Exploring Genetic Diversity and Population Structure of Australian Passion Fruit Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction
2.3. Genotyping and Data Processing
2.4. Marker Quality and Genetic Diversity Analysis
2.5. Population Structure Analysis
3. Results
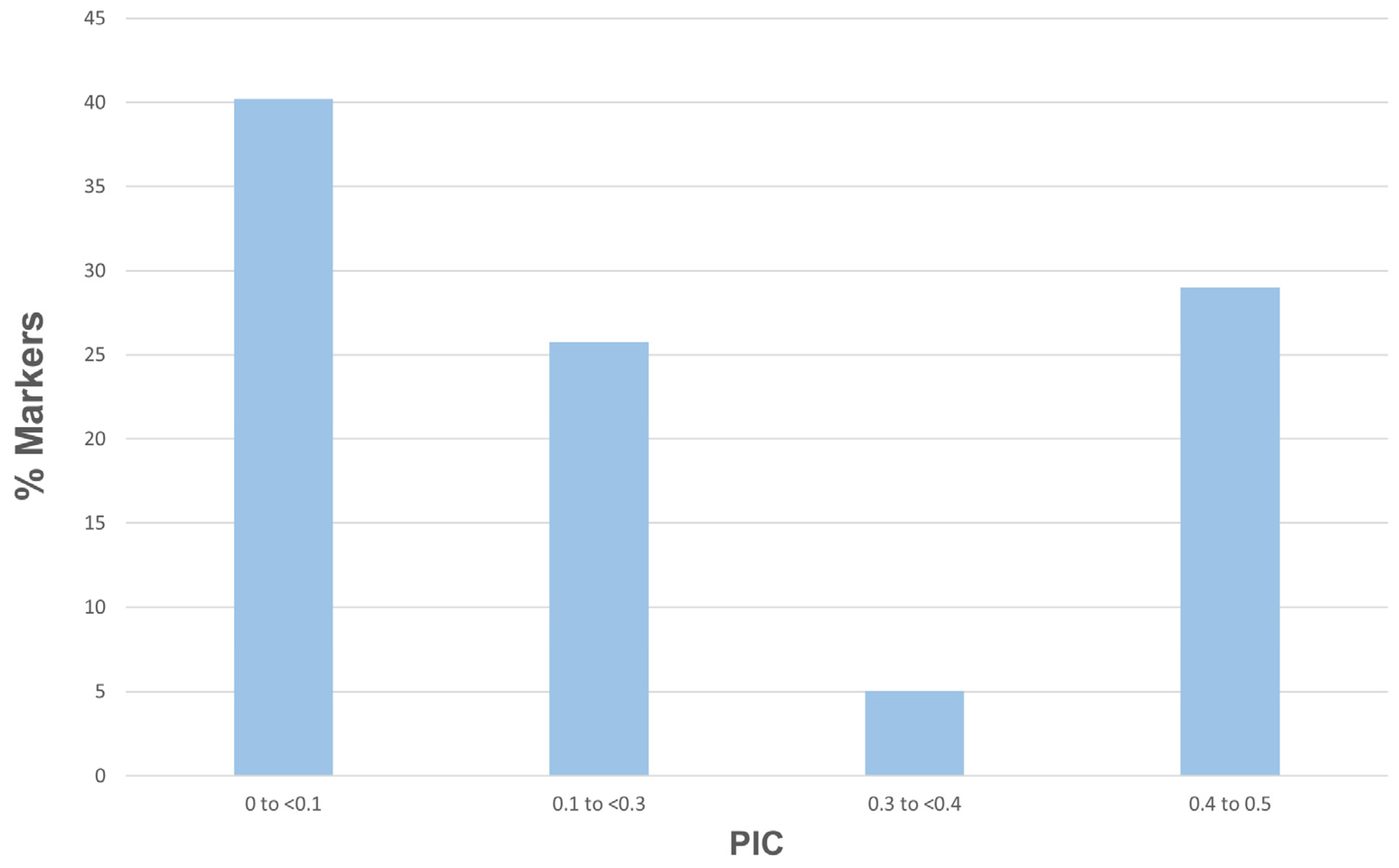
3.1. Genetic Properties of SNP Markers
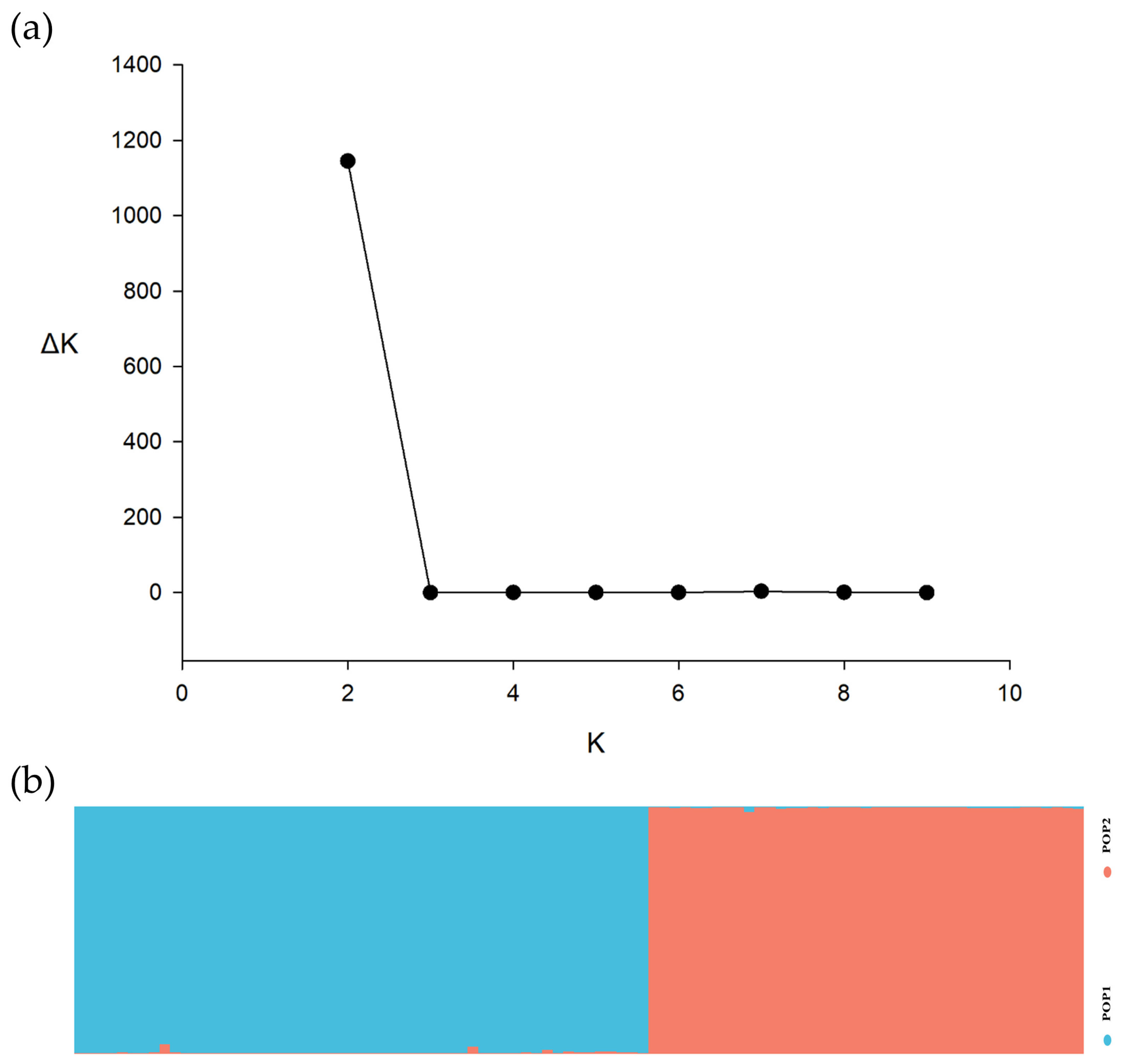
3.2. Population Structure and Genetic Diversity Analysis
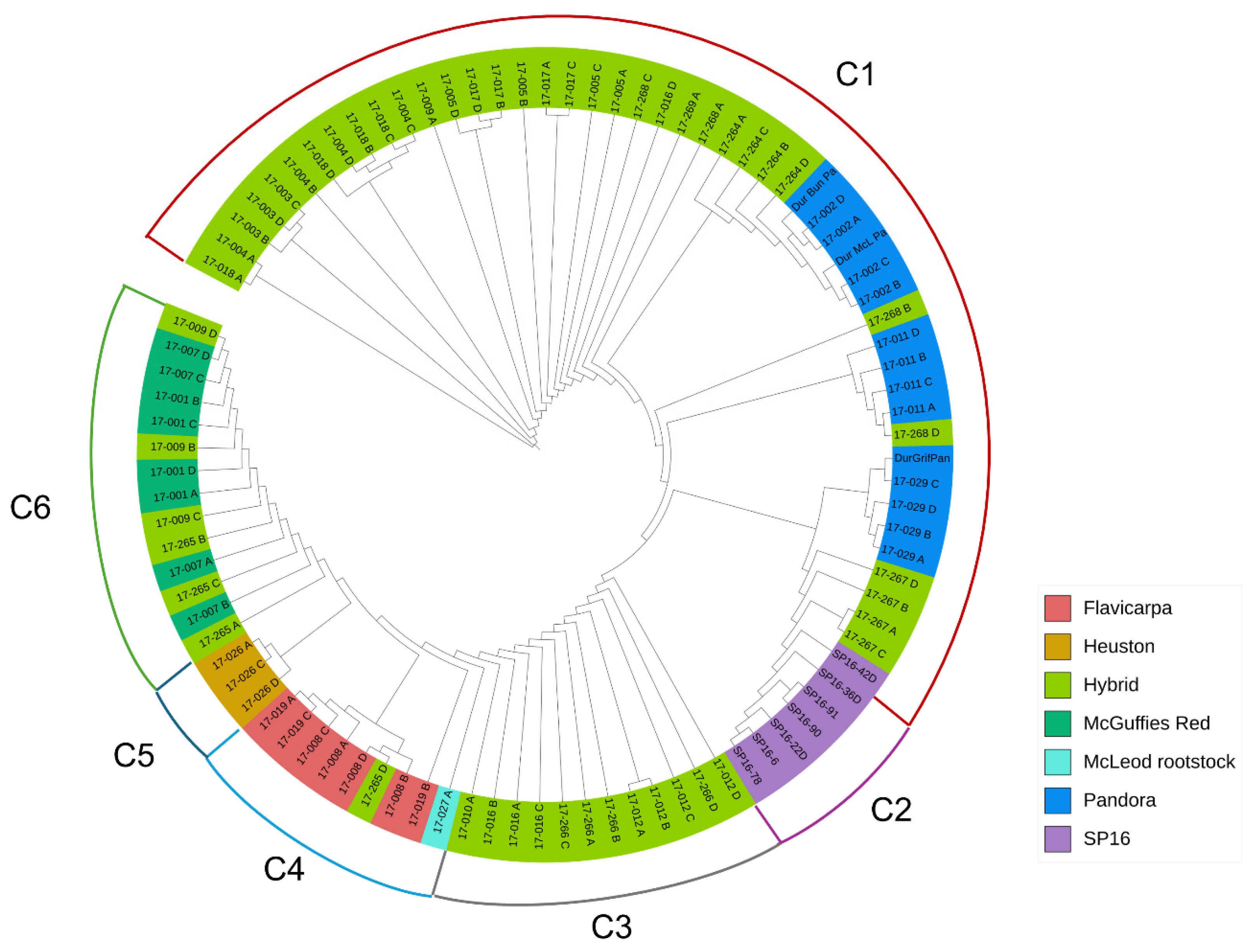
3.3. Genetic Relationships and Hierarchical Clustering
4. Discussion
4.1. DArT SNP Marker Platform in Characterization of Passiflora
4.2. Analysis of Population Structure and Genetic Diversity
4.3. Implications on Breeding Efforts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CTAB | Cetyltrimethylammonium bromide |
| DArT | Diversity Arrays Technology |
| ISSR | Inter Simple Sequence Repeat |
| RAPD | Random Amplified Polymorphic DNA |
| SNP | Single-nucleotide polymorphism |
| SSR | Simple sequence repeat |
References
- He, X.; Luan, F.; Yang, Y.; Wang, Z.; Zhao, Z.; Fang, J.; Wang, M.; Zuo, M.; Li, Y. Passiflora edulis: An insight into current researches on phytochemistry and pharmacology. Front. Pharmacol. 2020, 11, 617. [Google Scholar] [CrossRef] [PubMed]
- Oluoch, P.; Nyaboga, E.N.; Bargul, J.L. Analysis of genetic diversity of passion fruit (Passiflora edulis Sims) genotypes grown in Kenya by sequence-related amplified polymorphism (SRAP) markers. Ann. Agrar. Sci. 2018, 16, 367–375. [Google Scholar] [CrossRef]
- Weyya, G.; Belay, A.; Tadesse, E. Passion fruit (Passiflora edulis Sims) by-products as a source of bioactive compounds for non-communicable disease prevention: Extraction methods and mechanisms of action: A systematic review. Front. Nutr. 2024, 11, 1340511. [Google Scholar] [CrossRef]
- Australian Department of Agriculture, Fisheries and Forestry: Passionfruit from Vietnam. Available online: https://www.agriculture.gov.au/biosecurity-trade/policy/risk-analysis/plant/passionfruit-from-vietnam (accessed on 21 March 2025).
- Horticulture Innovation Australia: Passionfruit Strategic Investment Plan 2022–2026. Available online: https://www.horticulture.com.au/globalassets/hort-innovation/levy-fund-financial-and-management-documents/sip-2022-2026-pdfs/hort-innovation-sip-2022-26-passionfruit.pdf (accessed on 21 March 2025).
- Melo, N.J.d.A.; Negreiros, A.M.P.; Medeiros, H.L.d.S.; Sales Junior, R. Evaluation of Fusarium wilt disease in passion fruit species inoculated with Fusarium oxysporum f. sp. passiflorae. J. Phytopathol. 2020, 168, 81–87. [Google Scholar] [CrossRef]
- Greber, R. Passion-fruit woodiness virus as the cause of passion vine tip blight disease. Qld. J. Agric. Anim. Sci. 1966, 23, 533–538. [Google Scholar]
- de Oliveira Correia, A.; Alexandre, R.S.; Pfenning, L.H.; Cabanez, P.A.; Ferreira, A.; da Silva Ferreira, M.F.; de Lima, P.A.M.; de Mello, T.; Otoni, W.C.; Lopes, J.C. Passiflora mucronata, a passion fruit wild species resistant to fusariosis and a potential rootstock for commercial varieties. Sci. Hortic. 2022, 302, 111174. [Google Scholar] [CrossRef]
- Anderson, J. Genetic Improvement of Passionfruit to Achieve Improved Disease Resistance. Available online: https://era.dpi.qld.gov.au/id/eprint/2540/ (accessed on 21 March 2025).
- Rangare, N.; Bagare, V.; Kumar, R.; Kurmi, K.P.; Upadhyay, J.; Chowdhury, S.; Pathak, V.N.; Kumar, R. Rootstock Importance in Fruit Crops: A Comprehensive Review. Afr. J. Biomed. Res. 2024, 27, 1380–1386. [Google Scholar] [CrossRef]
- Ollat, N.; Peccoux, A.; Papura, D.; Esmenjaud, D.; Marguerit, E.; Tandonnet, J.P.; Bordenave, L.; Cookson, S.; Barrieu, F.; Rossdeutsch, L. Rootstocks as a component of adaptation to environment. In Grapevine in a Changing Environment: A Molecular and Ecophysiological Perspective; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 68–108. [Google Scholar]
- Rasool, A.; Mansoor, S.; Bhat, K.; Hassan, G.; Baba, T.R.; Alyemeni, M.N.; Alsahli, A.A.; El-Serehy, H.A.; Paray, B.A.; Ahmad, P. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants. Front. Plant Sci. 2020, 11, 590847. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.B.M.; Santos, E.S.; Jesus, O.N.; Vieira, J.G.; Mori, G.M.; Corrêa, R.X.; Souza, A.P. Molecular genetic variability of commercial and wild accessions of passion fruit (Passiflora spp.) targeting ex situ conservation and breeding. Int. J. Mol. Sci. 2014, 15, 22933–22959. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.; Cardoso-Silva, C.; Santos, E.; Conceição, L.; Pereira, A.; Oliveira, A.; Corrêa, R. Genetic diversity in wild species of passion fruit (Passiflora trintae) based on molecular markers. Genet. Mol. Res. 2010, 9, 2123–2130. [Google Scholar] [CrossRef]
- de Almeida Pereira, D.; Corrêa, R.X.; de Oliveira, A.C. Molecular genetic diversity and differentiation of populations of ‘somnus’ passion fruit trees (Passiflora setacea DC): Implications for conservation and pre-breeding. Biochem. Syst. Ecol. 2015, 59, 12–21. [Google Scholar] [CrossRef]
- de Moraes, M.C.; Geraldi, I.O.; de Pina Matta, F.; Vieira, M.L.C. Genetic and phenotypic parameter estimates for yield and fruit quality traits from a single wide cross in yellow passion fruit. HortScience 2005, 40, 1978–1981. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.; Jesus, O.; Oliveira, E.J.; Santos, E.S.; Souza, A.P. Characterization and selection of passion fruit (yellow and purple) accessions based on molecular markers and disease reactions for use in breeding programs. Euphytica 2015, 202, 345–359. [Google Scholar] [CrossRef]
- Castro, J.; Oliveira, E.; Jesus, O.; Soares, T.; Margarido, G.R.A. Molecular markers for conservation genetic resources of four Passiflora species. Sci. Hortic. 2016, 212, 251–261. [Google Scholar] [CrossRef]
- Chepkoech, E.; Rotich, F.; Alkamoi, B. Assessment of genetic variability of passion fruit using simple sequence repeat (SSR) markers. J. Agric. Sci. Pract. 2020, 5, 5. [Google Scholar] [CrossRef]
- Junior, E.M.; Rosado, L.D.S.; Costa, A.C.; Caixeta, E.T.; Dos Santos, C.E.M. Full-sib progenies show greater genetic diversity than half-sib progenies in sour passion fruit: An approach by ssr markers. Mol. Biol. Rep. 2023, 50, 4133–4144. [Google Scholar] [CrossRef]
- Ocampo, J.; Acosta-Barón, N.; Hernández-Fernández, J. Variability and genetic structure of yellow passion fruit (Passiflora edulis f. flavicarpa Degener) in Colombia using microsatellite DNA markers. Agron. Colomb. 2017, 35, 135–149. [Google Scholar] [CrossRef]
- Coronado, A.C.M.; Camargo, M.A.M.; Coronado, Y.M. Genetic diversity pattern of Passiflora spp. in Boyacá, Colombia. Pesqui. Agropecuária Bras. 2023, 58, e03062. [Google Scholar] [CrossRef]
- Costa, J.L.; Jesus, O.N.d.; Oliveira, G.A.F.; Oliveira, E.J.d. Effect of selection on genetic variability in yellow passion fruit. Crop Breed. Appl. Biotechnol. 2012, 12, 253–260. [Google Scholar] [CrossRef]
- Ho, V.; Ngo, T.; Phan, T.; Ta, T.; Tran, T. Genetic diversity among passion fruit (Passiflora edulis) accessions of Southern Vietnam using RAPD and ISSR markers. SABRAO J. Breed. Genet. 2021, 53, 1–14. [Google Scholar]
- Aukar, A.P.d.A.; Lemos, E.G.d.M.; Oliveira, J.C. Genetic variations among passion fruit species using RAPD markers. Rev. Bras. De Frutic. 2002, 24, 738–740. [Google Scholar] [CrossRef]
- Campos, A.V.S.; Peixoto, J.R.; Faleiro, F.G.; Vilela, M.S.; Pires, M. Molecular characterization and genetic diversity of yellow passion fruit based on RAPD markers. J. Agric. Sci. 2019, 11, 575–580. [Google Scholar] [CrossRef]
- Anderson, J.D.; Vidal, R.F.; Brym, M.; Stafne, E.T.; Resende Jr, M.F.; Viana, A.P.; Chambers, A.H. Genotyping-by-sequencing of passion fruit (Passiflora spp.) generates genomic resources for breeding and systematics. Genet. Resour. Crop Evol. 2022, 69, 2769–2786. [Google Scholar] [CrossRef]
- Rodriguez Castillo, N.C.; Wu, X.; Chacón, M.I.; Melgarejo, L.M.; Blair, M.W. Genetic diversity of purple passion fruit, Passiflora edulis f. edulis, based on single-nucleotide polymorphism markers discovered through genotyping by sequencing. Diversity 2021, 13, 144. [Google Scholar]
- Brhane, H.; Hammenhag, C. Genetic diversity and population structure analysis of a diverse panel of pea (Pisum sativum). Front. Genet. 2024, 15, 1396888. [Google Scholar] [CrossRef]
- Peterson, G.W.; Dong, Y.; Horbach, C.; Fu, Y.-B. Genotyping-by-sequencing for plant genetic diversity analysis: A lab guide for SNP genotyping. Diversity 2014, 6, 665–680. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C. Diversity arrays technology: A generic genome profiling technology on open platforms. In Data Production and Analysis in Population Genomics: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2012; pp. 67–89. [Google Scholar]
- Alam, M.; Neal, J.; O’Connor, K.; Kilian, A.; Topp, B. Ultra-high-throughput DArTseq-based silicoDArT and SNP markers for genomic studies in macadamia. PLoS ONE 2018, 13, e0203465. [Google Scholar] [CrossRef]
- Alemu, A.; Brantestam, A.K.; Chawade, A. Unraveling the genetic basis of key agronomic traits of wrinkled vining pea (Pisum sativum L.) for sustainable production. Front. Plant Sci. 2022, 13, 844450. [Google Scholar] [CrossRef]
- Mace, E.S.; Xia, L.; Jordan, D.R.; Halloran, K.; Parh, D.K.; Huttner, E.; Wenzl, P.; Kilian, A. DArT markers: Diversity analyses and mapping in Sorghum bicolor. BMC Genom. 2008, 9, 26. [Google Scholar] [CrossRef]
- Sánchez-Sevilla, J.F.; Horvath, A.; Botella, M.A.; Gaston, A.; Folta, K.; Kilian, A.; Denoyes, B.; Amaya, I. Diversity arrays technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria × ananassa). PLoS ONE 2015, 10, e0144960. [Google Scholar] [CrossRef]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef] [PubMed]
- Osundare, O.T.; Akinyele, B.O.; Odiyi, A.C.; Paliwal, R.; Oyatomi, O.A.; Abberton, M.T. Genetic diversity and population structure of some Nigerian accessions of Bambara groundnut (Vigna subterranea (L.) Verdc.) using DArT SNP markers. Genet. Resour. Crop Evol. 2023, 70, 887–901. [Google Scholar] [CrossRef]
- Gelaw, Y.M.; Eleblu, J.S.; Ofori, K.; Fenta, B.A.; Mukankusi, C.; Emam, E.A.; Offei, S. High-density DArTSeq SNP markers revealed wide genetic diversity and structured population in common bean (Phaseolus vulgaris L.) germplasm in Ethiopia. Mol. Biol. Rep. 2023, 50, 6739–6751. [Google Scholar] [CrossRef]
- Mason, A.S. SSR genotyping. In Plant Genotyping: Methods and Protocols; Springer Nature: Berlin/Heidelberg, Germany, 2015; pp. 77–89. [Google Scholar]
- Wu, F.; Cai, G.; Xi, P.; Guo, Y.; Xu, M.; Li, A. Genetic diversity analysis and fingerprint construction for 87 passionfruit (Passiflora spp.) germplasm accessions on the basis of SSR fluorescence markers. Int. J. Mol. Sci. 2024, 25, 10815. [Google Scholar] [CrossRef]
- Doyle, J. DNA protocols for plants. Molecular techniques in taxonomy. NATO ASI Ser. 1991, 57, 283–293. [Google Scholar]
- Wenzl, P.; Carling, J.; Kudrna, D.; Jaccoud, D.; Huttner, E.; Kleinhofs, A.; Kilian, A. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 2004, 101, 9915–9920. [Google Scholar] [CrossRef]
- Barilli, E.; Cobos, M.J.; Carrillo, E.; Kilian, A.; Carling, J.; Rubiales, D. A high-density integrated DArTseq SNP-based genetic map of Pisum fulvum and identification of QTLs controlling rust resistance. Front. Plant Sci. 2018, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Montilla-Bascón, G.; Rispail, N.; Sánchez-Martín, J.; Rubiales, D.; Mur, L.A.; Langdon, T.; Howarth, C.J.; Prats, E. Genome-wide association study for crown rust (Puccinia coronata f. sp. avenae) and powdery mildew (Blumeria graminis f. sp. avenae) resistance in an oat (Avena sativa) collection of commercial varieties and landraces. Front. Plant science 2015, 6, 103. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Padua, J.G.; Zucchi, M.I.; Camargo, L.E.A.; Fungaro, M.H.P.; Vieira, M.L.C. Development and characterization of microsatellite markers from the yellow passion fruit (Passiflora edulis f. flavicarpa). Mol. Ecol. Notes 2005, 5, 331–333. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Harris, A.M.; DeGiorgio, M. An Unbiased Estimator of Gene Diversity with Improved Variance for Samples Containing Related and Inbred Individuals of any Ploidy. G3-Genes Genomes Genet. 2017, 7, 671–691. [Google Scholar] [CrossRef]
- Coulon, A. GENHET: An easy-to-use R function to estimate individual heterozygosity. Mol. Ecol. Resour. 2010, 10, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Cortés, O.; Eusebi, P.; Dunner, S.; Sevane, N.; Cañón, J. Comparison of diversity parameters from SNP, microsatellites and pedigree records in the Lidia cattle breed. Livest. Sci. 2019, 219, 80–85. [Google Scholar] [CrossRef]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic diversity and population structure of a Camelina sativa spring panel. Front. Plant science 2019, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar] [PubMed]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010; Volume 1, p. 37. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Francis, R. pophelper: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Boca, S.M.; Rosenberg, N.A. Mathematical properties of Fst between admixed populations and their parental source populations. Theor. Popul. Biol. 2011, 80, 208–216. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Peeters, J.; Martinelli, J. Hierarchical cluster analysis as a tool to manage variation in germplasm collections. Theor. Appl. Genet. 1989, 78, 42–48. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Araya, S.; Martins, A.M.; Junqueira, N.T.; Costa, A.M.; Faleiro, F.G.; Ferreira, M.E. Microsatellite marker development by partial sequencing of the sour passion fruit genome (Passiflora edulis Sims). BMC Genom. 2017, 18, 549. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Salem, K.F.; Sallam, A. Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. Comptes Rendus Biol. 2016, 339, 1–9. [Google Scholar] [CrossRef]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.; Poland, J.; Baenziger, P.S. Genetic diversity and population structure of F3: 6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Shete, S.; Tiwari, H.; Elston, R.C. On estimating the heterozygosity and polymorphism information content value. Theor. Popul. Biol. 2000, 57, 265–271. [Google Scholar] [CrossRef]
- Ortiz, D.C.; Bohórquez, A.; Duque, M.C.; Tohme, J.; Cuéllar, D.; Mosquera Vásquez, T. Evaluating purple passion fruit (Passiflora edulis Sims f. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet. Resour. Crop Evol. 2012, 59, 1089–1099. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme diversity in cultivated crops. Crop Sci. 1997, 37, 26–30. [Google Scholar] [CrossRef]
- Morillo, A.; Muñoz, D.; Morillo, Y. Molecular characterization of Passiflora edulis f. Flavicarpa Degener with ISSRs markers. Braz. J. Biol. 2023, 83, e278167. [Google Scholar] [CrossRef] [PubMed]
- Asande, L.K.; Ombori, O.; Oduor, R.O.; Nchore, S.B.; Nyaboga, E.N. Occurrence of passion fruit woodiness disease in the coastal lowlands of Kenya and screening of passion fruit genotypes for resistance to passion fruit woodiness disease. BMC Plant Biol. 2023, 23, 544. [Google Scholar] [CrossRef] [PubMed]
- Fridman, E. Consequences of hybridization and heterozygosity on plant vigor and phenotypic stability. Plant Sci. 2015, 232, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.; Browne, C.; Guill, K.; Coles, N.; Eller, M.; Garcia, A.; Lepak, N.; Melia-Hancock, S.; Oropeza-Rosas, M.; Salvo, S. The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity 2012, 108, 490–499. [Google Scholar] [CrossRef]


| POP | Net Nucleotide Distance | Exp. Het | Mean Fst | Prop. Mem | ||||
|---|---|---|---|---|---|---|---|---|
| POP2 | POP3 | POP4 | POP5 | POP6 | ||||
| POP1 | 0.124 | 0.141 | 0.241 | 0.245 | 0.224 | 0.063 | 0.922 | 0.030 |
| POP2 | 0.121 | 0.158 | 0.095 | 0.030 | 0.386 | 0.014 | 0.003 | |
| POP3 | 0.241 | 0.205 | 0.217 | 0.015 | 0.960 | 0.184 | ||
| POP4 | 0.278 | 0.263 | 0.013 | 0.971 | 0.229 | |||
| POP5 | 0.123 | 0.028 | 0.934 | 0.492 | ||||
| POP6 | 0.328 | 0.252 | 0.062 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Bundock, P.; Mason, P.; Poudel, P.D.; Varshney, R.; Topp, B.; Alam, M. Exploring Genetic Diversity and Population Structure of Australian Passion Fruit Germplasm. BioTech 2025, 14, 37. https://doi.org/10.3390/biotech14020037
Sun X, Bundock P, Mason P, Poudel PD, Varshney R, Topp B, Alam M. Exploring Genetic Diversity and Population Structure of Australian Passion Fruit Germplasm. BioTech. 2025; 14(2):37. https://doi.org/10.3390/biotech14020037
Chicago/Turabian StyleSun, Xinhang, Peter Bundock, Patrick Mason, Pragya Dhakal Poudel, Rajeev Varshney, Bruce Topp, and Mobashwer Alam. 2025. "Exploring Genetic Diversity and Population Structure of Australian Passion Fruit Germplasm" BioTech 14, no. 2: 37. https://doi.org/10.3390/biotech14020037
APA StyleSun, X., Bundock, P., Mason, P., Poudel, P. D., Varshney, R., Topp, B., & Alam, M. (2025). Exploring Genetic Diversity and Population Structure of Australian Passion Fruit Germplasm. BioTech, 14(2), 37. https://doi.org/10.3390/biotech14020037








