Comparing Handcrafted Radiomics Versus Latent Deep Learning Features of Admission Head CT for Hemorrhagic Stroke Outcome Prediction
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients’ Datasets
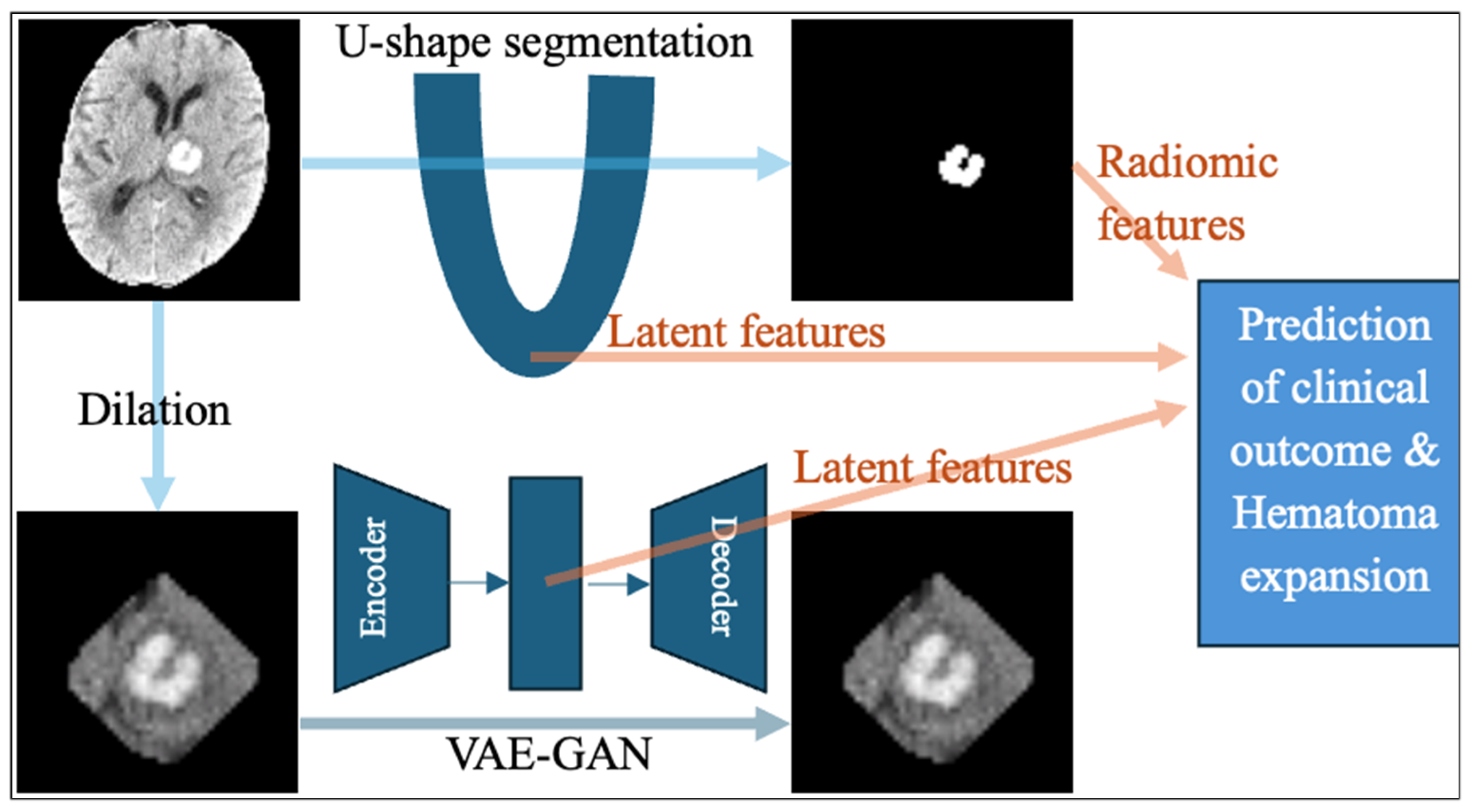
2.2. The U-Shaped Hematoma Segmentation Model
- Preprocessing: Standardized intensity normalization, resampling to isotropic voxel spacing, and automatic cropping based on region of interest.
- Network Architecture: A fully convolutional encoder–decoder model with residual blocks and deep supervision for improved gradient flow.
- Training Strategy: Dice loss and cross-entropy loss are combined to address class imbalance, ensuring accurate segmentation of small hematomas.
2.3. Extraction of Handcrafted Radiomic Features
- Shape-based Features: Quantifying the geometry of the region of interest (ROI), such as volume, surface area, and sphericity.
- First-order Statistics (Intensity-based): Quantifying the distribution of voxel intensities within the ROI, such as mean, median, variance, skewness, kurtosis, entropy, and energy.
- Texture Features (second-order and higher): Capturing spatial relationships between lesion voxels, such as Gray-Level Co-occurrence Matrix (GLCM), and Gray-Level Run Length Matrix (GLRLM).
- Wavelet/Filter-based Features: Applying transforms such as wavelet or Laplacian of Gaussian to reveal multiscale features.
2.4. Extraction of Latent Deep Learning Features from nnU-Net
2.5. Extraction of Latent Deep Features from a Generative Adversarial Network Autoencoder
2.6. Unsupervised Feature Selection
2.7. Machine Learning Prediction Models
2.8. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
3.2. Automated Hematoma Segmentation Performance
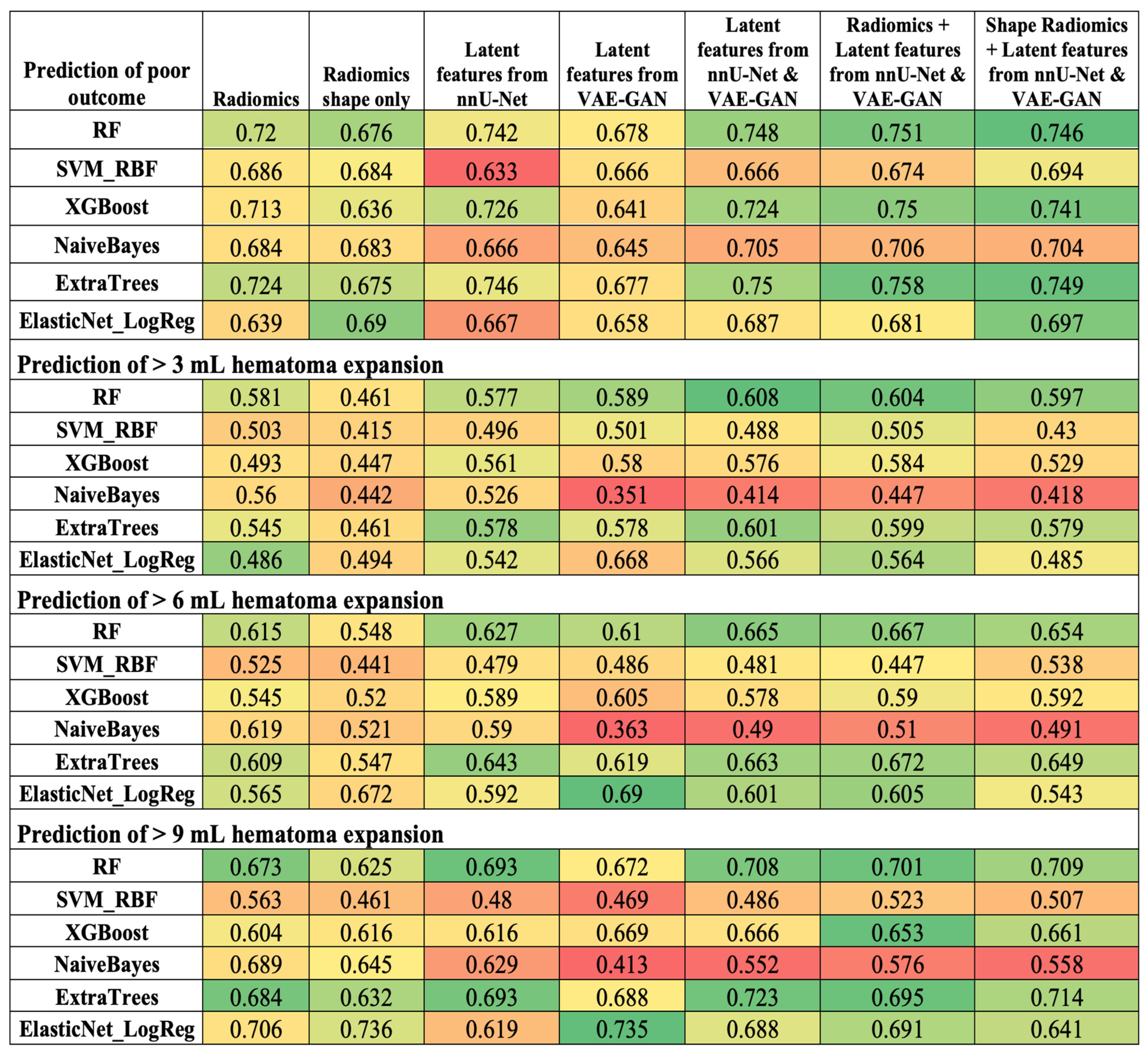
3.3. Comparison of Radiomics and Latent Deep Features in ICH Outcome Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATACH-2 | Antihypertensive Treatment of Acute Cerebral Hemorrhage II |
| AUC | Area under the curve |
| CNN | Convolutional Neural Networks |
| DALYs | Disability-adjusted life years |
| FDR | False Discovery Rate |
| GLRLM | Gray-Level Run Length Matrix |
| GLCM | Gray-Level Co-occurrence Matrix |
| ICH | Intracerebral hemorrhage |
| NMF | Non-negative Matrix Factorization |
| RBF | Radial basis function |
| ROC | Receiver operating characteristics |
| ROI | Region of interest |
| RF | Random Forest |
| SVM | Support Vector Machine |
| VAE-GAN | Variational Autoencoder–Generative Adversarial Network |
| SHAP | Shapley Additive Interpretation |
References
- Mariotti, F.; Agostini, A.; Borgheresi, A.; Marchegiani, M.; Zannotti, A.; Giacomelli, G.; Pierpaoli, L.; Tola, E.; Galiffa, E.; Giovagnoni, A. Insights into radiomics: A comprehensive review for beginners. Clin. Transl. Oncol. 2025, 27, 4091–4102. [Google Scholar] [CrossRef]
- Vrettos, K.; Triantafyllou, M.; Marias, K.; Karantanas, A.H.; Klontzas, M.E. Artificial intelligence-driven radiomics: Developing valuable radiomics signatures with the use of artificial intelligence. BJR Artif. Intell. 2024, 1, ubae011. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Chen, J.; Ye, Z.; Zhang, R.; Li, H.; Fang, B.; Zhang, L.B.; Wang, W. Medical image translation with deep learning: Advances, datasets and perspectives. Med. Image Anal. 2025, 103, 103605. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zheng, H.; Zou, H.; Luo, D.; Tang, H.; Li, L.; Jiang, B. A comprehensive review of deep learning for medical image segmentation. Neurocomputing 2025, 613, 128740. [Google Scholar] [CrossRef]
- Zhijin He, A.B.M. Comparative Evaluation of Radiomics and Deep Learning Models for Disease Detection in Chest Radiography. arXiv 2025, arXiv:2504.12249. [Google Scholar] [CrossRef]
- Shariaty, F.; Pavlov, V.; Baranov, M. AI-Driven Precision Oncology: Integrating Deep Learning, Radiomics, and Genomic Analysis for Enhanced Lung Cancer Diagnosis and Treatment. Signal Image Video Process. 2025, 19, 693. [Google Scholar] [CrossRef]
- Buvat, I.; Dutta, J.; Jha, A.K.; Siegel, E.; Yousefirizi, F.; Rahmim, A.; Bradshaw, T. Should end-to-end deep learning replace handcrafted radiomics? Eur. J. Nucl. Med. Mol. Imaging 2025, 52, 4360–4363. [Google Scholar] [CrossRef]
- Jain, A.; Pandey, M.; Sahu, S. A Deep Learning-Based Feature Extraction Model for Classification Brain Tumor. In Lecture Notes on Data Engineering and Communications Technologies, Proceedings of the Data Analytics and Management, Virtual, 25–26 June 2021; Springer Nature: Singapore, 2022; pp. 493–508. [Google Scholar]
- Sage, A.; Badura, P. Intracranial Hemorrhage Detection in Head CT Using Double-Branch Convolutional Neural Network, Support Vector Machine, and Random Forest. Appl. Sci. 2020, 10, 7577. [Google Scholar] [CrossRef]
- Ertuğrul, Ö.F.; Akıl, M.F. Detecting hemorrhage types and bounding box of hemorrhage by deep learning. Biomed. Signal Process. Control 2022, 71, 103085. [Google Scholar] [CrossRef]
- Bijari, S.; Sayfollahi, S.; Mardokh-Rouhani, S.; Bijari, S.; Moradian, S.; Zahiri, Z.; Rezaeijo, S.M. Radiomics and Deep Features: Robust Classification of Brain Hemorrhages and Reproducibility Analysis Using a 3D Autoencoder Neural Network. Bioengineering 2024, 11, 643. [Google Scholar] [CrossRef] [PubMed]
- Sasagasako, T.; Ueda, A.; Mineharu, Y.; Mochizuki, Y.; Doi, S.; Park, S.; Terada, Y.; Sano, N.; Tanji, M.; Arakawa, Y.; et al. Postoperative Karnofsky performance status prediction in patients with IDH wild-type glioblastoma: A multimodal approach integrating clinical and deep imaging features. PLoS ONE 2024, 19, e0303002. [Google Scholar] [CrossRef] [PubMed]
- Suero Molina, E.; Azemi, G.; Ozdemir, Z.; Russo, C.; Krahling, H.; Valls Chavarria, A.; Liu, S.; Stummer, W.; Di Ieva, A. Predicting intraoperative 5-ALA-induced tumor fluorescence via MRI and deep learning in gliomas with radiographic lower-grade characteristics. J. Neurooncol. 2025, 171, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Denes-Fazakas, L.; Kovacs, L.; Eigner, G.; Szilagyi, L. Enhancing Brain Tumor Diagnosis with L-Net: A Novel Deep Learning Approach for MRI Image Segmentation and Classification. Biomedicines 2024, 12, 2388. [Google Scholar] [CrossRef]
- Rai, H.M.; Yoo, J.; Dashkevych, S. Two-headed UNetEfficientNets for parallel execution of segmentation and classification of brain tumors: Incorporating postprocessing techniques with connected component labelling. J. Cancer Res. Clin. Oncol. 2024, 150, 220. [Google Scholar] [CrossRef]
- Lv, C.; Shu, X.J.; Liang, Q.; Qiu, J.; Xiong, Z.C.; Ye, J.B.; Li, S.B.; Liu, C.Q.; Niu, J.Z.; Chen, S.B.; et al. BrainTumNet: Multi-task deep learning framework for brain tumor segmentation and classification using adaptive masked transformers. Front. Oncol. 2025, 15, 1585891. [Google Scholar] [CrossRef]
- Kihira, S.; Mei, X.; Mahmoudi, K.; Liu, Z.; Dogra, S.; Belani, P.; Tsankova, N.; Hormigo, A.; Fayad, Z.A.; Doshi, A.; et al. U-Net Based Segmentation and Characterization of Gliomas. Cancers 2022, 14, 4457. [Google Scholar] [CrossRef]
- Cheng, J.; Gao, M.; Liu, J.; Yue, H.; Kuang, H.; Liu, J.; Wang, J. Multimodal Disentangled Variational Autoencoder with Game Theoretic Interpretability for Glioma Grading. IEEE J. Biomed. Health Inform. 2022, 26, 673–684. [Google Scholar] [CrossRef]
- Yathirajam, S.S.; Gutta, S. Efficient glioma grade prediction using learned features extracted from convolutional neural networks. J. Med. Artif. Intell. 2024, 7. Available online: https://jmai.amegroups.org/article/view/8452/html (accessed on 1 October 2025). [CrossRef]
- Abd El Kader, I.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Ahmad, I.S.; Kamhi, S. Brain Tumor Detection and Classification on MR Images by a Deep Wavelet Auto-Encoder Model. Diagnostics 2021, 11, 1589. [Google Scholar] [CrossRef]
- Ullah, M.S.; Khan, M.A.; Almujally, N.A.; Alhaisoni, M.; Akram, T.; Shabaz, M. BrainNet: A fusion assisted novel optimal framework of residual blocks and stacked autoencoders for multimodal brain tumor classification. Sci. Rep. 2024, 14, 5895. [Google Scholar] [CrossRef]
- Sandeep Waghere, S.; Prashant Shinde, J. A robust classification of brain tumor disease in MRI using twin-attention based dense convolutional auto-encoder. Biomed. Signal Process. Control 2024, 92, 106088. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, F.; Zhao, T.; Han, J.; Wang, Y.; Wu, H.; Zhang, K.; Qiu, H.; Ding, Y.; Zhu, H. Brain tumor intelligent diagnosis based on Auto-Encoder and U-Net feature extraction. PLoS ONE 2025, 20, e0315631. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Sun, J.; You, Q.; Palade, V.; Mao, Z. Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 2022, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Kordnoori, S.; Sabeti, M.; Shakoor, M.H.; Moradi, E. Deep multi-task learning structure for segmentation and classification of supratentorial brain tumors in MR images. Interdiscip. Neurosurg. 2024, 36, 101931. [Google Scholar] [CrossRef]
- Li, G.; Hui, X.; Li, W.; Luo, Y. Multitask Learning with Multiscale Residual Attention for Brain Tumor Segmentation and Classification. Mach. Intell. Res. 2023, 20, 897–908. [Google Scholar] [CrossRef]
- Parry-Jones, A.R.; Krishnamurthi, R.; Ziai, W.C.; Shoamanesh, A.; Wu, S.; Martins, S.O.; Anderson, C.S. World Stroke Organization (WSO): Global intracerebral hemorrhage factsheet 2025. Int. J. Stroke 2025, 20, 145–150. [Google Scholar] [CrossRef]
- Chen, S.; Fan, J.; Abdollahi, A.; Ashrafi, N.; Alaei, K.; Placencia, G.; Pishgar, M. Machine Learning-Based Prediction of ICU Readmissions in Intracerebral Hemorrhage Patients: Insights from the MIMIC Databases. Medrxiv 2025. [Google Scholar] [CrossRef]
- Yu, F.; Yang, M.; He, C.; Yang, Y.; Peng, Y.; Yang, H.; Lu, H.; Liu, H. CT radiomics combined with clinical and radiological factors predict hematoma expansion in hypertensive intracerebral hemorrhage. Eur. Radiol. 2025, 35, 6–19. [Google Scholar] [CrossRef]
- Zeng, W.; Chen, J.; Shen, L.; Xia, G.; Xie, J.; Zheng, S.; He, Z.; Deng, L.; Guo, Y.; Yang, J.; et al. Clinical, radiological, and radiomics feature-based explainable machine learning models for prediction of neurological deterioration and 90-day outcomes in mild intracerebral hemorrhage. BMC Med. Imaging 2025, 25, 184. [Google Scholar] [CrossRef]
- Dierksen, F.; Sommer, J.K.; Tran, A.T.; Lin, H.; Haider, S.P.; Maier, I.L.; Aneja, S.; Sanelli, P.C.; Malhotra, A.; Qureshi, A.I.; et al. Machine Learning Models for 3-Month Outcome Prediction Using Radiomics of Intracerebral Hemorrhage and Perihematomal Edema from Admission Head Computed Tomography (CT). Diagnostics 2024, 14, 2827. [Google Scholar] [CrossRef]
- Haider, S.P.; Qureshi, A.I.; Jain, A.; Tharmaseelan, H.; Berson, E.R.; Zeevi, T.; Werring, D.J.; Gross, M.; Mak, A.; Malhotra, A.; et al. Radiomic markers of intracerebral hemorrhage expansion on non-contrast CT: Independent validation and comparison with visual markers. Front. Neurosci. 2023, 17, 1225342. [Google Scholar] [CrossRef]
- Zaman, S.; Dierksen, F.; Knapp, A.; Haider, S.P.; Abou Karam, G.; Qureshi, A.I.; Falcone, G.J.; Sheth, K.N.; Payabvash, S. Radiomic Features of Acute Cerebral Hemorrhage on Non-Contrast CT Associated with Patient Survival. Diagnostics 2024, 14, 944. [Google Scholar] [CrossRef]
- Jain, A.; Malhotra, A.; Payabvash, S. Imaging of Spontaneous Intracerebral Hemorrhage. Neuroimaging Clin. N. Am. 2021, 31, 193–203. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, D.; Liu, J.; Zhang, M.; Xu, H.; Xiang, Y.; Zhan, C.; Zhang, Y.; Huang, S.; Yang, Y. Clinical-radiomics Nomogram for Risk Estimation of Early Hematoma Expansion after Acute Intracerebral Hemorrhage. Acad. Radiol. 2021, 28, 307–317. [Google Scholar] [CrossRef]
- Lu, M.; Wang, Y.; Tian, J.; Feng, H. Application of deep learning and radiomics in the prediction of hematoma expansion in intracerebral hemorrhage: A fully automated hybrid approach. Diagn. Interv. Radiol. 2024, 30, 299–312. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, Y.; Niyazi, T.; Wei, J.; Guocai, G.; Liu, J.; Liang, S.; Liang, F.; Yan, P.; Wang, K.; et al. Radiomics for predicting hematoma expansion in patients with hypertensive intraparenchymal hematomas. Eur. J. Radiol. 2019, 115, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Pszczolkowski, S.; Manzano-Patron, J.P.; Law, Z.K.; Krishnan, K.; Ali, A.; Bath, P.M.; Sprigg, N.; Dineen, R.A. Quantitative CT radiomics-based models for prediction of haematoma expansion and poor functional outcome in primary intracerebral haemorrhage. Eur. Radiol. 2021, 31, 7945–7959. [Google Scholar] [CrossRef]
- Xie, H.; Ma, S.; Wang, X.; Zhang, X. Noncontrast computer tomography-based radiomics model for predicting intracerebral hemorrhage expansion: Preliminary findings and comparison with conventional radiological model. Eur. Radiol. 2020, 30, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Melmed, K.R.; Frontera, J.; Zhu, W.; Huang, H.; Qureshi, A.I.; Maggard, A.; Steinhof, M.; Kuohn, L.; Kumar, A.; et al. Predicting hematoma expansion after intracerebral hemorrhage: A comparison of clinician prediction with deep learning radiomics models. Neurocrit. Care 2025, 43, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.B.L.; Sønderby, S.K.; Larochelle, H.; Winther, O. Autoencoding beyond pixels using a learned similarity metric. In Proceedings of the Proceedings of the 33rd International Conference on Machine Learning (ICML), New York, NY, USA, 19 June 2016; pp. 1558–1566. [Google Scholar]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Ge, H. TBUnet: A Pure Convolutional U-Net Capable of Multifaceted Feature Extraction for Medical Image Segmentation. J. Med. Syst. 2023, 47, 122. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Y.; Yao, L.; Adeli, E.; Zhang, Y.; Wang, X. Generative adversarial U-Net for domain-free few-shot medical diagnosis. Pattern Recognit. Lett. 2022, 157, 112–118. [Google Scholar] [CrossRef]
- Skandarani, Y.; Jodoin, P.M.; Lalande, A. GANs for Medical Image Synthesis: An Empirical Study. J. Imaging 2023, 9, 69. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Qureshi, A.I.; Palesch, Y.Y.; Barsan, W.G.; Hanley, D.F.; Hsu, C.Y.; Martin, R.L.; Moy, C.S.; Silbergleit, R.; Steiner, T.; Suarez, J.I.; et al. Intensive Blood-Pressure Lowering in Patients with Acute Cerebral Hemorrhage. N. Engl. J. Med. 2016, 375, 1033–1043. [Google Scholar] [CrossRef]
- Torres-Lopez, V.M.; Rovenolt, G.E.; Olcese, A.J.; Garcia, G.E.; Chacko, S.M.; Robinson, A.; Gaiser, E.; Acosta, J.; Herman, A.L.; Kuohn, L.R.; et al. Development and Validation of a Model to Identify Critical Brain Injuries Using Natural Language Processing of Text Computed Tomography Reports. JAMA Netw. Open 2022, 5, e2227109. [Google Scholar] [CrossRef]
- Su, R.; Zhang, D.; Liu, J.; Cheng, C. MSU-Net: Multi-Scale U-Net for 2D Medical Image Segmentation. Front. Genet. 2021, 12, 639930. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Avery, E.; Sanelli, P.C.; Aboian, M.; Payabvash, S. Radiomics: A Primer on Processing Workflow and Analysis. Semin. Ultrasound CT MR 2022, 43, 142–146. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Solorio-Fernández, S.; Carrasco-Ochoa, J.A.; Martínez-Trinidad, J.F. A review of unsupervised feature selection methods. Artif. Intell. Rev. 2019, 53, 907–948. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Tianqi Chen, C.G. XGBoost: A Scalable Tree Boosting System. In Proceedings of the Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Andrew McCallum, K.N. A Comparison of Event Models for Naive Bayes Text Classification. In Proceedings of the AAAI-98 Workshop on Learning for Text Categorization, Madison, WI, USA, 26–27 July 1998; pp. 41–48. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection Via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Meng, L.; Jiang, X.Y.; Liu, X.Y.; Fan, J.H.; Ren, H.R.; Guo, Y.; Diao, H.K.; Wang, Z.H.; Chen, C.; Dai, C.Y.; et al. User-Tailored Hand Gesture Recognition System for Wearable Prosthesis and Armband Based on Surface Electromyogram. IEEE Trans. Instrum. Meas. 2022, 71, 1–16. [Google Scholar] [CrossRef]
- Zeng, Z.; Tao, L.; Su, R.; Zhu, Y.; Meng, L.; Tuheti, A.; Huang, H.; Shu, F.; Chen, W.; Chen, C. Unsupervised Transfer Learning Approach with Adaptive Reweighting and Resampling Strategy for Inter-subject EOG-based Gaze Angle Estimation. IEEE J. Biomed. Health Inform. 2023, 28, 157–168. [Google Scholar] [CrossRef]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate-a Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, J.; Gong, X.; Li, F. Skin lesion segmentation with a multiscale input fusion U-Net incorporating Res2-SE and pyramid dilated convolution. Sci. Rep. 2025, 15, 7975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Jin, Y.; Xu, J.; Xu, X. MDU-Net: Multi-scale densely connected U-Net for biomedical image segmentation. Health Inform. Sci. Syst. 2023, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.R.; Na, H.; Jung, S.; Lee, S.; Jeon, J.; Ahn, S.J. Hematoma expansion prediction in patients with intracerebral hemorrhage using a deep learning approach. J. Med. Artif. Intell. 2024, 7, 10. [Google Scholar] [CrossRef]
- Teng, L.; Ren, Q.; Zhang, P.; Wu, Z.; Guo, W.; Ren, T. Artificial Intelligence Can Effectively Predict Early Hematoma Expansion of Intracerebral Hemorrhage Analyzing Noncontrast Computed Tomography Image. Front. Aging Neurosci. 2021, 13, 632138. [Google Scholar] [CrossRef]
- Zhong, J.W.; Jin, Y.J.; Song, Z.J.; Lin, B.; Lu, X.H.; Chen, F.; Tong, L.S. Deep learning for automatically predicting early haematoma expansion in Chinese patients. Stroke Vasc. Neurol. 2021, 6, 610–614. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.; Jang, J.; Hwang, I.; Choi, K.S.; Park, J.H.; Chung, J.W.; Choi, S.H. Predicting hematoma expansion in acute spontaneous intracerebral hemorrhage: Integrating clinical factors with a multitask deep learning model for non-contrast head CT. Neuroradiology 2024, 66, 577–587. [Google Scholar] [CrossRef]



| ATACH-2 (n = 866) | Yale (n = 645) | p Values | ||
|---|---|---|---|---|
| 3-month poor outcome | 316 (36.5%) | 301 (46.7%) | <0.001 | |
| >3 mL hematoma expansion | 98 (11.3%) | 163 (25.3%) | <0.001 | |
| >6 mL hematoma expansion | 79 (9.1%) | 122 (18.9%) | <0.001 | |
| >9 mL hematoma expansion | 53 (6.1%) | 97 (15.0%) | <0.001 | |
| Sex [male] | 528 (60.9%) | 354 (54.9%) | 0.020 | |
| Age [years] | 62.1 ± 12.9 | 69.6 ± 14.4 | <0.001 | |
| Hispanic | 69 (8.0%) | 329 (52.2%) | <0.001 | |
| Race | White | 241 | 440 | <0.001 |
| Black | 110 | 125 | ||
| Asian | 489 | 17 | ||
| Other | 26 | 63 | ||
| Systolic blood pressure [mmHg] | 175.2 ± 25.1 | 172.9 ± 32.9 | 0.147 | |
| History of hypertension | 690 (79.7%) | 548 (85.0%) | 0.010 | |
| History of diabetes mellitus | 166 (19.2%) | 173 (26.8%) | <0.001 | |
| History of hyperlipidemia | 213 (24.6%) | 346 (53.6%) | <0.001 | |
| History of atrial fibrillation | 29 (3.4%) | 139 (21.6%) | <0.001 | |
| Baseline Glasgow Coma Scale | 3–11 | 127 (14.7%) | 179 (27.8%) | <0.001 |
| 12–14 | 242 (27.9%) | 168 (26.1%) | ||
| 15 | 497 (57.4%) | 270 (41.9%) | ||
| unknown | 28 (4.3%) | |||
| Baseline NIH Stroke Scale score | 0–4 | 137 (15.8%) | 181 (28.1%) | <0.001 |
| 5–9 | 226 (26.1%) | 110 (17.1%) | ||
| 10–14 | 235 (27.1%) | 76 (11.8%) | ||
| 15–19 | 159 (18.4%) | 86 (13.3%) | ||
| 20–25 | 77 (8.9%) | 67 (10.4%) | ||
| >25 | 27 (3.1%) | 38 (5.9%) | ||
| unknown | 5 (0.6%) | 87 (13.5%) | ||
| Baseline hematoma volume [mL] | 13.1 ± 12.6 | 18.7 ± 20.7 | <0.001 | |
| Follow-up hematoma volume [mL] | 15.8 ± 16.7 | 23.0 ± 25.9 | <0.001 | |
| CT scans | Slice thickness [mm] | 5.3 ± 1.8 | 4.8 ± 0.7 | <0.001 |
| Min axial image matrix [n x n] | [418 × 418] | [472 × 472] | ||
| Max axial matrix [n x n] | [512 × 734] | [1024 × 1024] | ||
| Number of slices | 31.0 ± 18.0 | 35.1 ± 11.5 | <0.001 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, A.T.; Wen, J.; Abou Karam, G.; Zeevi, D.; Qureshi, A.I.; Malhotra, A.; Majidi, S.; Valizadeh, N.; Murthy, S.B.; Sabuncu, M.R.; et al. Comparing Handcrafted Radiomics Versus Latent Deep Learning Features of Admission Head CT for Hemorrhagic Stroke Outcome Prediction. BioTech 2025, 14, 87. https://doi.org/10.3390/biotech14040087
Tran AT, Wen J, Abou Karam G, Zeevi D, Qureshi AI, Malhotra A, Majidi S, Valizadeh N, Murthy SB, Sabuncu MR, et al. Comparing Handcrafted Radiomics Versus Latent Deep Learning Features of Admission Head CT for Hemorrhagic Stroke Outcome Prediction. BioTech. 2025; 14(4):87. https://doi.org/10.3390/biotech14040087
Chicago/Turabian StyleTran, Anh T., Junhao Wen, Gaby Abou Karam, Dorin Zeevi, Adnan I. Qureshi, Ajay Malhotra, Shahram Majidi, Niloufar Valizadeh, Santosh B. Murthy, Mert R. Sabuncu, and et al. 2025. "Comparing Handcrafted Radiomics Versus Latent Deep Learning Features of Admission Head CT for Hemorrhagic Stroke Outcome Prediction" BioTech 14, no. 4: 87. https://doi.org/10.3390/biotech14040087
APA StyleTran, A. T., Wen, J., Abou Karam, G., Zeevi, D., Qureshi, A. I., Malhotra, A., Majidi, S., Valizadeh, N., Murthy, S. B., Sabuncu, M. R., Roh, D., Falcone, G. J., Sheth, K. N., & Payabvash, S. (2025). Comparing Handcrafted Radiomics Versus Latent Deep Learning Features of Admission Head CT for Hemorrhagic Stroke Outcome Prediction. BioTech, 14(4), 87. https://doi.org/10.3390/biotech14040087







