahmad, G.; Hossain, F.; Liu, Y.; Wang, X.; Dennis, J.; Freedman, B.; Witting, P.K. High-Density Lipoprotein (HDL) Inhibits Serum Amyloid A (SAA)-Induced Vascular and Renal Dysfunctions in Apolipoprotein E-Deficient Mice. Int. J. Mol. Sci. 2020, 21, 1316. [Google Scholar] [CrossRef]
- Jayaraman, S.; Fandrich, M.; Gursky, O. Synergy between serum amyloid A and secretory phospholipase A2. eLife 2019, 8, e46630. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, Y.; Chen, W.; Li, W.; Wang, A.; Wong, S.; Bao, G.; Li, J.; Yang, H.; Tracey, K.J.; et al. High-Density Lipoprotein (HDL) Counter-Regulates Serum Amyloid A (SAA)-Induced sPLA2-IIE and sPLA2-V Expression in Macrophages. PLoS ONE 2016, 11, e0167468. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.M.; Lauring, A.S. Complexities of Viral Mutation Rates. J. Virol. 2018, 92, e01031-17. [Google Scholar] [CrossRef]
- Kang, L.; He, G.; Sharp, A.K.; Wang, X.; Brown, A.M.; Michalak, P.; Weger-Lucarelli, J. A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell 2021, 184, 4392–4400.e4. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, B.M. More Severe Obesity Leads to More Severe COVID-19 in Study. JAMA 2021, 325, 1603. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Song, L.; Feng, S.; Li, L.; Yu, H.; Wang, Q.; Wang, X.; Hou, Z.; Li, X.; Li, Y.; et al. Fatty Acid Metabolism is Associated With Disease Severity After H7N9 Infection. EBioMedicine 2018, 33, 218–229. [Google Scholar] [CrossRef]
- Ohno, M.; Sekiya, T.; Nomura, N.; Daito, T.J.; Shingai, M.; Kida, H. Influenza virus infection affects insulin signaling, fatty acid-metabolizing enzyme expressions, and the tricarboxylic acid cycle in mice. Sci. Rep. 2020, 10, 10879. [Google Scholar] [CrossRef]
- Limsuwat, N.; Boonarkart, C.; Phakaratsakul, S.; Suptawiwat, O.; Auewarakul, P. Influence of cellular lipid content on influenza A virus replication. Arch. Virol. 2020, 165, 1151–1161. [Google Scholar] [CrossRef]
- Sieber, J.; Jehle, A.W. Free Fatty acids and their metabolism affect function and survival of podocytes. Front. Endocrinol. 2014, 5, 186. [Google Scholar] [CrossRef]
- Piccolis, M.; Bond, L.M.; Kampmann, M.; Pulimeno, P.; Chitraju, C.; Jayson, C.B.; Vaites, L.P.; Boland, S.; Lai, Z.W.; Gabriel, K.R.; et al. Probing the Global Cellular Responses to Lipotoxicity Caused by Saturated Fatty Acids. Mol. Cell 2019, 74, 32–44.e8. [Google Scholar] [CrossRef]
- Boden, G. Obesity and free fatty acids. Endocrinol. Metab. Clin. N. Am. 2008, 37, 635–646. [Google Scholar] [CrossRef]
- Kozłowska, D.; Harasim-Symbor, E.; Myśliwiec, H.; Milewska, A.; Chabowski, A.; Flisiak, I. Lipid profile disturbances may predispose psoriatic patients to liver dysfunction. Adv. Dermatol. Allergol. 2021, 38, 310–318. [Google Scholar] [CrossRef]
- Tanner, J.E.; Alfieri, C. The Fatty Acid Lipid Metabolism Nexus in COVID-19. Viruses 2021, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Christie, W.W.; Harwood, J.L. Oxidation of polyunsaturated fatty acids to produce lipid mediators. Essays Biochem. 2020, 64, 401–421. [Google Scholar] [PubMed]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef]
- Lone, A.M.; Taskén, K. Proinflammatory and Immunoregulatory Roles of Eicosanoids in T Cells. Front. Immunol. 2013, 4, 130. [Google Scholar] [CrossRef]
- Alvarez, Y.; Valera, I.; Municio, C.; Hugo, E.; Padrón, F.; Blanco, L.; Rodríguez, M.; Fernández, N.; Crespo, M.S. Eicosanoids in the Innate Immune Response: TLR and Non-TLR Routes. Mediat. Inflamm. 2010, 2010, 201929. [Google Scholar] [CrossRef]
- Ripon, M.A.R.; Bhowmick, D.R.; Amin, M.T.; Hossain, M.S. Role of arachidonic cascade in COVID-19 infection: A review. Prostaglandins Other Lipid Mediat. 2021, 154, 106539. [Google Scholar] [CrossRef] [PubMed]
- Chilosi, M.; Poletti, V.; Ravaglia, C.; Rossi, G.; Dubini, A.; Piciucchi, S.; Pedica, F.; Bronte, V.; Pizzolo, G.; Martignoni, G.; et al. The pathogenic role of epithelial and endothelial cells in early-phase COVID-19 pneumonia: Victims and partners in crime. Mod. Pathol. 2021, 34, 1444–1455. [Google Scholar] [CrossRef]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Hammock, B.D.; Wang, W.; Gilligan, M.M.; Panigrahyyz, D. Eicosanoids: The Overlooked Storm in Coronavirus Disease 2019 (COVID-19)? Am. J. Pathol. 2020, 190, 1782–1788. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome P450 research and The Journal of Biological Chemistry. J. Biol. Chem. 2019, 294, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Isopi, E.; Mattoscio, D.; Codagnone, M.; Mari, V.C.; Lamolinara, A.; Patruno, S.; D’Aurora, M.; Cianci, E.; Nespoli, A.; Franchi, S.; et al. Resolvin D1 Reduces Lung Infection and Inflammation Activating Resolution in Cystic Fibrosis. Front. Immunol. 2020, 11, 581. [Google Scholar] [CrossRef]
- Dunlap, N.E.; Fulmer, J.D. Corticosteroid therapy in asthma. Clin. Chest Med. 1984, 5, 669–683. [Google Scholar] [CrossRef]
- Sichitiu, J.; Fakhouri, F.; Desseauve, D. Antenatal corticosteroid therapy and COVID-19: Pathophysiological considerations. Acta Obstet. Gynecol. Scand. 2020, 99, 952. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wang, Y.; Colunga-Lozano, L.E.; Prasad, M.; Tangamornsuksan, W.; Rochwerg, B.; Yao, L.; Motaghi, S.; Couban, R.J.; Ghadimi, M.; et al. Efficacy and safety of corticosteroids in COVID-19 based on evidence for COVID-19, other coronavirus infections, influenza, community-acquired pneumonia and acute respiratory distress syndrome: A systematic review and meta-analysis. Can. Med. Assoc. J. 2020, 192, E756–E767. [Google Scholar] [CrossRef] [PubMed]
- Penttilä, P.A.; van Gassen, S.; Panovska, D.; Vanderbeke, L.; van Herck, Y.; Quintelier, K.; Emmaneel, A.; Filtjens, J.; Malengier-Devlies, B.; Ahmadzadeh, K.; et al. High dimensional profiling identifies specific immune types along the recovery trajectories of critically ill COVID19 patients. Experientia 2021, 78, 3987–4002. [Google Scholar] [CrossRef] [PubMed]
- Boer, M.M.B.-D.; Van Wetten, M.-L.; Pruimboom, L. Chronic inflammatory diseases are stimulated by current lifestyle: How diet, stress levels and medication prevent our body from recovering. Nutr. Metab. 2012, 9, 32. [Google Scholar] [CrossRef]
- Chen, J.; Qi, T.; Liu, L.; Ling, Y.; Qian, Z.; Li, T.; Li, F.; Xu, Q.; Zhang, Y.; Xu, S.; et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Infect. 2020, 80, e1–e6. [Google Scholar] [CrossRef]
- Li, C.; Wu, X.; Liu, S.; Shen, D.; Zhu, J.; Liu, K. Role of Resolvins in the Inflammatory Resolution of Neurological Diseases. Front. Pharmacol. 2020, 11, 612. [Google Scholar] [CrossRef]
- Freire, M.O.; Dalli, J.; Serhan, C.N.; Van Dyke, T.E. Neutrophil Resolvin E1 Receptor Expression and Function in Type 2 Diabetes. J. Immunol. 2016, 198, 718–728. [Google Scholar] [CrossRef]
- Tian, H.; Lu, Y.; Shah, S.P.; Hong, S. Autacoid 14S,21R-Dihydroxy-Docosahexaenoic Acid Counteracts Diabetic Impairment of Macrophage Prohealing Functions. Am. J. Pathol. 2011, 179, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Lu, Y. Omega-3 fatty acid-derived resolvins and protectins in inflammation resolution and leukocyte functions: Targeting novel lipid mediator pathways in mitigation of acute kidney injury. Front. Immunol. 2013, 4, 13. [Google Scholar] [CrossRef]
- Arnardottir, H.; Pawelzik, S.-C.; Wistbacka, U.; Artiach, G.; Hofmann, R.; Reinholdsson, I.; Braunschweig, F.; Tornvall, P.; Religa, D.; Bäck, M. Stimulating the Resolution of Inflammation Through Omega-3 Polyunsaturated Fatty Acids in COVID-19: Rationale for the COVID-Omega-F Trial. Front. Physiol. 2021, 11, 624657. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.J.; Brokstad, K.A. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020, 20, 581–582. [Google Scholar] [CrossRef]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371. [Google Scholar] [CrossRef]
- Quast, I.; Tarlinton, D. B cell memory: Understanding COVID-19. Immunity 2021, 54, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Sherina, N.; Piralla, A.; Du, L.; Wan, H.; Kumagai-Braesch, M.; Andréll, J.; Braesch-Andersen, S.; Cassaniti, I.; Percivalle, E.; Sarasini, A.; et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6–8 months after the infection. Med 2021, 2, 281–295.e4. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Miller, L.M.; Barnes, L.F.; Raab, S.A.; Draper, B.E.; El-Baba, T.J.; Lutomski, C.A.; Robinson, C.V.; Clemmer, D.E.; Jarrold, M.F. Heterogeneity of Glycan Processing on Trimeric SARS-CoV-2 Spike Protein Revealed by Charge Detection Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 3959–3966. [Google Scholar] [CrossRef]
- Roberts, D.S.; Mann, M.W.; Melby, J.A.; Larson, E.J.; Zhu, Y.; Brasier, A.R.; Jin, S.; Ge, Y. Structural O-Glycoform Heterogeneity of the SARS-CoV-2 Spike Protein Receptor-Binding Domain Revealed by Top-Down Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 12014–12024. [Google Scholar] [CrossRef]
- Cho, B.G.; Gautam, S.; Peng, W.; Huang, Y.; Goli, M.; Mechref, Y. Direct Comparison of N-Glycans and Their Isomers Derived from Spike Glycoprotein 1 of MERS-CoV, SARS-CoV-1, and SARS-CoV-2. J. Proteome Res. 2021, 20, 4357–4365. [Google Scholar] [CrossRef]
- Antonopoulos, A.; Broome, S.; Sharov, V.; Ziegenfuss, C.; Easton, R.L.; Panico, M.; Dell, A.; Morris, H.R.; Haslam, S.M. Site-specific characterization of SARS-CoV-2 spike glycoprotein receptor-binding domain. Glycobiology 2021, 31, 181–187. [Google Scholar] [CrossRef]
- Witkowska, D. Mass Spectrometry and Structural Biology Techniques in the Studies on the Coronavirus-Receptor Interaction. Molecules 2020, 25, 4133. [Google Scholar] [CrossRef]
- Krishnan, S.; Krishnan, G.P. N-Glycosylation Network Construction and Analysis to Modify Glycans on the Spike (S) Glycoprotein of SARS-CoV-2. Front. Bioinform. 2021, 1. [Google Scholar] [CrossRef]
- Watanabe, Y.; Berndsen, Z.T.; Raghwani, J.; Seabright, G.E.; Allen, J.D.; Pybus, O.; McLellan, J.S.; Wilson, I.A.; Bowden, T.A.; Ward, A.B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef] [PubMed]
- Petrović, T.; Alves, I.; Bugada, D.; Pascual, J.; Vučković, F.; Skelin, A.; Gaifem, J.; Villar-Garcia, J.; Vicente, M.M.; Fernandes, Â.; et al. Composition of the immunoglobulin G glycome associates with the severity of COVID-19. Glycobiology 2020, 31, 372–377. [Google Scholar]
- Trkulja, V.; Kodvanj, I.; Homolak, J. Immunoglobulin G glycome and severity of COVID-19: More likely a quantification of bias than a true association. A comment on Petrović et al., “Composition of the immunoglobulin G glycome associates with the severity of COVID-19”. Glycobiology 2021, 31, 713–716. [Google Scholar] [CrossRef]
- Hou, H.; Yang, H.; Liu, P.; Huang, C.; Wang, M.; Li, Y.; Zhu, M.; Wang, J.; Xu, Y.; Wang, Y.; et al. Profile of Immunoglobulin G N-Glycome in COVID-19 Patients: A Case-Control Study. Front. Immunol. 2021, 12, 748566. [Google Scholar] [CrossRef]
- Rosenbalm, K.E.; Tiemeyer, M.; Wells, L.; Aoki, K.; Zhao, P. Glycomics-informed glycoproteomic analysis of site-specific glycosylation for SARS-CoV-2 spike protein. STAR Protoc. 2020, 1, 100214. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 spike protein glycan shield: Implications for immune recognition. Sci. Rep. 2020, 10, 14991. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Praissman, J.L.; Grant, O.C.; Cai, Y.; Xiao, T.; Rosenbalm, K.E.; Aoki, K.; Kellman, B.P.; Bridger, R.; Barouch, D.H.; et al. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28, 586–601.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, H.; Wang, H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front. Mol. Biosci. 2021, 8, 629873. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Z.; Hu, W.; Hao, P.; Yang, S. Impact of Expressing Cells on Glycosylation and Glycan of the SARS-CoV-2 Spike Glycoprotein. ACS Omega 2021, 6, 15988–15999. [Google Scholar] [CrossRef]
- Kasuga, Y.; Zhu, B.; Jang, K.-J.; Yoo, J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef]
- Bourgeois, C.; Kuchler, K. Fungal pathogens—A sweet and sour treat for toll-like receptors. Front. Cell. Infect. Microbiol. 2012, 2, 142. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Choubey, D. Type I interferon (IFN)-inducible Absent in Melanoma 2 proteins in neuroinflammation: Implications for Alzheimer’s disease. J. Neuroinflamm. 2019, 16, 236. [Google Scholar] [CrossRef]
- Chiang, H.-S.; Liu, H.M. The Molecular Basis of Viral Inhibition of IRF- and STAT-Dependent Immune Responses. Front. Immunol. 2019, 9, 3086. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.M.; Shalagina, M.N.; Protopopov, V.A.; Sergeev, V.G.; Ovechkin, S.V.; Ovchinina, N.G.; Sekunov, A.V.; Zefirov, A.L.; Zakirjanova, G.F.; Bryndina, I.G. Changes in Membrane Ceramide Pools in Rat Soleus Muscle in Response to Short-Term Disuse. Int. J. Mol. Sci. 2019, 20, 4860. [Google Scholar] [CrossRef]
- Khan, S.A.; Goliwas, K.F.; Deshane, J.S. Sphingolipids in Lung Pathology in the Coronavirus Disease Era: A Review of Sphingolipid Involvement in the Pathogenesis of Lung Damage. Front. Physiol. 2021, 12, 1757. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sarkar, P.; Chattopadhyay, A. Metabolic Depletion of Sphingolipids Reduces Cell Surface Population of the Human Serotonin1A Receptor due to Impaired Trafficking. ACS Chem. Neurosci. 2021, 12, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Asahara, T.; Achiwa, K.; Hoshino, H. Synthesis of N-Acetylglucosaminyl- and N-Acetylgalactosaminylceramides as Cerebroside Analogs and Their Anti-human Immunodeficiency Virus Type 1 Activities. Chem. Pharm. Bull. 1997, 45, 402–405. [Google Scholar] [CrossRef][Green Version]
- Bieberich, E. Sphingolipids and lipid rafts: Novel concepts and methods of analysis. Chem. Phys. Lipids 2018, 216, 114–131. [Google Scholar] [CrossRef]
- Stephenson, E.; Reynolds, G.; Botting, R.A.; Calero-Nieto, F.J.; Morgan, M.D.; Tuong, Z.K.; Bach, K.; Sungnak, W.; Worlock, K.B.; Yoshida, M.; et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021, 27, 904–916. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, J.; Liu, D.; Sun, Y.; Wu, C. An integrative multiomics analysis identifies putative causal genes for COVID-19 severity. Genet. Med. 2021, 23, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020, 12, 23–40.e7. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Andrade, B.S.; Weener, M.E.; Góes-Neto, A.; Azevedo, V.; Ghosh, P.; Blum, K.; Ganguly, N.K. A novel multi-omics-based highly accurate prediction of symptoms, comorbid conditions, and possible long-term complications of COVID-19. Mol. Omics 2021, 17, 317–337. [Google Scholar] [CrossRef]
- Aggarwal, S.; Acharjee, A.; Mukherjee, A.; Baker, M.S.; Srivastava, S. Role of Multiomics Data to Understand Host–Pathogen Interactions in COVID-19 Pathogenesis. J. Proteome Res. 2021, 20, 1107–1132. [Google Scholar] [CrossRef] [PubMed]
- Sen, R. High-throughput approaches of diagnosis and therapies for COVID-19: Antibody panels, proteomics and metabolomics. Future Drug Discov. 2021, 3, FDD55. [Google Scholar] [CrossRef]
- Singh, R.; Singh, P.K.; Kumar, R.; Kabir, T.; Kamal, M.A.; Rauf, A.; Albadrani, G.M.; Sayed, A.A.; Mousa, S.A.; Abdel-Daim, M.M.; et al. Multi-Omics Approach in the Identification of Potential Therapeutic Biomolecule for COVID-19. Front. Pharmacol. 2021, 12, 652335. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ren, X.; Wen, W.; Fan, X.; Hou, W.; Su, B.; Cai, P.; Li, J.; Liu, Y.; Tang, F.; Zhang, F.; et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021, 184, 1895–1913.e19. [Google Scholar] [CrossRef] [PubMed]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef]
- Sarma, A.; Christenson, S.A.; Byrne, A.; Mick, E.; Pisco, A.O.; DeVoe, C.; Deiss, T.; Ghale, R.; Zha, B.S.; Tsitsiklis, A.; et al. Tracheal aspirate RNA sequencing identifies distinct immunological features of COVID-19 ARDS. Nat. Commun. 2021, 12, 5152. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef]
- Liu, T.; Jia, P.; Fang, B.; Zhao, Z. Differential Expression of Viral Transcripts From Single-Cell RNA Sequencing of Moderate and Severe COVID-19 Patients and Its Implications for Case Severity. Front. Microbiol. 2020, 11, 603509. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Kornberg, R.D. Structure of chromatin. Annu. Rev. Biochem. 1977, 46, 931–954. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [CrossRef]
- Robertson, G.; Hirst, M.; Bainbridge, M.; Bilenky, M.; Zhao, Y.; Zeng, T.; Euskirchen, G.; Bernier, B.; Varhol, R.; Delaney, A.; et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 2007, 4, 651–657. [Google Scholar] [CrossRef]
- Wang, A.; Chiou, J.; Poirion, O.B.; Buchanan, J.; Valdez, M.J.; Verheyden, J.M.; Hou, X.; Kudtarkar, P.; Narendra, S.; Newsome, J.M.; et al. Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife 2020, 9, e62522. [Google Scholar] [CrossRef]
- Li, S.; Wu, B.; Ling, Y.; Guo, M.; Qin, B.; Ren, X.; Wang, C.; Yang, H.; Chen, L.; Liao, Y.; et al. Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients. Front. Immunol. 2021, 12, 625881. [Google Scholar] [CrossRef]
- de Moura, M.C.; Davalos, V.; Planas-Serra, L.; Alvarez-Errico, D.; Arribas, C.; Ruiz, M.; Aguilera-Albesa, S.; Troya, J.; Valencia-Ramos, J.; Vélez-Santamaria, V.; et al. Epigenome-wide association study of COVID-19 severity with respiratory failure. EBioMedicine 2021, 66, 103339. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.J.; Pang, A.P.; Dody, K.; Mudd, P.A.; Patterson, B.K.; Seethamraju, H.; Bram, Y.; Peluso, M.J.; Torres, L.; Iyer, N.S.; et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J. Leukoc. Biol. 2021, 110, 21–26. [Google Scholar] [CrossRef]
- Balnis, J.; Madrid, A.; Hogan, K.J.; Drake, L.A.; Chieng, H.C.; Tiwari, A.; Vincent, C.E.; Chopra, A.; Vincent, P.A.; Robek, M.D.; et al. Blood DNA methylation and COVID-19 outcomes. Clin. Epigenet. 2021, 13, 118. [Google Scholar] [CrossRef]
- Atlante, S.; Mongelli, A.; Barbi, V.; Martelli, F.; Farsetti, A.; Gaetano, C. The epigenetic implication in coronavirus infection and therapy. Clin. Epigenet. 2020, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Garbati, M.R.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Saksena, N.; Bonam, S.R.; Miranda-Saksena, M. Epigenetic Lens to Visualize the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) Infection in COVID-19 Pandemic. Front. Genet. 2021, 12, 581726. [Google Scholar] [CrossRef]
- You, M.; Chen, L.; Zhang, D.; Zhao, P.; Chen, Z.; Qin, E.-Q.; Gao, Y.; Davis, M.M.; Yang, P. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat. Cell Biol. 2021, 23, 620–630. [Google Scholar] [CrossRef]
- Ho, J.S.Y.; Mok, B.W.Y.; Campisi, L.; Jordan, T.; Yildiz, S.; Parameswaran, S.; Wayman, J.A.; Gaudreault, N.N.; Meekins, D.A.; Indran, S.V.; et al. TOP1 inhibition therapy protects against SARS-CoV-2-induced lethal inflammation. Cell 2021, 184, 2618–2632.e17. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- van Berkum, N.L.; Lieberman-Aiden, E.; Williams, L.; Imakaev, M.; Gnirke, A.; Mirny, L.A.; Dekker, J.; Lander, E.S. Hi-C: A method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010, 39, e1869. [Google Scholar] [CrossRef]
- Belton, J.-M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Furlan-Magaril, M.; Mifsud, B.; Tavares-Cadete, F.; Sugar, R.; Javierre, B.-M.; Nagano, T.; Katsman, Y.; Sakthidevi, M.; Wingett, S.W.; et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015, 25, 582–597. [Google Scholar] [CrossRef]
- Nagano, T.; Lubling, Y.; Varnai, C.; Dudley, C.; Leung, W.; Baran, Y.; Cohen, N.M.; Wingett, S.; Fraser, P.; Tanay, A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017, 547, 61–67. [Google Scholar] [CrossRef]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef]
- Ying, H.; Ebrahimi, M.; Keivan, M.; Khoshnam, S.E.; Salahi, S.; Farzaneh, M. miRNAs; a novel strategy for the treatment of COVID-19. Cell Biol. Int. 2021, 45, 2045–2053. [Google Scholar] [CrossRef]
- McDonald, J.T.; Enguita, F.J.; Taylor, D.; Griffin, R.J.; Priebe, W.; Emmett, M.R.; McGrath, M.; Sajadi, M.; Harris, A.D.; Clement, J.; et al. The Great Deceiver: miR-2392’s Hidden Role in Driving SARS-CoV-2 Infection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Farr, R.; Rootes, C.; Rowntree, L.; Nguyen, T.; Hensen, L.; Kedzierski, L.; Cheng, A.; Kedzierska, K.; Au, G.; Marsh, G.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.; Lipovich, L. miRCOVID-19: Potential Targets of Human miRNAs in SARS-CoV-2 for RNA-Based Drug Discovery. Non-Coding RNA 2021, 7, 18. [Google Scholar] [CrossRef]
- Arora, S.; Singh, P.; Dohare, R.; Jha, R.; Syed, M.A. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene 2020, 762, 145057. [Google Scholar] [CrossRef]
- Chang, C.; Kong, W.; Mou, X.; Wang, S. Investigating the correlation between DNA methylation and immune-associated genes of lung adenocarcinoma based on a competing endogenous RNA network. Mol. Med. Rep. 2020, 22, 3173–3182. [Google Scholar] [CrossRef] [PubMed]
- Ala, U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, L.; López-Royo, T.; Calvo, A.; Toivonen, J.; De La Torre, M.; Moreno-Martínez, L.; Molina, N.; Aparicio, P.; Zaragoza, P.; Manzano, R.; et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9582. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, M.; Du, S.; Feng, W.; Zhang, K.; Zhang, L.; Liu, H.; Jia, G.; Wu, L.; Hu, X.; et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 2019, 10, 1637. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).