Exploring DNA Damage and Repair Mechanisms: A Review with Computational Insights
Abstract
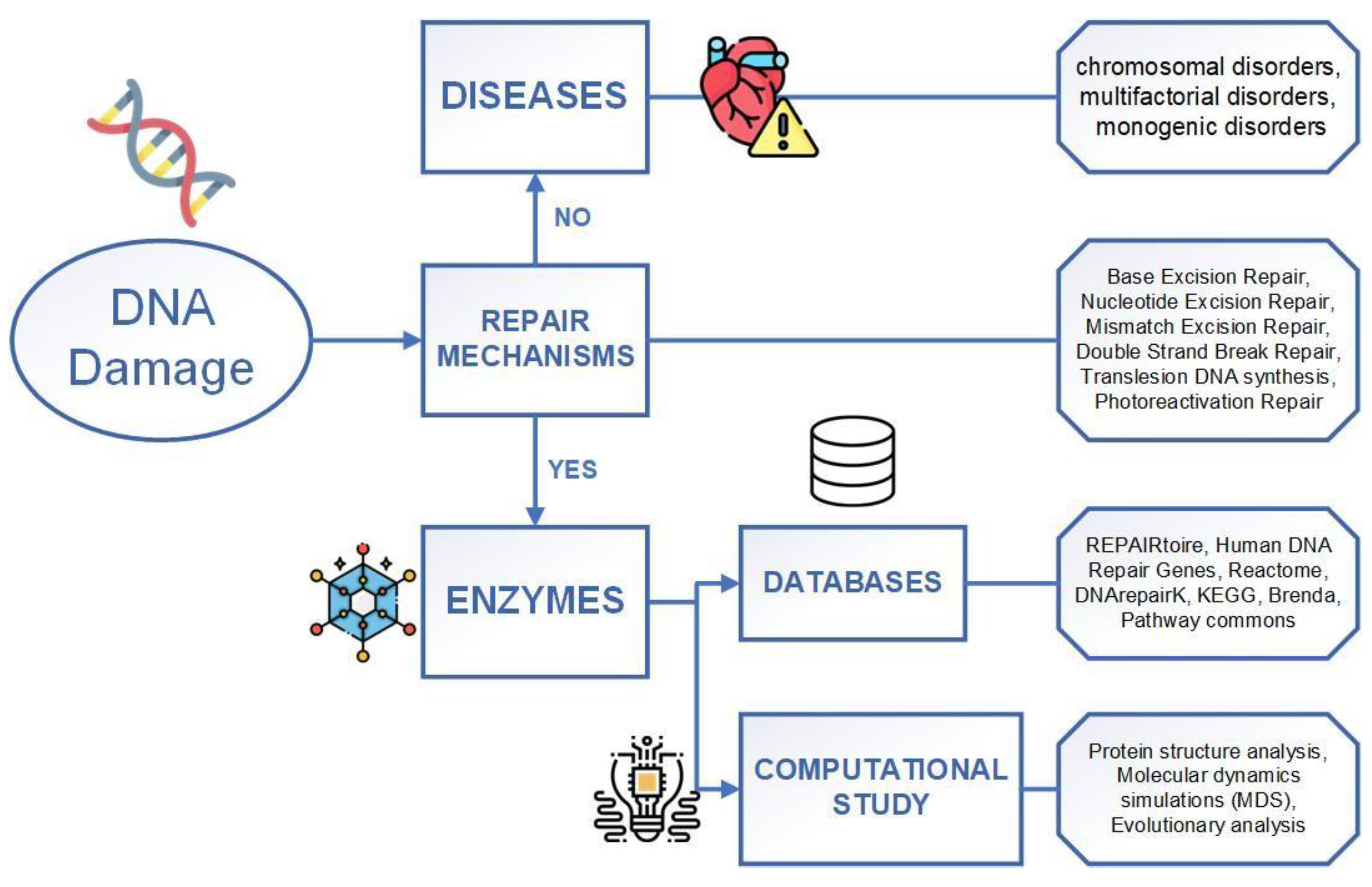
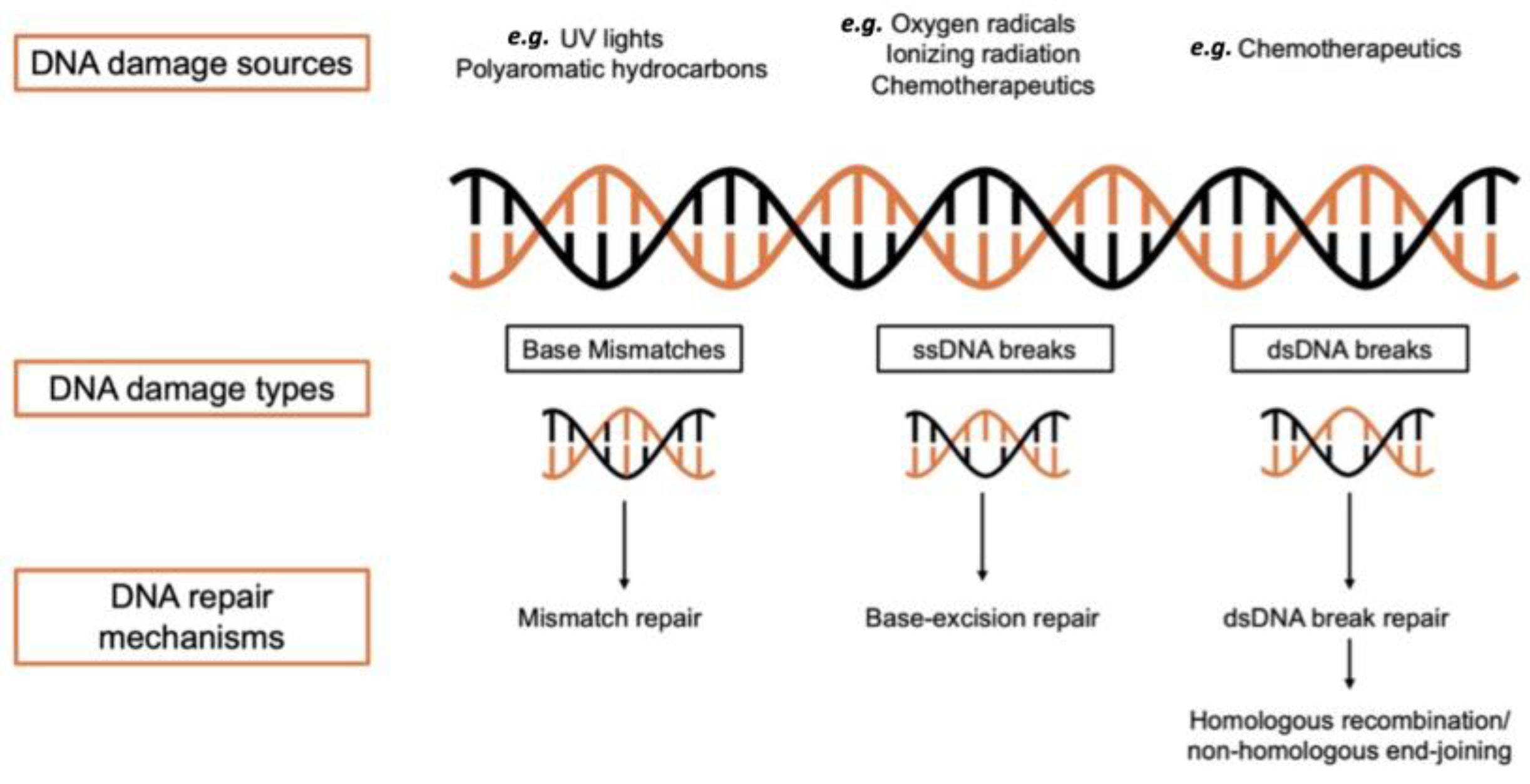
1. Introduction
2. DNA Repair Mechanisms
2.1. BER Mechanism
2.2. NER Mechanism
2.3. MMR Mechanism
3. DNA-Related Database
3.1. REPAIRtoire
3.2. Human DNA Repair Genes
3.3. Reactome
3.4. DNArepairK
3.5. KEGG
3.6. Brenda
3.7. Pathway Commons
4. DNA Repair Computational Research Methods
4.1. Protein Structure Analysis
4.2. Molecular Dynamics Simulations
- -
- Performing molecular dynamics simulations to study the dynamics of proteins involved in DNA repair processes, such as simulating the movement and interactions of DNA repair enzymes;
- -
- Studying the effects of genetic variations on the structures and functions of DNA repair proteins, such as simulating the impact of SNPs on the structure and function of DNA repair enzymes;
- -
- Predicting the binding process between small molecules and proteins involved in DNA repair to identify potential drug candidates for treating DNA repair-related diseases;
- -
- Identifying potential drug candidates for treating and studying the interactions between proteins involved in DNA repair processes, such as simulating the interactions between DNA repair enzymes and DNA damage response proteins.
4.3. Evolutionary Analysis
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Yang, Z.; Zhou, X.; Jin, M.; Dai, Z.; Ming, D.; Zhang, Z.; Zhu, L.; Jiang, L. Sequence, structure, and function of the Dps DNA-binding protein from Deinococcus wulumuqiensis R12. Microb. Cell Fact. 2022, 21, 132. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, Y.; Wei, Q.; Lin, H.; Li, X.; Yu, M.E.; Wang, J.; Huang, Z.; Xue, D. Structure and function encoding of a bidirectional activatable synergetic DNA machine for speeded and ultrasensitive determination of microRNAs. Talanta 2022, 238 Pt 1, 123037. [Google Scholar] [CrossRef]
- Trasvina-Arenas, C.H.; Demir, M.; Lin, W.J.; David, S.S. Structure, function and evolution of the Helix-hairpin-Helix DNA glycosylase superfamily: Piecing together the evolutionary puzzle of DNA base damage repair mechanisms. DNA Repair 2021, 108, 103231. [Google Scholar] [CrossRef]
- Said, H.; Kaji, I.; Kaunitz, J.D. Gastroduodenal mucosal defense mechanisms. Curr. Opin. Gastroenterol. 2015, 31, 486–491. [Google Scholar] [CrossRef]
- Li, Y.; Tang, W.; Guo, M. The Cell as Matter: Connecting Molecular Biology to Cellular Functions. Matter 2021, 4, 1863–1891. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef]
- Bian, K.; Lenz, S.A.P.; Tang, Q.; Chen, F.; Qi, R.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Wetmore, S.D.; Li, D. DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro. Nucleic Acids Res. 2019, 47, 5522–5529. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Andrew, S.E.; Peters, A.C. DNA instability and human disease. Am. J. Pharmacogenom. 2001, 1, 21–28. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef]
- Rass, U.; Ahel, I.; West, S.C. Defective DNA repair and neurodegenerative disease. Cell 2007, 130, 991–1004. [Google Scholar] [CrossRef]
- Mazurek, A.; Berardini, M.; Fishel, R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 2002, 277, 8260–8266. [Google Scholar] [CrossRef]
- Best, B.P. Nuclear DNA damage as a direct cause of aging. Rejuvenation Res. 2009, 12, 199–208. [Google Scholar] [CrossRef]
- Giacco, F.; Brownlee, M. Oxidative stress and diabetic complications. Circ. Res. 2010, 107, 1058–1070. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Meyenberg, M.; da Silva, J.F.; Loizou, J.I. Tissue Specific DNA Repair Outcomes Shape the Landscape of Genome Editing. Front. Genet. 2021, 12, 728520. [Google Scholar] [CrossRef]
- Sudeshna, N.; Amy, B.; Andris, Z.; Peteris, R.; Karlis, P.; Martins, O.; Edgars, C.; Juris, V.; Maria, K.; McCarthy, M.I.; et al. PASSIM—An open source software system for managing information in biomedical studies. BMC Bioinform. 2007, 8, 52. [Google Scholar]
- Grimes, S.M.; Ji, H.P. MendeLIMS: A web-based laboratory information management system for clinical genome sequencing. BMC Bioinform. 2014, 15, 290. [Google Scholar] [CrossRef]
- Alexandrov, N.; Alexandrov, V. Computational Science Research Methods for Science Education at PG Level. Procedia Comput. Sci. 2015, 51, 1685–1693. [Google Scholar] [CrossRef][Green Version]
- Milanowska, K.; Rother, K.; Bujnicki, J.M. Databases and bioinformatics tools for the study of DNA repair. Mol. Biol. Int. 2011, 2011, 475718. [Google Scholar] [CrossRef] [PubMed]
- Krwawicz, J.; Arczewska, K.D.; Speina, E.; Maciejewska, A.; Grzesiuk, E. Bacterial DNA repair genes and their eukaryotic homologues: 1. Mutations in genes involved in base excision repair (BER) and DNA-end processors and their implication in mutagenesis and human disease. Acta Biochim. Pol. 2007, 54, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Lindahl, T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef]
- Zentout, S.; Smith, R.; Jacquier, M.; Huet, S. New Methodologies to Study DNA Repair Processes in Space and Time Within Living Cells. Front. Cell Dev. Biol. 2021, 9, 730998. [Google Scholar] [CrossRef] [PubMed]
- Egly, J.M.; Coin, F. A history of TFIIH: Two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair 2011, 10, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.R. The molecular biology of nucleotide excision repair and double-strand break repair in eukaryotes. Genet. Eng. 1995, 17, 1–19. [Google Scholar]
- Wood, R.D.; Shivji, M.K. Which DNA polymerases are used for DNA-repair in eukaryotes? Carcinogenesis 1997, 18, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Leibeling, D.; Laspe, P.; Emmert, S. Nucleotide excision repair and cancer. J. Mol. Histol. 2006, 37, 225–238. [Google Scholar] [CrossRef]
- Hanawalt, P.C. Subpathways of nucleotide excision repair and their regulation. Oncogene 2002, 21, 8949–8956. [Google Scholar] [CrossRef]
- Gillet, L.C.; Scharer, O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006, 106, 253–276. [Google Scholar] [CrossRef]
- Saxowsky, T.T.; Doetsch, P.W. RNA polymerase encounters with DNA damage: Transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 2006, 106, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Gaul, L.; Svejstrup, J.Q. Transcription-coupled repair and the transcriptional response to UV-Irradiation. DNA Repair 2021, 107, 103208. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Kisker, C. Damage recognition in nucleotide excision DNA repair. Curr. Opin. Struct. Biol. 2012, 22, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K. The xeroderma pigmentosum group C protein complex and ultraviolet-damaged DNA-binding protein: Functional assays for damage recognition factors involved in global genome repair. Methods Enzymol. 2006, 408, 171–188. [Google Scholar]
- Tang, J.Y.; Hwang, B.J.; Ford, J.M.; Hanawalt, P.C.; Chu, G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 2000, 5, 737–744. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, D.; van der Weegen, Y.; Boer, D.E.C.; Ogi, T.; Luijsterburg, M.S. Transcription-Coupled DNA Repair: From Mechanism to Human Disorder. Trends Cell Biol. 2021, 31, 359–371. [Google Scholar] [CrossRef]
- Petruseva, I.O.; Evdokimov, A.N.; Lavrik, O.I. Molecular mechanism of global genome nucleotide excision repair. Acta Naturae 2014, 6, 23–34. [Google Scholar] [CrossRef] [PubMed]
- de Laat, W.L.; Appeldoorn, E.; Sugasawa, K.; Weterings, E.; Jaspers, N.G.; Hoeijmakers, J.H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes. Dev. 1998, 12, 2598–2609. [Google Scholar] [CrossRef]
- Sijbers, A.M.; de Laat, W.L.; Ariza, R.R.; Biggerstaff, M.; Wei, Y.F.; Moggs, J.G. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 1996, 86, 811–822. [Google Scholar] [CrossRef]
- Matsunaga, T.; Park, C.H.; Bessho, T.; Mu, D.; Sancar, A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 1996, 271, 11047–11050. [Google Scholar] [CrossRef]
- Scherly, D.; Nouspikel, T.; Corlet, J.; Ucla, C.; Bairoch, A.; Clarkson, S.G. Complementation of the DNA repair defect in xeroderma pigmentosum group G cells by a human cDNA related to yeast RAD2. Nature 1993, 363, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.S.; Ryu, E.; Lee, S.W.; Park, J.; Ha, N.Y.; Ra, J.S. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun. 2019, 10, 2420. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mo, J.; Rodriguez-Belmonte, E.M.; Lee, M.Y. Identification of a fourth subunit of mammalian DNA polymerase delta. J. Biol. Chem. 2000, 275, 18739–18744. [Google Scholar] [CrossRef]
- Mennecier, S.; Coste, G.; Servant, P.; Bailone, A.; Sommer, S. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genom. 2004, 272, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.J.; Hsieh, P. DNA mismatch repair: Molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003, 57, 579–608. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Kadyrov, F.A.; Dzantiev, L.; Constantin, N.; Modrich, P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell 2006, 126, 297–308. [Google Scholar] [CrossRef]
- Ramilo, C.; Gu, L.; Guo, S.; Zhang, X.; Patrick, S.M.; Turchi, J.J.; Li, G.M. Partial reconstitution of human DNA mismatch repair in vitro: Characterization of the role of human replication protein A. Mol. Cell Biol. 2002, 22, 2037–2046. [Google Scholar] [CrossRef]
- Kolodner, R.D.; Putnam, C.D.; Myung, K. Maintenance of genome stability in Saccharomyces cerevisiae. Science 2002, 297, 552–557. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Schrader, C.E.; Guikema, J.E.; Linehan, E.K.; Selsing, E.; Stavnezer, J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J. Immunol. 2007, 179, 6064–6071. [Google Scholar] [CrossRef] [PubMed]
- Milanowska, K.; Krwawicz, J.; Papaj, G.; Kosinski, J.; Poleszak, K.; Lesiak, J. REPAIRtoire—A database of DNA repair pathways. Nucleic Acids Res. 2011, 39, D788–D792. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.D.; Mitchell, M.; Sgouros, J.; Lindahl, T. Human DNA repair genes. Science 2001, 291, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Cutts, R.J.; Gadaleta, E.; Hahn, S.A.; Crnogorac-Jurcevic, T.; Lemoine, N.R.; Chelala, C. The Pancreatic Expression database: 2011 update. Nucleic Acids Res. 2011, 39, D1023–D1028. [Google Scholar] [CrossRef] [PubMed]
- Babukov, Y.; Aleksandrov, R.; Ivanova, A.; Atemin, A.; Stoynov, S. DNArepairK: An Interactive Database for Exploring the Impact of Anticancer Drugs onto the Dynamics of DNA Repair Proteins. Biomedicines 2021, 9, 1238. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, L.; Wan, J.; Dang, K.; Lin, Y.; Meng, S. Dynamic patterns of gene expression and regulatory variation in the maize seed coat. BMC Plant Biol. 2023, 23, 82. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, I.; Chang, A.; Hofmann, O.; Ebeling, C.; Ehrentreich, F.; Schomburg, D. BRENDA: A resource for enzyme data and metabolic information. Trends Biochem. Sci. 2002, 27, 54–56. [Google Scholar] [CrossRef]
- Rodchenkov, I.; Babur, O.; Luna, A.; Aksoy, B.A.; Wong, J.V.; Fong, D. Pathway Commons 2019 Update: Integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020, 48, D489–D497. [Google Scholar] [CrossRef]
- Vollebergh, M.A.; Lips, E.H.; Nederlof, P.M.; Wessels, L.F.; Wesseling, J.; Vd Vijver, M.J. Genomic patterns resembling BRCA1- and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014, 16, R47. [Google Scholar] [CrossRef]
- Xie, Y.; Du, D.; Karki, C.B.; Guo, W.; Lopez-Hernandez, A.E.; Sun, S. Revealing the mechanism of SARS-CoV-2 spike protein binding with ACE2. Comput. Sci. Eng. 2020, 22, 21–29. [Google Scholar] [CrossRef]
- Xie, Y.; Karki, C.B.; Du, D.; Li, H.; Wang, J.; Sobitan, A. Spike Proteins of SARS-CoV and SARS-CoV-2 Utilize Different Mechanisms to Bind With Human ACE2. Front. Mol. Biosci. 2020, 7, 591873. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Moult, J. SNPs, protein structure, and disease. Hum. Mutat. 2001, 17, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Chasman, D.; Adams, R.M. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J. Mol. Biol. 2001, 307, 683–706. [Google Scholar] [CrossRef] [PubMed]
- Sunyaev, S.; Ramensky, V.; Koch, I.; Lathe, W.; Kondrashov, A.S.; Bork, P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001, 10, 591–597. [Google Scholar] [CrossRef]
- wwPDB Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc. 2007, 2007, pdb.top17. [Google Scholar] [CrossRef]
- Perez, A.; Luque, F.J.; Orozco, M. Frontiers in molecular dynamics simulations of DNA. Acc. Chem. Res. 2012, 45, 196–205. [Google Scholar] [CrossRef]
- Sampoli, B.B.; Barbati, Z.R.; Arora, K.; Bogdanovic, J.; Schlick, T. How DNA polymerase X preferentially accommodates incoming dATP opposite 8-oxoguanine on the template. Biophys. J. 2013, 105, 2559–2568. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salsbury, F.R.; Clodfelter, J.E.; Gentry, M.B.; Hollis, T.; Scarpinato, K.D. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Res. 2006, 34, 2173–2185. [Google Scholar] [CrossRef]
- Minko, I.G.; Vartanian, V.L.; Tozaki, N.N.; Coskun, E.; Coskun, S.H.; Jaruga, P. Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1. DNA Repair 2020, 85, 102741. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Karki, C.B.; Chen, J.; Liu, D.; Li, L. Computational Study on DNA Repair: The Roles of Electrostatic Interactions Between Uracil-DNA Glycosylase (UDG) and DNA. Front. Mol. Biosci. 2021, 8, 718587. [Google Scholar] [CrossRef] [PubMed]
- Xi, T.; Jones, I.M.; Mohrenweiser, H.W. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 2004, 83, 970–979. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002, 12, 436–446. [Google Scholar] [CrossRef]
- Zadok, E. Legislative and Ethical Questions regarding DNA and Other Forensic “Biometric” Databases. In Proceedings of the International Conference on Ethics and Policy of Biometrics, Hong Kong, China, 3–4 January 2010; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Rothman, J.E. Starting at Go: Protein structure prediction succumbs to machine learning. Proc. Natl. Acad. Sci. USA 2023, 120, e2311128120. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Jiang, X.; Trozzi, F.; Xiao, S.; Larson, E.C.; Tao, P. Explore protein conformational space with variational autoencoder. Front. Mol. Biosci. 2021, 8, 781635. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Dror, R.O.; Salmon, J.K.; Grossman, J.; Mackenzie, K.M.; Bank, J.A. Millisecond-scale molecular dynamics simulations on Anton. In Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis, Portland, OR, USA, 13–19 November 2009. [Google Scholar]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Carugo, O.; Djinović-Carugo, K. Structural biology: A golden era. PLoS Biol. 2023, 21, e3002187. [Google Scholar] [CrossRef]
- Huang, B.; Kong, L.; Wang, C.; Ju, F.; Zhang, Q.; Zhu, J. Protein Structure Prediction: Challenges, Advances, and the Shift of Research Paradigms. Genom. Proteom. Bioinform. 2023, in press. [Google Scholar] [CrossRef]
| Database | Size | Feature | Function | Last Update |
|---|---|---|---|---|
| REPAIRtoire | Proteins and DNA damage Diseases: 429 | Multi-organism support; gene search | Identify and analyze pathways involved in DNA repair | October 2010 |
| Human DNA Repair Genes | Genes: 256 | Categorized DNA repair datasets | Related gene activity and chromosome location | June 2020 |
| Reactome | Curated human protein: 11,350 | Visualize biological processes online | Visualization, interpretation, and analysis | November 2022 |
| DNArepairK | Proteins: 72 | Animation of DNA repair protein kinetics | Dynamics of DNA repair proteins at the sites of DNA lesions | Unknown |
| KEGG | 45, 822, 810 | Large-scale integrated database | Molecular networks and network variants | November 2020 |
| Brenda | Enzymes: 8423 | Collection of gene data and enzymes | Searching enzymes | January 2023 |
| Pathway commons | 5772 Pathways; 2.3 million interaction data | Multiple databases to collect data | Data downloads, BioPAX web services, and data visualization | January 2020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Potlapalli, R.; Quan, H.; Chen, L.; Xie, Y.; Pouriyeh, S.; Sakib, N.; Liu, L.; Xie, Y. Exploring DNA Damage and Repair Mechanisms: A Review with Computational Insights. BioTech 2024, 13, 3. https://doi.org/10.3390/biotech13010003
Chen J, Potlapalli R, Quan H, Chen L, Xie Y, Pouriyeh S, Sakib N, Liu L, Xie Y. Exploring DNA Damage and Repair Mechanisms: A Review with Computational Insights. BioTech. 2024; 13(1):3. https://doi.org/10.3390/biotech13010003
Chicago/Turabian StyleChen, Jiawei, Ravi Potlapalli, Heng Quan, Lingtao Chen, Ying Xie, Seyedamin Pouriyeh, Nazmus Sakib, Lichao Liu, and Yixin Xie. 2024. "Exploring DNA Damage and Repair Mechanisms: A Review with Computational Insights" BioTech 13, no. 1: 3. https://doi.org/10.3390/biotech13010003
APA StyleChen, J., Potlapalli, R., Quan, H., Chen, L., Xie, Y., Pouriyeh, S., Sakib, N., Liu, L., & Xie, Y. (2024). Exploring DNA Damage and Repair Mechanisms: A Review with Computational Insights. BioTech, 13(1), 3. https://doi.org/10.3390/biotech13010003







