Abstract
Extracellular vesicles (EVs) are nano-sized particles released from cells and transferring molecules (proteins, lipids and nucleic acids such as mRNA, tRNA and miRNA) to recipient cells. Surface antigens and components are important for the functions as cell-to-cell communication of EVs. Thus, EVs are useful biomarkers for various diseases including leukemias and other types of malignancies. We evaluated whether miRNAs in EVs released from chronic myelogenous leukemia (CML) cells could be used for diagnosis. Microarray analysis of miRNAs in EVs obtained from the culture supernatants of two CML cell lines showed that miR-494 and miR-373-5p were significantly decreased by tyrosine kinase inhibitor for BCR-ABL1. Validation analysis with Taqman-based qRT-PCR of whole serum obtained patients with CML in the chronic phase (n = 5) did not show a significant difference in miR-494 levels compared to the CML accelerated phase and blast crisis patients (n = 5). However, the levels of miR-494 were 2.9-fold higher in the accelerated phase or blast crisis than in the chronic phase (p < 0.05). These results indicate that it is important to measure miR-494 using only EVs rather than whole serum. Our data suggest that EV-miR-494 is a useful biomarker of CML progression and evaluation of response to tyrosine kinase inhibitors.
1. Introduction
Chronic myelogenous leukemia (CML) is characterized by the presence of the chimeric gene, BCR-ABL1. It produces a protein with constitutively enhanced tyrosine kinase activity which promotes cell proliferation. Tyrosine kinase inhibitors (TKIs) which recognize the constitutional structure of BCR-ABL1 proteins specifically inhibit the activity of BCR-ABL1 proteins and suppress the proliferation of CML cells [1]. Imatinib (IM) is the first generation TKI and used for CML patients in chronic phase (CP). The second generation TKIs, dasatinib (DS) and nilotinib, have been approved as effective for Imatinib-resistant CML. They have been approved as first-line treatments for over 10 years [2], and in addition, nilotinib and dasatinib are recommended as first-line agents [3]. The mutations in the kinase domain of BCR-ABL1 gene, especially T315I, cause the resistance to the first and second generation TKIs. The third generation TKIs, ponatinib and bosutinib, are the second line drugs. Ponatinib is effective on BCR-ABL1 with T315I mutation. Bosutinib has favorable tolerability and acts on various types of mutated BCR-ABL1 except T315I. Early detection of relapse of leukemia is very important for CML patients with TKI treatment. Quantitative analysis using qRT-PCR is the standard method to monitor leukemic cells. As this method needs mRNAs extracted from CML cells for detection, it is occasionally difficult to quantify BCR-ABL1 mRNA for the patients who have minimal residual disease, although it is a sensitive enough method to monitor MR5.0 (BCR-ABL1 transcripts/ABL or internal control transcript ≤ 0.001%).
Extracellular vesicles (EVs) are heterogenous particles of 50–150 nm size in diameter and are released from various cells [4]. On their surface membrane, EV-specific antigens (CD63, CD9, CD81) and tissue-specific antigens are co-expressed [5]. Furthermore, EV contains mRNAs, which are small non-coding RNAs and proteins [6]. These molecules are derived from original tissues or cells. In healthy individuals, EVs function as components for cell-to-cell interaction [6] and immune systems [7], therefore EVs are important for maintaining homeostasis. On the other hand, EVs derived from tumor cells are involved in the progress of pathological conditions. For examples, EVs from breast cancer [8], lung cancer [9] and gastric cancer [10] promote metastasis, and EVs from acute leukemia cells stimulate angiogenesis [11] and suppress normal hematopoiesis. EVs derived from CML cells have been reported to mediate cell proliferation [12] and communicate with other cells [13,14]. Since EVs exist stably as stable forms in body fluids (e.g., plasma, pleural effusion, saliva, urine), they are potential biomarkers of various diseases.
MiRNAs are noncoding RNAs that interact with the mRNA 3′-untranslated region and regulate a variety of biological processes by suppressing transcription [15]. Microvesicles, which are released by budding from the plasma membrane, are relatively large sized EVs that include molecules in cytoplasm [16]. On the other hand, exosomes have smaller sizes and contain abundant miRNAs [6]. MiRNAs are selectively included in EVs, especially in exosomes, and are actively released from cells. Non-EV miRNAs which bind to Argonaute2 proteins or lipids [17,18] exist in plasma as another stable form despite the coexistence of RNase [19]. These non-EV miRNAs are partly released by tumor lysis, necrosis and apoptosis available for diagnosis [20], but they are also released by inflammation and organ damage associated with the primary disease.
We focused on EV-miRNAs in the serum of CML patients or supernatants of cultured CML cell lines, and examined whether the miRNAs in these EVs reflect the pathophysiology of CML using array analysis and the qRT-PCR method and can be used for clinical diagnosis. We demonstrated the change of signature of EV-miRNAs from supernatants of CML cell lines after TKIs treatment. Moreover, the level of miR-494 in EVs derived from CML patients’ serum was significantly changed with TKI treatment.
2. Materials and Methods
2.1. Cell Culture and TKIs
The K562 and KU812 cell lines were purchased from the RIKEN Cell Bank (Ibaraki, Japan). Both cell lines are Philadelphia chromosome positive and produce BCR-ABL1 protein, but K562 is characterized as erythroleukemia [21], whereas KU812 is characterized as a basophilic precursor [22]. Although BCR-ABL1’s transcript level in KU812 is lower than K562 [23], KU812 has high-risk additional chromosomal abnormalities (e.g., i(17q), +19, +der(9)t(9;22)) [24]. Cells were plated at 2 × 105 cells/mL and maintained in RPMI 1640 medium (GIBCO, Brooklyn, NY, USA) supplemented with 10% fetal bovine serum (GIBCO), and stored at 37 °C in a humidified atmosphere containing 5% CO2. These cells were treated with 1 µM IM or 10 nM DS (both drugs were purchased from Cayman Chemical (Ann Arbor, MI, USA)) for 72 h. These concentrations of TKIs inhibit the cell proliferation adequately and did not induce excess cell death (Supplementary Figure S1).
2.2. Patients and Healthy Control Samples
After informed consent was obtained, all serums were collected from patients in Takagi hospital, and healthy volunteers of the International University of Health and Welfare, in accordance with the Declaration of Helsinki. This study complied with the institutional policies of the Takagi hospital and The International University of Health and Welfare and approved by the ethical committee. Five patients were in the chronic phase of CML (CML-CP) and were treated with IM (400 mg/day), and another five patients were diagnosed with the accelerated phase or blast crisis of CML (CML-AP/BC). All patients diagnosed as CML-AP/BC had never received treatments for any hematologic diseases. Blood samples from these five patients used in this study were obtained at the first visit without history of TKIs treatment. According to European LeukemiaNet (ELN) classification, the phase of each CML patient was defined. Clinical data of each group are shown in Table 1. Healthy volunteers were confirmed by medical interview and physical examination. Nine milliliters of peripheral blood were collected without any anticoagulants. After blood coagulation and centrifugation (3500× g, 10 min), serum was obtained and stored at −80 °C until extraction of miRNA.

Table 1.
Summary of healthy volunteer and CML patients.
2.3. Purification and Detection of Extracellular Vesicles
After in vitro culture for 72 h, culture media were centrifuged at 3000× g (remove cells) and 15,000× g (remove debris). The supernatants were obtained after filtration by a Minisart® 0.2 μm filter (Sartorius, Goettingen, Germany). EVs from these supernatants were purified by using a miRCURY Exosome Cell/Urine/CSF Kit (Qiagen, Venlo, The Netherlands). The EVs collection from three different media was performed to extract miRNA.
Serum samples were centrifuged at 15,000× g (remove debris and residual blood cells) and filtered by 0.2 μm filter. To purify EVs from serum samples, an ExoQuick® (System Biosciences, Palo Alto, CA, USA) was used according to the manufacturer’s protocol. Briefly, 63 µL of ExoQuick® reagent was added to the filtered serum, and this solution was incubated at 4 °C for 30 min. The EV pellet was then obtained by centrifugation (1500× g, 30 min).
The purity of EVs was evaluated by flowcytometry (FACSAria II; BD Biosciences, San Jose, CA, USA) and anti-CD63 PE-conjugated antibody (Thermo Fisher Scientific, Waltham, MA, USA). The quantity of EVs was measured by western blotting and ImageJ software. EVs from 40 mL supernatant of cell cultures or 250 µL serum were lysed by a 50 µL RIPA buffer (Thermo Fisher). Each 12 µL lysed EV was then electrophoresed in 10% SDS polyacrylamide gel. The primary antibody for CD63 (1:1000; GeneTex, Irvine, CA, USA) and the secondary anti-rabbit IgG antibody (1:1000; Abcam, Cambridge, UK) were used for western blotting. The images obtained from western blotting were analyzed by ImageJ software (NIH, Bethesda, MA, USA).
2.4. miRNA Microarray Assay
miRNAs were isolated from all EVs and the whole serum using NucleoSpin® miRNA Plasma kit (Macherey-Nagel, Düren, Germany) according to the manufacturer protocol. One fmol cel-miR-39 (Qiagen, Hilden, Germany) was spiked-in during the protein denaturing step as a normalizer. miRNAs were eluted in 100 µL of RNase free water, then 50 µL miRNA solution was used for microarray assay. MiRNAs for microarray were labeled by a Nucleic Acid Labeling Kit, PlatinumBright 647 Infrared (KREATECH, Amsterdam, The Netherlands). Labeled-MiRNAs were mixed with a hybridization buffer (Nuclease-Free Water, 1 M Tris-HCl,1 M NaCl, 0.5% Tween 20) and the Genopal®-MICH14 DNA chips (Mitsubishi Chemical, Tokyo, Japan), on which 228 oligonucleotide DNA probes were installed for detection of human tumor specific miRNAs. After hybridization for 16 h at 50 °C, the DNA chips were first washed with solution A (0.24 M Tris-HCl, 0.24 M NaCl, 0.05% Tween 20) and then with solution B (0.24 M Tris-HCl, 0.24 M NaCl). Each chip was measured by a Biochip reader (Yokogawa Electric Corporation, Tokyo, Japan).
2.5. Quantification of miRNA by qRT-PCR
MiRNAs for qRT-PCR were transcribed to cDNAs using a TaqMan™ MicroRNA Reverse Transcription Kit (Thermo Fisher), and 5 µL of the extracted miRNAs solution. Looped RT-primers specific for each miRNA were purchased (TaqMan MicroRNA Assays Kit, Thermo Fisher) and used for the reverse transcription reaction. For detecting miRNAs, the amplification was done using TaqMan™ Fast Advanced Master Mix (Thermo Fisher), and ABI 7500 FAST (Thermo Fisher). The amounts of each miRNA were normalized with the level of spiked-in cel-miR-39.
2.6. Static Analysis
The “DESeq2” R package was used to normalize the signal intensity of the microarray. The heatmap was generated from the normalized microarray data using the “heatmap” R package for log2-fold change. Hierarchical clustering was performed simultaneously with heat mapping using the Ward criterion. Volcano plots were visualized by using “EnhancedVolcano”. Benjamini–Hochberg-adjusted p-value < 0.05 and log2-fold change ≥1.5 were set as criteria. The statistical analysis for qRT-PCR was performed by a Student’s paired two-tailed t-test, and p-values < 0.05 were considered to have statistical significance.
3. Results
3.1. Effects of TKI on Extracellular Vesicle Release from CML Cell Lines
EVs in supernatant of CML cell lines (K562 and KU812) were collected by miRCURY Exosome Cell/Urine/CSF Kit. By flowcytometric analysis of surface CD63 antigen, the purity of EVs was almost 90% (Figure 1A). The signal intensities of CD63 shown by western blotting were not different between cultures with or without TKIs, and showed enough quantity collected (Figure 1B). Thus, the TKIs did not change the amount of extracellular vesicle released from CML cell lines.
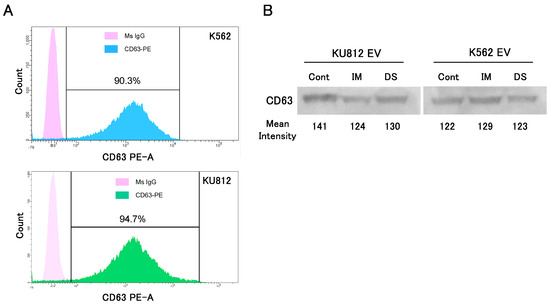
Figure 1.
The purification and quantification of EV from the supernatant of cell cultures. (A) The purity of EVs was confirmed by surface CD63 antigen of EV by flowcytometric analysis. The upper histogram is the EVs from the K562 cell, and the lower histogram is EVs from KU812 cells. (B) EVs from each cell line were quantified by western blotting. The mean intensity of each band was measured by ImageJ software. (IM: imatinib, DS: dasatinib).
3.2. Microarray Analysis of EV-miRNAs Derived from CML Cells
After extraction of miRNA from EVs, the signature of 228 miRNAs was analyzed by the microarray method. Results showed abundant 10 miRNAs (miR-191*, 197, 198, 210, 296, 320, 373-5p, 409-3p, 494, 574) in CML-EVs. Up-regulation of these miRNAs were common in two cell lines. TKIs treatment for 72 h changed the signatures of EV-miRNA. Hierarchical cluster analysis showed that six miRNAs (miR-191*, 198, 320, 373*, 409-3p, 494) were independent cluster (Figure 2A). As to the expression of cellular miR-494, there was no difference between the control and TKI treatments by microarray analysis (1.06 (IM) and 1.26 (DS) in KU812, 1.03 (IM) and 1.04 (DS) in K562).
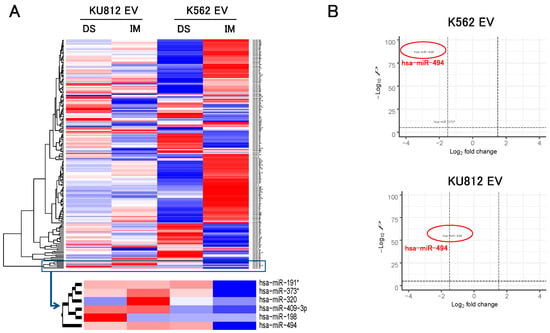
Figure 2.
MiRNA microarray analysis in EV from two cell lines. (A) Heatmap and hierarchical clustering after TKIs treatments. Red or blue color indicate increased or decreased miRNA in EVs after TKIs treatment, respectively. Six miRNAs were clustered as the independent group. (IM: imatinib, DS: dasatinib) (B) Volcano plots from miRNA microarray analysis of two cell lines. For drawing volcano plots, the integrated data from both TKIs was used to detect commonly fluctuating miRNAs.
Volcano plots indicated TKI-induced significant reduction of EV-miR-494: IM (K562: 0.12-fold, KU812: 0.28-fold) and DS (K562: 0.13-fold, KU812: 0.31-fold) (Figure 2B). This reduction was much larger than that of miR-373-5p, which was the second reduced miRNA.
3.3. Validation of the Level of miRNAs by qRT-PCR
Microarray analysis showed that miR-494 is the most down-regulated miRNA with in vitro TKIs treatment, and the level was validated with the qRT-PCR method. MiR-373-5p was also validated simultaneously because the volcano plot of K562 was suggested as a candidate of altered miRNA after TKIs.
In K562-derived EVs, the relative levels of miR-494 were 0.16 (IM, p < 0.01) and 0.41-fold (DS, p < 0.05) in comparison with non-treated cultures, and that of miR-373-5p was 0.47 (IM, p < 0.05) and 0.56 (DS, p < 0.05) folds. As for KU812 cells, the relative levels of miR-494 were 0.29 (IM, p < 0.05) and 0.11 (DS, p < 0.01) folds, and that of miR-373-5p were 0.41 (IM, p < 0.05) and 0.53 (DS, p > 0.05) folds (Figure 3). In both cell lines, the quantity of the two miRNAs in EVs were significantly decreased.
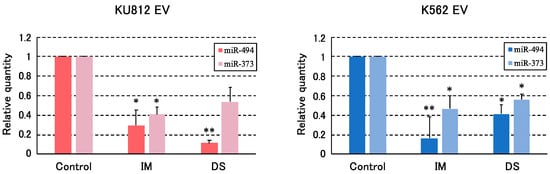
Figure 3.
The validations of two miRNA by qRT-PCR (n = 3). miR-494 and miR-373-5p were validated by qRT-PCR using TaqMan probe. The expressions of miRNAs were normalized by spiked cel-miR-39 and shown as relative expressions to control. Each value represents the mean ± SEM (n = 3). Asterisks indicate statistical significance (* p < 0.05, ** p < 0.01). (IM: imatinib, DS: dasatinib).
3.4. The Level of EV-miRNAs Serum of Patient with CML
We observed a change of miR-373-5p and miR-494 in whole serum and purified EVs. Western blotting analysis showed that the same amounts of EVs were obtained from each serum (Figure 4A). MiR-373-5p was abundant in EVs of CML cell lines but it was not detected in serum and purified EVs (data not shown). Figure 4B (left panel) shows the level of miR-494 in serum of CML patients. Its median level was 0.031 in healthy controls against 0.016 in CML patients in AP or BC phases, and 0.030 in the chronic phase. There were no significant differences the miR-494 levels. On the other hand, the levels of EV-miR-494 were higher at the AP or BC phases (0.027) than in CP (0.0093), which is almost equal to the level in the healthy control (0.0090) (Figure 4B (right panel)).
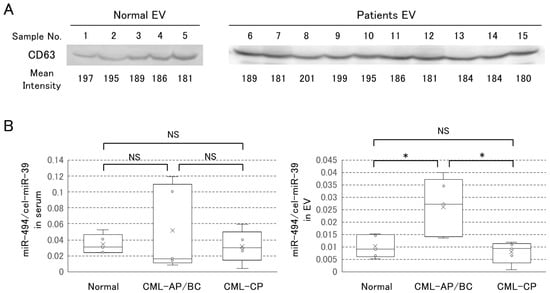
Figure 4.
The analysis of EV in serum. (A) The EVs from serum were recovered by ExoQuick® reagent and quantified by western blotting of CD63 protein. Sample number indicate; 1–5 (Normal), 6–10 (CML-AP/BC), 11–15 (CML-CP). The mean intensity of each band is shown below the bands. (B) qRT-PCR analysis of miR-494. The black line in each box plot indicates the median. The lower and upper edges of the box indicate the 25% and 75% distribution percentiles, respectively. (B) shows in whole serum (left panel) and in only EVs (right panel). Asterisks indicate statistical significance and NS means “Not Significant” (* p < 0.05).
4. Discussion
Various scoring methods and guidelines for CML risk assessment have been proposed. For prognosis prediction, the Sokal score (Age, Spleen size, Platelets (×10⁹/L), blasts (% peripheral blood)) or the Euro (Hasford) score (eosinophils (%), basophils (%) added to the Sokal score) have been used from the days when IFN-γ treatment was the mainstay, and EUTOS (The European Treatment Outcome Study) score (7 × basophils + 4 × spleen size) was used for patients treated with TKIs; furthermore, the EUTOS long-term survival (ELTS) score was reported to be superior to these scoring systems in predicting the long-term prognosis of CML patients. [25,26,27]. In practical guidelines for hematological malignancies published by the Japanese Society of Hematology, the Sokal, Euro (Hasford) and EUTOS scores are listed for prognosis of CML. The evidence-based choice of TKIs using these scores is important for the treatment of CML patients. The concept of CML treatment is to control BCR-ABL1 positive CML cells and to avoid the progression to the CML stage. The effectiveness of treatment in the chronic phase of CML is evaluated by the quantification of BCR-ABL1 transcripts on the international scale (BCR-ABL1IS) and according to the criterion of ELN (European LeukemiaNet recommendations) 2013 and 2020 [3]. As CML treatments using TKIs aim to achieve Major Molecular Response (BCR-ABL1IS ≤ 0.1%) or Deep Molecular Response (MR4.0; BCR-ABL1IS ≤ 0.01%; MR4.5; BCR-ABL1IS ≤ 0.032%; MR5.0; BCR-ABL1IS ≤ 0.001%) [3]. Although this means the evaluation of treatments requires the measurement of BCR-ABL1 mRNA by qRT-PCR, it should be considered that the quantification of BCR-ABL1 mRNA is not correlated with CML conditions in some cases. Yuda et al. reported BCR-ABLIns35bp that are BCR-ABL1 mRNA positive but functionally dead. They also reported that quite a few CML patients are estimated of BCR-ABL1IS due to BCR-ABLIns35bp [28]. The RT-PCR method may be negative if the breakpoint of BCR or ABL is outside the primer setting range or if it is atypical of CML. Furthermore, when it comes to measure BCR-ABL1 mRNA using peripheral blood, the stability of RNA is drastically decreased more than MR4.5 [29]. There is no doubt of the importance of BCR-ABL1 mRNA measurement; however, it is necessary to consider other measurement methods in consideration of these detection limits.
Liquid biopsy is the new approach to detect EVs, circulating tumor cells and cell-free DNA/RNA in body fluids. As various tissues secrete EVs to circulating blood, we can obtain those materials from small amounts of bodily fluids. EVs are carrying the same membrane molecules as the originated cells. Thus, we can identify and purify specified cell-origin EVs. Since EVs contain protein and nucleic acids including miRNAs, EVs are expected to be ideal biomarkers for use in many clinical applications [4,30,31]. Since tumor cells secrete EVs more than normal cells, there are many tumor-derived EVs in plasma of patients diagnosed with tumor [32]. In terms of function of EVs in vivo, they are involved in metastasis, angiogenesis and drug resistance, so detection of EVs from various tumors are clinically valuable.
EVs released from CML cells have an important role in cell-to-cell communication. Umezu et al. showed that EVs from K562 cells enhanced endothelial cell migration and tube formation by transporting miR-92a into the endothelial cell [33]. EVs from K562 cells also transfer miR-365, which induces drug-resistance in CML cells [34]. Our microarray study did not show significant changes in the two miRNAs after TKI treatments (Figure 2A). On the other hand, our data using qRT-PCR also showed the drastic down-regulation of EV-miR-494 of CML cell lines after in vitro treatment with TKIs (Figure 4), but the amount of secreted EVs did not change by TKIs (Figure 1). Furthermore, the amounts of miRNA in cells did not significantly change with TKIs treatment. It was considered that the level of EV-miR-494 reflect the change of selective inclusion capability of miRNA to EVs. This means the inclusion of miR-494 to EVs may be decreased by the inhibition of tyrosine kinase activity and correlated with the proliferation of CML cells. Our data also showed that miR-494 was up-regulated in patients with an accelerated or blast crisis phase, suggesting that miR-494 correlates with progression of the disease. MiR-494 was reported to target PTEN [35] and to induce cell proliferation, invasion and migration in solid tumor [36,37]. In hematopoietic malignancy, overexpressed miR-494 suppresses drug resistance in acute myeloid leukemia by downregulating c-Myc [38]. Salati et al. reported that the down-regulation of miR-494 leads to c-Myc up-regulation and decreases apoptosis by TKI in CML cells [39]. PTEN inhibits AKT activation via PI3K, and PTEN inhibition by miR-494 is thought to cause an increase in AKT activation, which leads to cell proliferation. Furthermore, c-Myc increases the miR-17-92 cluster, which inhibits PTEN [40]. Taken together, these reports suggest that miR-494 is involved in PTEN/AKT/c-Myc signaling. This is the first report about the correlation between hematopoietic malignancy and miR-494 in EVs, although they have been reported in cardiac disease [41] and malignant melanoma [42]. MiR-373-5p is one of the miR-371/372/373 clusters on chromosome 19 and is reported as a tumor suppressor miRNA targeting NF-κB and TGF-β signaling [43], DNMT1A [44] or oncogenes in testicular germ cell tumors [45], esophageal cancer [46]. miR-373-5p is seen to act either as an oncogene or as a tumor suppressor [47]. In our study, although miR-373-5p in CML-EVs was decreased after TKIs, its reduction ratio was almost 0.5-fold compared to non TKIs-treated EVs (Figure 3).
Abnormal profiles of circulating miRNAs have been reported in various human diseases including metabolic diseases such as diabetes, cardiovascular disease, and various tumors. Its availability is considered unquestionable, and clinical trials with the therapeutic agents have already been conducted [48,49,50]. Changes of circulating miRNAs have been reported in various hematopoietic malignancies: acute myeloid leukemia [51], acute lymphoblastic leukemia [52,53], chronic myeloid leukemia [54,55,56], chronic lymphocytic leukemia [57,58], and multiple myeloma [59]. Although miR-155 in serum is a candidate of biomarker to diagnosis of various hematopoietic malignancy, it also changes at diabetes [60] and inflammation [61]. If we analyze the whole miRNA in serum, including EVs-miRNAs and non-EV- miRNAs, it is important to consider the heterogeneity of miRNA origins, namely various tissues in addition to tumor cells. Our data suggest that EV-miRNAs are more sensitive biomarkers than miRNAs in the whole serum (Figure 4B). This approach may be a potential tool for using miRNAs as a biomarker.
The amount of released EVs depends on the cell type, but it is reported to be 2–3 × 1010 particles/mL in the supernatant of various cells cultured in 100 mm culture dishes (<10 × 108 cells at confluent) [62]. This means that one cell in culture releases hundreds to thousands of EVs for a few days. Even if the patient is in a deep remission state, EVs circulating throughout the body including bone marrow might capture the pathological changes of patients. In our results, it was possible to distinguish between CML-AP/BC and CML-CP (MR4.0–MR5.0) patients with EVs obtained from 250 µL serum (Figure 4), although further study about sample volume may be needed to determine whether MR4.0, MR4.5, and MR5.0 can be differentiated from each other. Tissue or cell-lineage-specific purified EVs and the analysis of miRNAs may be recommended for more accurate and early diagnosis. Caivano et al. [63] and Cerisoli et al. [64] reported increased CD13-positive EVs in some myeloid tumors, including CML. They also reported that CD19-positive EVs are increased in B cell lymphoma and that CD30-positive EVs are increased in Hodgkin lymphoma. Thus, it may be possible to increase sensitivity by collecting these EVs with magnetic bead conjugated specific antibodies and measure the characteristic EV-miRNAs. Although we showed that miR-494 was significantly changed in the total serum EVs of CML patients, the analysis may be more sensitive by using myeloid-EVs.
In conclusion, our data suggest that miR-494 in circulating EVs may be a useful biomarker for monitoring CML. Further studies are necessary to decide the cut-off value of miR-494 and confirm whether its value changes in other diseases by using larger cohorts of CML clinical samples.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/hemato3020026/s1, Figure S1: Cell viability analysis of imatinib or dasatinib treatment.
Author Contributions
Conceptualization, T.S., T.U.; methodology and software, T.S.; validation, H.S., A.F., S.T.; formal analysis, H.S., A.F., S.T.; investigation, T.S.; resources, T.S., Y.T. and T.U; data curation, H.S., A.F., S.T.; writing—original draft preparation, T.S.; writing—review and editing, T.U.; visualization, T.S.; supervision, T.S.; project administration, T.S.; funding acquisition, T.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by JSPS KAKENHI Grant Number 17H07059 and 17K09020.
Institutional Review Board Statement
This study has been complied with the institutional policies of The International University of Health and Welfare, Japan (Project No.:20-Ifh-053).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Druker, B.J.; Talpaz, M.; Resta, D.J.; Peng, B.; Buchdunger, E.; Ford, J.M.; Lydon, N.B.; Kantarjian, H.; Capdeville, R.; Ohno-Jones, S.; et al. Efficacy and Safety of a Specific Inhibitor of the BCR-ABL Tyrosine Kinase in Chronic Myeloid Leukemia. N. Engl. J. Med. 2001, 344, 1031–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Rafiyath, S.; Liu, D. First-line treatment for chronic myeloid leukemia: Dasatinib, nilotinib, or imatinib. J. Hematol. Oncol. 2010, 3, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.; Giusti, I.; D’Ascenzo, S.; Pizzorno, L.; Dolo, V. Breast Cancer Derived Extracellular Vesicles in Bone Metastasis Induction and Their Clinical Implications as Biomarkers. Int. J. Mol. Sci. 2020, 21, 3573. [Google Scholar] [CrossRef]
- Saviana, M.; Romano, G.; Le, P.; Acunzo, M.; Nana-Sinkam, P. Extracellular Vesicles in Lung Cancer Metastasis and Their Clinical Applications. Cancers 2021, 13, 5633. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Wei, S.; He, Z.; Huang, X.; Wang, L.; Xia, Y.; Xu, Z.; Li, Z.; Wang, W.; et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018, 9, 854. [Google Scholar] [CrossRef] [Green Version]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a src-dependent fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafarzadeh, N.; Safari, Z.; Pornour, M.; Amirizadeh, N.; Moghadam, M.F.; Sadeghizadeh, M. Alteration of cellular and immune-related properties of bone marrow mesenchymal stem cells and macrophages by K562 chronic myeloid leukemia cell derived exosomes. J. Cell. Physiol. 2019, 234, 3697–3710. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, H.; Umezu, T.; Ohyashiki, K.; Hirano, T.; Ohyashiki, J.H. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J. Biol. Chem. 2013, 288, 34343–34351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yip, K.W.; Spence, T.; Liu, F.-F. MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J. Hum. Genet. 2017, 62, 67–74. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.-L.; Cao, Y.; Liu, D.; Xu, M.; Liu, H.; Tang, R.-N.; Ma, K.-L.; Liu, B.-C. Isolation and Quantification of MicroRNAs from Urinary Exosomes/Microvesicles for Biomarker Discovery. Int. J. Biol. Sci. 2013, 9, 1021–1031. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015, 6, 8474–8490. [Google Scholar] [CrossRef] [Green Version]
- Lozzio, C.; Lozzio, B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 1975, 45, 321–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi, K. A new leukemia cell line with philadelphia chromosome characterized as basophil precursors. Leuk. Res. 1985, 9, 381–390. [Google Scholar] [CrossRef]
- Deregowska, A.; Pepek, M.; Pruszczyk, K.; Machnicki, M.M.; Wnuk, M.; Stoklosa, T. Differential Regulation of Telomeric Complex by BCR-ABL1 Kinase in Human Cellular Models of Chronic Myeloid Leukemia—From Single Cell Analysis to Next-Generation Sequencing. Genes 2020, 11, e1145. [Google Scholar] [CrossRef]
- Hehlmann, R.; Voskanyan, A.; Lauseker, M.; Pfirrmann, M.; Kalmanti, L.; Rinaldetti, S.; Kohlbrenner, K.; Haferlach, C.; Schlegelberger, B.; Fabarius, A.; et al. Correction: High-risk additional chromosomal abnormalities at low blast counts herald death by CML. Leukemia 2020, 34, 2823. [Google Scholar] [CrossRef] [PubMed]
- Pfirrmann, M.; Clark, R.E.; Prejzner, W.; Lauseker, M.; Baccarani, M.; Saussele, S.; Guilhot, F.; Heibl, S.; Hehlmann, R.; Faber, E.; et al. The EUTOS long-term survival (ELTS) score is superior to the Sokal score for predicting survival in chronic myeloid leukemia. Leukemia 2020, 34, 2138–2149. [Google Scholar] [CrossRef] [PubMed]
- Hehlmann, R. The New ELN Recommendations for Treating CML. J. Clin. Med. 2020, 9, e3671. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Iriyama, N.; Tokuhira, M.; Takaku, T.; Ishikawa, M.; Nakazato, T.; Sugimoto, K.; Fujita, H.; Kimura, Y.; Fujioka, I.; et al. The EUTOS long-term survival score predicts disease-specific mortality and molecular responses among patients with chronic myeloid leukemia in a practice-based cohort. Cancer Med. 2020, 9, 8931–8939. [Google Scholar] [CrossRef]
- Yuda, J.; Odawara, J.; Minami, M.; Muta, T.; Kohno, K.; Tanimoto, K.; Eto, T.; Shima, T.; Kikushige, Y.; Kato, K.; et al. Tyrosine kinase inhibitors induce alternative spliced BCR-ABL Ins35bp variant via inhibition of RNA polymerase II on genomic BCR-ABL. Cancer Sci. 2020, 111, 2361–2373. [Google Scholar] [CrossRef]
- Brown, J.T.; Beldorth, I.J.; Laosinchai-Wolf, W.; Fahey, M.E.; Jefferson, K.L.; Ruskin, A.K.; Roth, J.J.; Cai, L.; Watt, C.D.; Press, R.D.; et al. Analytical Validation of a Highly Sensitive, Multiplexed Chronic Myeloid Leukemia Monitoring System Targeting BCR-ABL1 RNA. J. Mol. Diagn. 2019, 21, 718–733. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Tableros, V.; Gomez, Y.; Camussi, G.; Brizzi, M.F. Extracellular Vesicles: New Players in Lymphomas. Int. J. Mol. Sci. 2018, 20, 41. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Ruhen, O.; Meehan, K. Tumor-Derived Extracellular Vesicles as a Novel Source of Protein Biomarkers for Cancer Diagnosis and Monitoring. Proteomics 2019, 19, e1800155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezu, T.; Ohyashiki, K.; Kuroda, M.I.; Ohyashiki, J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 2013, 32, 2747–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, Q.-H.; Wang, X.-Z.; Zhang, J.; Chen, Q.-G.; Li, S.-Q.; Liu, X.-Q.; Li, J.; Yang, W.-M.; Jiang, Y.-H.; Xu, Y.-M.; et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp. Cell Res. 2018, 362, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, G.; Wang, L.; Wang, X.; Jin, X.; Bo, W. miR-494 promotes progression of retinoblastoma via PTEN through PI3K/AKT signaling pathway. Oncol. Lett. 2020, 20, 1952–1960. [Google Scholar] [CrossRef]
- Shan, G.; Tang, T.; Xia, Y.; Qian, H. MEG3 interacted with miR-494 to repress bladder cancer progression through targeting PTEN. J. Cell. Physiol. 2020, 235, 1120–1128. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhao, M.; Lin, H.; Wang, W.; Li, D.; Cui, W.; Zhou, C.; Zhong, J.; Huang, C. MiR-494 acts as a tumor promoter by targeting CASP2 in non-small cell lung cancer. Sci. Rep. 2019, 9, 3008. [Google Scholar] [CrossRef]
- Tian, C.; Zheng, G.; Zhuang, H.; Li, X.; Hu, D.; Zhu, L.; Wang, T.; You, M.J.; Zhang, Y. MicroRNA-494 Activation Suppresses Bone Marrow Stromal Cell-Mediated Drug Resistance in Acute Myeloid Leukemia Cells. J. Cell. Physiol. 2017, 232, 1387–1395. [Google Scholar] [CrossRef]
- Salati, S.; Salvestrini, V.; Carretta, C.; Genovese, E.; Rontauroli, S.; Zini, R.; Rossi, C.; Ruberti, S.; Bianchi, E.; Barbieri, G.; et al. Deregulated expression of miR-29a-3p, miR-494-3p and miR-660-5p affects sensitivity to tyrosine kinase inhibitors in CML leukemic stem cells. Oncotarget 2017, 8, 49451–49469. [Google Scholar] [CrossRef] [Green Version]
- Benhamou, D.; Labi, V.; Getahun, A.; Benchetrit, E.; Dowery, R.; Rajewsky, K.; Cambier, J.C.; Melamed, D. The c-Myc/miR17-92/PTEN Axis Tunes PI3K Activity to Control Expression of Recombination Activating Genes in Early B Cell Development. Front. Immunol. 2018, 9, 2715. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Yuan, J.; Gao, W.; Zhong, X.; Yao, K.; Lin, L.; Ge, J. Dendritic cell-derived exosomal miR-494-3p promotes angiogenesis following myocardial infarction. Int. J. Mol. Med. 2020, 47, 315–325. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Wang, S.; Li, P.; Zheng, C.; Zhou, X.; Tao, Y.; Chen, X.; Sun, L.; Wang, A.; et al. Blockage of transferred exosome-shuttled miR-494 inhibits melanoma growth and metastasis. J. Cell. Physiol. 2019, 234, 15763–15774. [Google Scholar] [CrossRef] [PubMed]
- Keklikoglou, I.; Koerner, C.; Schmidt, C.; Zhang, J.D.; Heckmann, D.; Shavinskaya, A.; Allgayer, H.; Guckel, B.; Fehm, T.F.; Schneeweiss, A.; et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 2011, 31, 4150–4163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Estève, P.-O.; Chin, H.G.; Terragni, J.; Dai, N.; Corrêa, I.R.; Pradhan, S. Small RNA-mediated DNA (cytosine-5) methyltransferase 1 inhibition leads to aberrant DNA methylation. Nucleic Acids Res. 2015, 43, 6112–6124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.-P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A Genetic Screen Implicates miRNA-372 and miRNA-373 As Oncogenes in Testicular Germ Cell Tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef] [Green Version]
- Gillis, A.J.M.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W.; Sun, Y.; Chen, C.; Guenther, S.; Sherlock, J.; Veltman, I.; Baeten, J.; et al. High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 2007, 213, 319–328. [Google Scholar] [CrossRef]
- Shah, J.A.; Khattak, S.; Rauf, M.A.; Cai, Y.; Jin, J. Potential Biomarkers of miR-371–373 Gene Cluster in Tumorigenesis. Life 2021, 11, 984. [Google Scholar] [CrossRef]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef]
- Yoshida, K.; Yokoi, A.; Kato, T.; Ochiya, T.; Yamamoto, Y. The clinical impact of intra- and extracellular miRNAs in ovarian cancer. Cancer Sci. 2020, 111, 3435–3444. [Google Scholar] [CrossRef]
- Robelin, P.; Tod, M.; Colomban, O.; Lachuer, J.; Ray-Coquard, I.; De Rauglaudre, G.; Joly, F.; Chevalier-Place, A.; Combe, P.; Lortholary, A.; et al. Comparative analysis of predictive values of the kinetics of 11 circulating miRNAs and of CA125 in ovarian cancer during first line treatment (a GINECO study). Gynecol. Oncol. 2020, 159, 256–263. [Google Scholar] [CrossRef]
- Zhi, F.; Cao, X.; Xie, X.; Wang, B.; Dong, W.; Gu, W.; Ling, Y.; Wang, R.; Yang, Y.; Liu, Y. Identification of Circulating MicroRNAs as Potential Biomarkers for Detecting Acute Myeloid Leukemia. PLoS ONE 2013, 8, e56718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, S.; Lu, J.; Sun, M.; Li, Z.; Zhang, H.; Neilly, M.B.; Wang, Y.; Qian, Z.; Jin, J.; Zhang, Y.; et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luna-Aguirre, C.M.; Martinez-Fierro, M.D.L.L.; Mar-Aguilar, F.; Garza-Veloz, I.; Treviño-Alvarado, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomarkers 2015, 15, 299–310. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Y.; Han, X.; Roy, M.; Liu, W.; Zhao, X.; Liu, J. Differential expression profiles and functional analysis of plasma miRNAs associated with chronic myeloid leukemia phases. Future Oncol. 2019, 15, 763–776. [Google Scholar] [CrossRef]
- Ohyashiki, K.; Umezu, T.; Katagiri, S.; Kobayashi, C.; Azuma, K.; Tauchi, T.; Okabe, S.; Fukuoka, Y.; Ohyashiki, J.H. Downregulation of Plasma miR-215 in Chronic Myeloid Leukemia Patients with Successful Discontinuation of Imatinib. Int. J. Mol. Sci. 2016, 17, 570. [Google Scholar] [CrossRef]
- Keramati, F.; Jafarian, A.; Soltani, A.; Javandoost, E.; Mollaei, M.; Fallah, P. Circulating miRNAs can serve as potential diagnostic biomarkers in chronic myelogenous leukemia patients. Leuk. Res. Rep. 2021, 16, 100257. [Google Scholar] [CrossRef] [PubMed]
- Casabonne, D.; Benavente, Y.; Seifert, J.; Costas, L.; Armesto, M.; Arestin, M.; Besson, C.; Hosnijeh, F.S.; Duell, E.J.; Weiderpass, E.; et al. Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int. J. Cancer 2020, 147, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Farzadfard, E.; Kalantari, T.; Tamaddon, G. Serum Expression of Seven MicroRNAs in Chronic Lymphocytic Leukemia Patients. J. Blood Med. 2020, 11, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Akhbari, M.; Khalili, M.; Shahrabi-Farahani, M.; Biglari, A.; Bandarian, F. Expression Level of Circulating Cell Free miR-155 Gene in Serum of Patients with Diabetic Nephropathy. Clin. Lab. 2019, 65, 169–174. [Google Scholar] [CrossRef]
- Tili, E.; Croce, C.M.; Michaille, J.-J. miR-155: On the Crosstalk Between Inflammation and Cancer. Int. Rev. Immunol. 2009, 28, 264–284. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, E.M.; Vestad, B.; Steffensen, L.A.; Aass, H.C.D.; Saeed, M.; Øvstebø, R.; Costea, D.-E.; Galtung, H.K.; Søland, T.M. Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS ONE 2018, 13, e0204276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caivano, A.; Laurenzana, I.; De Luca, L.; La Rocca, F.; Simeon, V.; Trino, S.; D’Auria, F.; Traficante, A.; Maietti, M.; Izzo, T.; et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumor Biol. 2015, 36, 9739–9752. [Google Scholar] [CrossRef] [PubMed]
- Cerisoli, S.; Busilacchi, E.M.; Mattiucci, D.; Rossi, E.; Mariani, M.; Guescini, M.; Pugnaloni, A.; Olivieri, F.; Olivieri, A.; Poloni, A. The exosomal surface phenotype and inflamma-miR cargo correlate with MDS diagnosis. Br. J. Haematol. 2021, 192, e4–e7. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).