Bacterial Proteases as Potentially Exploitable Modulators of SARS-CoV-2 Infection: Logic from the Literature, Informatics, and Inspiration from the Dog
Abstract
1. Introduction
- Contrast between high-risk versus low-risk segments of the human population;
- Genomic and structural biological rationalization through informatics and simulations performed herein.
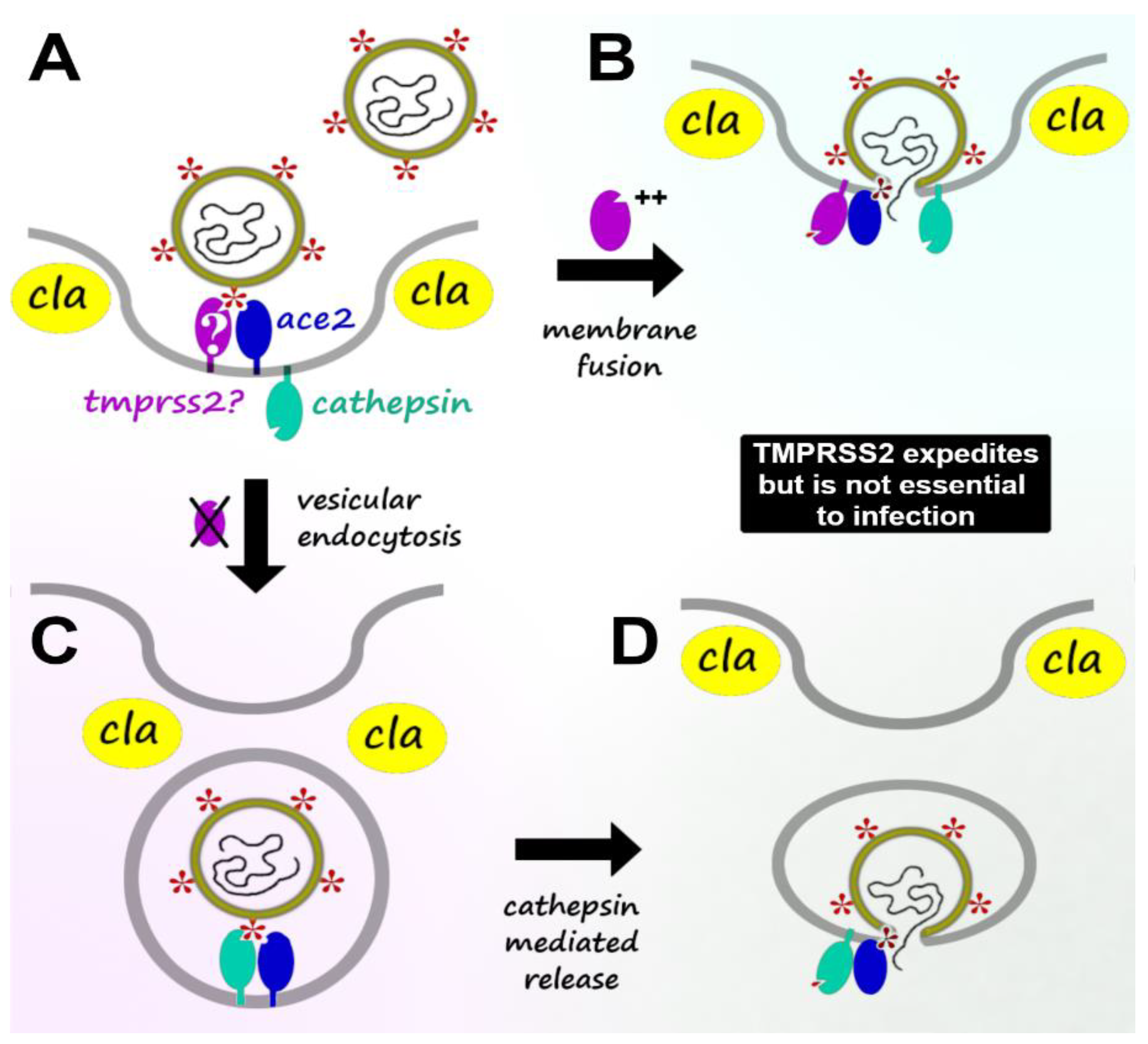
2. Viral Host Infection: TMPRSS2-Mediated vs. TMPRSS2-Independent Mechanisms
3. Role of Protease/Antiprotease Balance in Immunopathology
- Can we propose key players within the proteolytic machinery with major influences on SARS-CoV-2 adaptation and the determination of host vulnerability or resistance to the virus?
- What are the implications for future SARS-CoV-2 differentiation based on the assumption of PAB as a key evolutionary factor?
- Can this information be exploited for future health benefits?
4. Host Serpins and Proteases as Modulators of Virus Infection
5. Microbiotic Mimicry and Amplification of Protease/Antiprotease Dynamics
6. Vulnerability Trends in Comparative Biology and Human Population
7. Prospective Biochemical Rationalizations
8. Future Directions
9. Synopsis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riley, S.; Ainslie, K.E.C.; Eales, O.; Walters, C.E.; Wang, H.; Atchison, C.; Fronterre, C.; Diggle, P.J.; Ashby, D.; Donnelly, C.A.; et al. Resurgence of SARS-CoV-2: Detection by community viral surveillance. Science 2021, 372, 990–995. [Google Scholar] [CrossRef]
- Fernandes, R.S.; de Oliveira Silva, J.; Gomes, K.B.; Azevedo, R.B.; Townsend, D.M.; de Paula Sabino, A.; Branco de Barros, A.L. Recent advances in point of care testing for COVID-19 detection. Biomed. Pharmacother. 2022, 153, 113538. [Google Scholar] [CrossRef]
- Kwok, H.F. The significance of advanced COVID-19 diagnostic testing in pandemic control measures. Int. J. Biol. Sci. 2022, 18, 4610–4617. [Google Scholar] [CrossRef] [PubMed]
- Alhama, J.; Maestre, J.P.; Martín, M.Á.; Michán, C. Monitoring COVID-19 through SARS-CoV-2 quantification in wastewater: Progress, challenges and prospects. Microb. Biotechnol. 2022, 15, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Pepera, G.; Tribali, M.-S.; Batalik, L.; Petrov, I.; Papathanasiou, J. Epidemiology, risk factors and prognosis of cardiovascular disease in the Coronavirus Disease 2019 (COVID-19) pandemic era: A systematic review. Rev. Cardiovasc. Med. 2022, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Bahmer, T.; Borzikowsky, C.; Lieb, W.; Horn, A.; Krist, L.; Fricke, J.; Scheibenbogen, C.; Rabe, K.F.; Maetzler, W.; Maetzler, C.; et al. Severity, predictors and clinical correlates of Post-COVID syndrome (PCS) in Germany: A prospective, multi-centre, population-based cohort study. eClinicalMedicine 2022, 51, 101549. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, N.; Ingraham, N.E.; Nguyen, N.; Siegel, L.; Silverman, G.; Sahoo, H.S.; Pakhomov, S.; Morse, L.R.; Billings, J.; Usher, M.G.; et al. Predictors of Postacute Sequelae of COVID-19 Development and Rehabilitation: A Retrospective Study. Arch. Phys. Med. Rehabil. 2022, 103, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Lushington, G. Preface to CCHTS Volume 24: The imperative of host-directed pharmacology in the aftermath of the COVID-19 pandemic. Comb. Chem. High. Throughput Screen. 2021, 24, 1–2. [Google Scholar] [CrossRef]
- Su, Y.; Yuan, D.; Chen, D.G.; Ng, R.H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, 881–895.e20. [Google Scholar] [CrossRef]
- Di Ciaula, A.; Bonfrate, L.; Portincasa, P.; Appice, C.; Belfiore, A.; Binetti, M.; Cafagna, G.; Campanale, G.; Carrieri, A.; Cascella, G.; et al. Nitrogen dioxide pollution increases vulnerability to COVID-19 through altered immune function. Environ. Sci. Pollut. Res. 2022, 29, 44404–44412. [Google Scholar] [CrossRef]
- Ravindra, K.; Singh, T.; Vardhan, S.; Shrivastava, A.; Singh, S.; Kumar, P.; Mor, S. COVID-19 pandemic: What can we learn for better air quality and human health? J. Infect Public Health 2022, 15, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Al Rifai, M.; Jain, V.; Khan, S.U.; Bk, A.; Mahar, J.H.; Krittanawong, C.; Mishra, S.R.; Dani, S.S.; Petersen, L.A.; Virani, S.S. State-Level Social Vulnerability Index and Healthcare Access: The Behavioral Risk Factor Surveillance System Survey. Am. J. Prev. Med. 2022, 63, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Bruce, C.; Gearing, M.E.; DeMatteis, J.; Levin, K.; Mulcahy, T.; Newsome, J.; Wivagg, J. Financial vulnerability and the impact of COVID-19 on American households. PLoS ONE 2022, 17, e0262301. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M. Merck’s COVID Pill Loses Its Lustre: What That Means for the Pandemic. Nature News, 2021. Available online: https://www.nature.com/articles/d41586-021-03667-0 (accessed on 12 March 2023).
- Cohen, J.; Kupferschmidt, K. The ‘Very, Very Bad Look’ of Remdesivir, the First FDA-Approved COVID-19 Drug. Available online: https://www.science.org/content/article/very-very-bad-look-remdesivir-first-fda-approved-covid-19-drug (accessed on 28 October 2020).
- Kow, C.S.; Javed, A.; Ramachandram, D.; Hasan, S.S. Clinical outcomes of sofosbuvir-based antivirals in patients with COVID-19: A systematic review and meta-analysis of randomized trials. Expert. Rev. Anti Infect. Ther. 2022, 20, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Al-Taie, A.; Denkdemir, F.R.; Sharief, Z.; Buyuk, A.S.; Sardas, S. The Long View on COVID-19 Theranostics and Oral Antivirals: Living with Endemic Disease and Lessons from Molnupiravir. OMICS J. Integr. Biol. 2022, 26, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Vegivinti, C.T.R.; Evanson, K.W.; Lyons, H.; Akosman, I.; Barrett, A.; Hardy, N.; Kane, B.; Keesari, P.R.; Pulakurthi, Y.S.; Sheffels, E.; et al. Efficacy of antiviral therapies for COVID-19: A systematic review of randomized controlled trials. BMC Infect. Dis. 2022, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Self, W.H.; Semler, M.W.; Leither, L.M.; Casey, J.D.; Angus, D.C.; Brower, R.G.; Chang, S.Y.; Collins, S.P.; Eppensteiner, J.C.; Filbin, M.R.; et al. Effect of Hydroxychloroquine on Clinical Status at 14 Days in Hospitalized Patients With COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.C.L.; Hor, C.P.; Tay, K.H.; Mat Jelani, A.; Tan, W.H.; Ker, H.B.; Chow, T.S.; Zaid, M.; Cheah, W.K.; Lim, H.H.; et al. Efficacy of Ivermectin Treatment on Disease Progression Among Adults with Mild to Moderate COVID-19 and Comorbidities: The I-TECH Randomized Clinical Trial. JAMA Intern. Med. 2022, 182, 426–435. [Google Scholar] [CrossRef]
- Relman, D.A.; Hamburg, M.A.; Choffnes, E.R.; Mack, A. Infectious Disease Emergence: Past, Present, and Future; National Academies Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Anston, F.; Bayfield, O.; Bardy, P. Developing Antiviral Drugs Is Not Easy—Here’s Why. The Conversation. Available online: https://theconversation.com/developing-antiviral-drugs-is-not-easy-heres-why-159512 (accessed on 12 March 2023).
- Howes, L. Why Are Antivirals so Hard to Develop? Chem. Eng. News 2021, 99, 32–35. Available online: https://cen.acs.org/pharmaceuticals/drug-discovery/antiviral-drug-development-covid-19/99/i19 (accessed on 12 March 2023). [CrossRef]
- Center for Disease Control. Influenza Antiviral Drug Resistance. 2021. Available online: https://www.cdc.gov/flu/treatment/antiviralresistance.htm (accessed on 12 March 2023).
- Dance, A. The Challenges of Antiviral Treatments. 2021. Available online: https://knowablemagazine.org/article/health-disease/2021/challenges-antiviral-treatments (accessed on 12 March 2023).
- Irwin, K.K.; Renzette, N.; Kowalik, T.F.; Jensen, J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016, 2, vew014. [Google Scholar] [CrossRef]
- Hiscox, J.A.; Khoo, S.H.; Stewart, J.P.; Owen, A. Shutting the gate before the horse has bolted: Is it time for a conversation about SARS-CoV-2 and antiviral drug resistance? J. Antimicrob. Chemother. 2021, 76, 2230–2233. [Google Scholar] [CrossRef]
- Kumar, M.; Mazumder, P.; Mohapatra, S.; Kumar Thakur, A.; Dhangar, K.; Taki, K.; Mukherjee, S.; Kumar Patel, A.; Bhattacharya, P.; Mohapatra, P.; et al. A chronicle of SARS-CoV-2: Seasonality, environmental fate, transport, inactivation, and antiviral drug resistance. J. Hazard. Mater. 2021, 405, 124043. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, A. Sars-Cov-2 and risk of antiviral drug resistance. Ir. J. Med. Sci. 2022, 191, 2367–2368. [Google Scholar] [CrossRef] [PubMed]
- Mari, A.; Roloff, T.; Stange, M.; Søgaard, K.K.; Asllanaj, E.; Tauriello, G.; Alexander, L.T.; Schweitzer, M.; Leuzinger, K.; Gensch, A.; et al. Global Genomic Analysis of SARS-CoV-2 RNA Dependent RNA Polymerase Evolution and Antiviral Drug Resistance. Microorganisms 2021, 9, 1094. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Marsh, M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research 2019, 8, F1000 Faculty Rev-1628. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The Importance of Glycans of Viral and Host Proteins in Enveloped Virus Infection. Front. Immunol. 2021, 12, 638573. [Google Scholar] [CrossRef] [PubMed]
- Kalaichandran, A. A Lucky Few Seem “Resistant” to COVID-19. Scientists Want to Know Why. STAT. 2021. Available online: https://www.statnews.com/2021/08/23/lucky-few-seem-resistant-to-covid19-scientists-want-to-know-why-2/ (accessed on 12 March 2023).
- Broadbent, L. Haven’t Had COVID Yet? It Could Be More Than Just Luck. Available online: https://economictimes.indiatimes.com/news/how-to/havent-had-covid-yet-it-could-be-more-than-just-luck/articleshow/91665022.cms (accessed on 12 March 2023).
- Scientists Are Narrowing in on Why Some People Keep Avoiding COVID. BA.5 Could End that Luck. NBC News. Available online: https://theconversation.com/havent-had-covid-yet-it-could-be-more-than-just-luck-181708 (accessed on 12 March 2023).
- Browne, G. (Seot. 12, 2022) The Mystery of Why Some People Don’t Get COVID. Wired. Available online: https://www.wired.com/story/the-mystery-of-why-some-people-dont-get-covid/ (accessed on 12 March 2023).
- Muller, M. (March 30, 2022) Are Some People ‘Super-Immune’ to COVID? Bloomberg.com. Available online: https://www.bloomberg.com/news/newsletters/2022-03-30/are-some-people-super-immune-to-covid (accessed on 12 March 2023).
- Andreakos, E.; Abel, L.; Vinh, D.C.; Kaja, E.; Drolet, B.A.; Zhang, Q.; O’Farrelly, C.; Novelli, G.; Rodríguez-Gallego, C.; Haerynck, F.; et al. A global effort to dissect the human genetic basis of resistance to SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e16. [Google Scholar] [CrossRef]
- Tripathi, D.; Sodani, M.; Gupta, P.K.; Kulkarni, S. Host directed therapies: COVID-19 and beyond. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100058. [Google Scholar] [CrossRef]
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022, 601, 110–117. [Google Scholar] [CrossRef]
- Fulkerson, P.C.; Lussier, S.J.; Bendixsen, C.G.; Castina, S.M.; Gebretsadik, T.; Marlin, J.S.; Russell, P.B.; Seibold, M.A.; Everman, J.L.; Moore, C.M.; et al. Human Epidemiology and RespOnse to SARS-CoV-2 (HEROS): Objectives, Design and Enrollment Results of a 12-City Remote Observational Surveillance Study of Households with Children using Direct-to-Participant Methods. medRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Bravaccini, S.; Nicolini, F.; Balzi, W.; Azzali, I.; Calistri, A.; Parolin, C.; Vitiello, A.; Biasolo, M.A.; Mazzotti, L.; Gaimari, A.; et al. Tamoxifen Protects Breast Cancer Patients from COVID-19: First Evidence from Real World Data. 2021. preprint. Available online: https://www.researchsquare.com/article/rs-598923/v1 (accessed on 12 March 2023).
- Bravaccini, S.; Fonzi, E.; Tebaldi, M.; Angeli, D.; Martinelli, G.; Nicolini, F.; Parrella, P.; Mazza, M. Estrogen and Androgen Receptor Inhibitors: Unexpected Allies in the Fight Against COVID-19. Cell Transpl. 2021, 30, 0963689721991477. [Google Scholar] [CrossRef] [PubMed]
- The COVID-19 pandemic in 2023: Far from over. Lancet 2023, 401, 14–20.
- Lorenz, J. Study Confirms Benefits of COVID-19 Vaccines, Shows Effectiveness Wanes Over Time. Available online: https://www.contagionlive.com/view/study-confirms-benefits-of-covid-19-vaccines-shows-effectiveness-wanes-over-time (accessed on 12 March 2023).
- von Delft, A.; Hall, M.D.; Kwong, A.D.; Purcell, L.A.; Saikatendu, K.S.; Schmitz, U.; Tallarico, J.A.; Lee, A.A. Accelerating antiviral drug discovery: Lessons from COVID-19. Nat. Rev. Drug Discov. 2023, 22, 585–603. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.A.A.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life 2020, 72, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Zurabov, F.M.; Chernevskaya, E.A.; Beloborodova, N.V.; Zurabov, A.Y.; Petrova, M.V.; Yadgarov, M.Y.; Popova, V.M.; Fatuev, O.E.; Zakharchenko, V.E.; Gurkova, M.M.; et al. Bacteriophage Cocktails in the Post-COVID Rehabilitation. Viruses 2022, 14, 2614. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liao, B.; Cheng, L.; Peng, X.; Xu, X.; Li, Y.; Hu, T.; Li, J.; Zhou, X.; Ren, B. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 7777–7785. [Google Scholar] [CrossRef]
- Lai, C.-C.; Yu, W.-L. COVID-19 associated with pulmonary aspergillosis: A literature review. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan Ran Za Zhi 2021, 54, 46–53. [Google Scholar] [CrossRef]
- Yamamoto, S.; Saito, M.; Tamura, A.; Prawisuda, D.; Mizutani, T.; Yotsuyanagi, H. The human microbiome and COVID-19: A systematic review. PLoS ONE 2021, 16, e0253293. [Google Scholar] [CrossRef]
- Bomar, L.; Brugger, S.D.; Lemon, K.P. Bacterial Microbiota of the Nasal Passages Across the Span of Human Life. Curr. Opin. Microbiol. 2018, 41, 8–14. [Google Scholar] [CrossRef]
- Yu, X.; Wang, L.; Zheng, X.; Wen, Y.; Zhang, Z.; Fan, L.; Zhou, Q.; Yang, X.; Xue, B.; Lin, Y. Moraxella occupied the largest proportion in the nasal microbiome in healthy children, which potential protect them from COVID-19. Microb. Pathog. 2022, 170, 105685. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.M.; Prigge, A.D.; Anekalla, K.R.; Shukla, A.; Do-Umehara, H.C.; Setar, L.; Chavez, J.; Abdala-Valencia, H.; Politanska, Y.; Markov, N.S.; et al. Immune response to SARS-CoV-2 in the nasal mucosa in children and adults. medRxiv, 2021; preprint. [Google Scholar]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Porter, S.M.; Hartwig, A.E.; Bielefeldt-Ohmann, H.; Bosco-Lauth, A.M.; Root, J.J. Susceptibility of Wild Canids to SARS-CoV-2. Emerg. Infect Dis. 2022, 28, 1852–1855. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, M.; Damalanka, V.C.; Tartell, M.A.; Chung, D.; hee Lourenço, A.L.; Pwee, D.; Mayer Bridwell, A.E.; Hoffmann, M.; Voss, J.; Karmakar, P.; et al. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2108728118. [Google Scholar] [CrossRef] [PubMed]
- Hatesuer, B.; Bertram, S.; Mehnert, N.; Bahgat, M.M.; Nelson, P.S.; Pöhlman, S.; Schughart, K. Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice. PLoS Pathog. 2013, 9, e1003774. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Heurich, A.; Lavender, H.; Gierer, S.; Danisch, S.; Perin, P.; Lucas, J.M.; Nelson, P.S.; Pöhlmann, S.; Soilleux, E.J. Influenza and SARS-Coronavirus Activating Proteases TMPRSS2 and HAT Are Expressed at Multiple Sites in Human Respiratory and Gastrointestinal Tracts. PLoS ONE 2012, 7, e35876. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.W.; Mao, H.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie 2017, 142, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Limburg, H.; Harbig, A.; Bestle, D.; Stein, D.A.; Moulton, H.M.; Jaeger, J.; Janga, H.; Hardes, K.; Koepke, J.; Schulte, L.; et al. TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 2019, 93, e00649-19. [Google Scholar] [CrossRef]
- Matarese, A.; Gambardella, J.; Sardu, C.; Santulli, G. miR-98 Regulates TMPRSS2 Expression in Human Endothelial Cells: Key Implications for COVID-19. Biomedicines 2020, 8, 462. [Google Scholar] [CrossRef]
- Mahmoud, I.S.; Jarrar, Y.B. Targeting the intestinal TMPRSS2 protease to prevent SARS-CoV-2 entry into enterocytes-prospects and challenges. Mol. Biol. Rep. 2021, 48, 4667–4675. [Google Scholar] [CrossRef]
- Sakamoto, A.; Kawakami, R.; Kawai, K.; Gianatti, A.; Pellegrini, D.; Kutys, R.; Guo, L.; Mori, M.; Cornelissen, A.; Sato, Y.; et al. ACE2 (Angiotensin-Converting Enzyme 2) and TMPRSS2 (Transmembrane Serine Protease 2) Expression and Localization of SARS-CoV-2 Infection in the Human Heart. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 542–544. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, L.; Chew, K.Y.; Stocks, C.J.; Yordanov, T.E.; Essebier, P.; Kulasinghe, A.; Monkman, J.; dos Santos Miggiolaro, A.F.R.; Cooper, C.; de Noronha, L.; et al. Endothelial cells are not productively infected by SARS-CoV-2. Clin. Transl. Immunol. 2021, 10, e1350. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Karlsson, M.J.; Hober, A.; Svensson, A.-S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The human secretome. Sci. Signal 2019, 12, eaaz0274. [Google Scholar] [CrossRef] [PubMed]
- Bilinska, K.; Jakubowska, P.; Von Bartheld, C.S.; Butowt, R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020, 11, 1555–1562. [Google Scholar] [CrossRef] [PubMed]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Wettstein, L.; Kirchhoff, F.; Münch, J. The Transmembrane Protease TMPRSS2 as a Therapeutic Target for COVID-19 Treatment. Int. J. Mol. Sci. 2022, 23, 1351. [Google Scholar] [CrossRef]
- Simmons, G.; Zmora, P.; Gierer, S.; Heurich, A.; Pöhlmann, S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013, 100, 605–614. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alahmed, S.H.; Bazzi, A.M.; Alhani, H.M. A review of candidate therapies for Middle East respiratory syndrome from a molecular perspective. J. Med. Microbiol. 2017, 66, 1261–1274. [Google Scholar] [CrossRef]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Wettstein, L.; Knaff, P.M.; Kersten, C.; Müller, P.; Weil, T.; Conzelmann, C.; Müller, J.A.; Brückner, M.; Hoffmann, M.; Pöhlmann, S.; et al. Peptidomimetic inhibitors of TMPRSS2 block SARS-CoV-2 infection in cell culture. Commun. Biol. 2022, 5, 681. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, G.R. SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe 2021, 2, e488–e489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mann, M.; Syed, Z.A.; Reynolds, H.M.; Tian, E.; Samara, N.L.; Zeldin, D.C.; Tabak, L.A.; Ten Hagen, K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. USA 2021, 118, e2109905118. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Zhan, S.H. The Emergence of the Spike Furin Cleavage Site in SARS-CoV-2. Mol. Biol. Evol. 2022, 39, msab327. [Google Scholar] [CrossRef] [PubMed]
- Jocher, G.; Grass, V.; Tschirner, S.K.; Riepler, L.; Breimann, S.; Kaya, T.; Oelsner, M.; Hamad, M.S.; Hofmann, L.I.; Blobel, C.P.; et al. ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion. EMBO Rep. 2022, 23, e54305. [Google Scholar] [CrossRef]
- Yamamoto, M.; Gohda, J.; Kobayashi, A.; Tomita, K.; Hirayama, Y.; Koshikawa, N.; Seiki, M.; Semba, K.; Akiyama, T.; Kawaguchi, Y.; et al. Metalloproteinase-dependent and TMPRSS2-independnt cell surface entry pathway of SARS-CoV-2 requires the furin-cleavage site and the S2 domain of spike protein. Mbio 2022, 13, e00519-22. [Google Scholar] [CrossRef]
- Bosch, B.J.; Bartelink, W.; Rottier, P.J.M. Cathepsin L Functionally Cleaves the Severe Acute Respiratory Syndrome Coronavirus Class I Fusion Protein Upstream of Rather than Adjacent to the Fusion Peptide. J. Virol. 2008, 82, 8887–8890. [Google Scholar] [CrossRef]
- Scarcella, M.; d’Angelo, D.; Ciampa, M.; Tafuri, S.; Avallone, L.; Pavone, L.M.; De Pasquale, V. The Key Role of Lysosomal Protease Cathepsins in Viral Infections. Int. J. Mol. Sci. 2022, 23, 9089. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Chiaravalli, J.; Gellenoncourt, S.; Brownridge, P.; Bryne, D.P.; Daly, L.A.; Grauslys, A.; Walter, M.; Agou, F.; Chakrabarti, L.A.; et al. Characterising proteolysis during SARS-CoV-2 infection identifies viral cleavage sites and cellular targets with therapeutic potential. Nat. Commun. 2021, 12, 5553. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Tang, H.; Gao, L.; Wu, Z.; Meng, F.; Yan, R.; Qiao, S.; An, J.; Wang, C.; Qin, F.X.-F. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct. Target Ther. 2022, 7, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chan, J.F.-W.; Liu, H.; Liu, Y.; Chai, Y.; Shi, J.; Shuai, H.; Hou, Y.; Huang, X.; Yuen, T.T.-T.; et al. Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2. Emerg. Microbes Infect. 2022, 11, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lu, L.; Peng, Z.; Chen, L.-L.; Meng, X.; Zhang, C.; Ip, J.D.; Chan, W.-M.; Chu, A.W.-H.; Chan, K.-H.; et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022, 11, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, P.; Dixit, N. Modelling how increased Cathepsin B/L and decreased TMPRSS2 usage for cell entry by the SARS-CoV-2 Omicron variant may affect the efficacy and synergy of TMPRSS2 and Cathepsin B/L inhibitors. J. Theor. Biol. 2023, 572, 111568. [Google Scholar] [CrossRef] [PubMed]
- Berdowska, I.; Matusiewicz, M. Cathepsin L, transmembrane peptidase/serine subfamily member 2/4, and other host proteases in COVID-19 pathogenesis—With impact on gastrointestinal tract. World J. Gastroenterol. 2021, 27, 6590–6600. [Google Scholar] [CrossRef]
- Migueres, M.; Dimeglio, C.; Mansuy, J.M.; Abravanel, F.; Raymond, S.; Latour, J.; Jeanne, N.; Ranger, N.; Lhomme, S.; Saune, K.; et al. Influence of Nasopharyngeal Viral Load on the Spread of the Omicron BA. 2 Variant. Clin. Infect. Dis. 2022, 76, ciac563. [Google Scholar] [CrossRef]
- Kozlov, M. How does Omicron spread so fast? A high viral load isn’t the answer. Nature, 2022; ahead of print. [Google Scholar] [CrossRef]
- Lapid, N. Omicron Thrives in Airways, Not Lungs; New Data on Asymptomatic Cases. Reuters. 2021. Available online: https://www.reuters.com/business/healthcare-pharmaceuticals/omicron-thrives-airways-not-lungs-new-data-asymptomatic-cases-2021-12-15/ (accessed on 12 March 2023).
- Dance, A. Omicron’s lasting mysteries: Four questions scientists are racing to answer. Nature 2022, 603, 22–24. [Google Scholar] [CrossRef]
- Karimian, A.; Behjati, M.; Karimian, M. Molecular mechanisms involved in anosmia induced by SARS-CoV-2, with a focus on the transmembrane serine protease TMPRSS2. Arch Virol. 2022, 167, 1931–1946. [Google Scholar] [CrossRef]
- Butowt, R.; Bilińska, K.; von Bartheld, C. Why Does the Omicron Variant Largely Spare Olfactory Function? Implications for the Pathogenesis of Anosmia in Coronavirus Disease 2019. J. Infect Dis. 2022, 226, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R.; Mallik, B. Omicron (B.1.1.529)—A new heavily mutated variant: Mapped location and probable properties of its mutations with an emphasis on S-glycoprotein. Int. J. Biol. Macromol. 2022, 219, 980–997. [Google Scholar] [CrossRef] [PubMed]
- Stockley, R.A. Chronic bronchitis: The antiproteinase/proteinase balance and the effect of infection and corticosteroids. Clin. Chest Med. 1988, 9, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Gadek, J.E.; Pacht, E.R. The protease-antiprotease balance within the human lung: Implications for the pathogenesis of emphysema. Lung 1990, 168, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, D.C. The rôle of proteases and antiproteases in bronchial secretions. Eur. J. Respir. Dis. Suppl. 1987, 153, 78–85. [Google Scholar] [PubMed]
- Misz, M.; Fülep, E. Pathophysiological relations and clinical significance of serine protease inhibitors (serpins). Orv. Hetil. 1990, 131, 1851–1852, 1855–1859. [Google Scholar]
- Meyer, M.; Jaspers, I. Respiratory protease/antiprotease balance determines susceptibility to viral infection and can be modified by nutritional antioxidants. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 308, L1189–L1201. [Google Scholar] [CrossRef]
- Goel, K.; Serban, K.A. The multifaceted protease-antiprotease imbalance in COVID-19. EBioMedicine 2022, 78, 103973. [Google Scholar] [CrossRef]
- Oriano, M.; Amati, F.; Gramegna, A.; De Soyza, A.; Mantero, M.; Sibila, O.; Chotirmall, S.H.; Voza, A.; Marchisio, P.; Blasi, F.; et al. Protease–Antiprotease Imbalance in Bronchiectasis. Int. J. Mol. Sci. 2021, 22, 5996. [Google Scholar] [CrossRef]
- Chakraborti, S.; Sarkar, J.; Pramanik, P.K.; Chakraborti, T. Role of Proteases in Lung Disease: A Brief Overview. In Proteases in Human Diseases; Chakraborti, S., Chakraborti, T., Dhalla, N.S., Eds.; Springer: Singapore, 2017; pp. 333–374. [Google Scholar]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, J.; Wang, J.; Li, S.; Fukunaga, A.; Yodoi, J.; Tian, H. Progress in the mechanism targeted drug therapy for, C.O.P.D. Signal Transduct Target. Ther. 2020, 5, 201. [Google Scholar]
- Li, X.; Liu, H.; Tong, Y. Concerns on cross-species transmission of SARS-CoV-2 between pets and humans. Front. Microbiol. 2022, 13, 985528. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Shan, K.-J.; Wang, W.; Zhang, S.; Huan, Q.; Qian, W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genom. 2021, 48, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; López-Otín, C. A Genomic Analysis of Rat Proteases and Protease Inhibitors. Genome Res. 2004, 14, 609. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Sánchez, L.M.; Gutiérrez-Fernández, A.; Velasco, G.; López-Otín, C. A genomic view of the complexity of mammalian proteolytic systems. Biochem. Soc. Trans. 2005, 33, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Natalini, J.G.; Singh, S.; Segal, L.N. The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 2022, 21, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef]
- Garrigues, Q.; Apper, E.; Chastant, S.; Mila, H. Gut microbiota development in the growing dog: A dynamic process influenced by maternal, environmental and host factors. Front. Vet. Sci. 2022, 9, 964649. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Weinstein, S.B.; Martínez-Mota, R.; Stapleton, T.E.; Klure, D.M.; Greenhalgh, R.; Orr, T.J.; Dale, C.; Kohl, K.D.; Dearing, M.D. Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2108787118. [Google Scholar] [CrossRef]
- Carli, G.; Cecchi, L.; Stebbing, J.; Parronchi, P.; Farsi, A. Is asthma protective against COVID-19? Allergy 2021, 76, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Farne, H.; Singanayagam, A. Why asthma might surprisingly protect against poor outcomes in COVID-19. Eur. Respir J. 2020, 56, 2003045. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Xu, J.; Li, Y.; Wang, Y.; Yang, H. The Association of Asthma With COVID-19 Mortality: An Updated Meta-Analysis Based on Adjusted Effect Estimates. J. Allergy Clin. Immunol. Pract. 2021, 9, 3944–3968.e5. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.R.; Busse, W.; Holweg, C.T.J.; Rajput, Y.; Raimundo, K.; Meyer, C.S.; Seetasith, A.; Gupta, S.; Iqbal, A.; Kaner, R.J. Patients with allergic asthma have lower risk of severe COVID-19 outcomes than patients with nonallergic asthma. BMC Pulm. Med. 2022, 22, 418. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Agache, I.; Akdis, M.; Nadeau, K.; Klimek, L.; Jutel, M.; Akdis, C.A. The effect of allergy and asthma as a comorbidity on the susceptibility and outcomes of COVID-19. Int. Immunol. 2021, 34, dxab107. [Google Scholar] [CrossRef] [PubMed]
- CDC: Corticosteroids, Antibiotics Generally not Recommended for COVID-19 Outpatients|AHA News. Available online: https://www.aha.org/news/headline/2022-04-26-cdc-corticosteroids-antibiotics-generally-not-recommended-covid-19 (accessed on 12 March 2023).
- Singh, A. COVID-19 and Corticosteroids News-Medicalnet. 2022. Available online: https://www.news-medical.net/health/COVID-19-and-Corticosteroids.aspx (accessed on 12 March 2023).
- Ebrahimi Chaharom, F.; Pourafkari, L.; Ebrahimi Chaharom, A.A.; Nader, N.D. Effects of corticosteroids on Covid-19 patients: A systematic review and meta-analysis on clinical outcomes. Pulm. Pharmacol. Ther. 2022, 72, 102107. [Google Scholar] [CrossRef]
- Bradley, M.C.; Perez-Vilar, S.; Chillarige, Y.; Dong, D.; Martinez, A.I.; Weckstein, A.R.; Dal Pan, G.J. Systemic Corticosteroid Use for COVID-19 in US Outpatient Settings from April 2020 to August 2021. JAMA 2022, 327, 2015–2018. [Google Scholar] [CrossRef]
- Pandey, K.C.; De, S.; Mishra, P.K. Role of Proteases in Chronic Obstructive Pulmonary Disease. Front. Pharmacol. 2017, 8, 512. [Google Scholar] [CrossRef]
- Dey, T.; Kalita, J.; Weldon, S.; Taggart, C.C. Proteases and Their Inhibitors in Chronic Obstructive Pulmonary Disease. J. Clin. Med. 2018, 7, 244. [Google Scholar] [CrossRef]
- Kesic, M.J.; Hernandez, M.; Jaspers, I. Airway protease/antiprotease imbalance in atopic asthmatics contributes to increased Influenza A virus cleavage and replication. Respir. Res. 2012, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Balenga, N.A.; Klichinsky, M.; Xie, Z.; Chan, E.C.; Zhao, M.; Jude, J.; Laviolette, M.; Panettieri, R.A.; Druey, K.M. A fungal protease allergen provokes airway hyperresponsiveness in asthma. Nat. Commun. 2015, 6, 6763. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Li, Z.; She, Y.; Xie, B. Increased Expression of SERPINB10 Associated with Postoperative Recurrence in Chronic Rhinosinusitis with Nasal Polyps. Dis. Markers 2022, 2022, e7164318. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Zhang, K.; Feng, Y.; Yi, L.; Liang, Y.; Wu, W.; Zhao, J.; Zhang, Z.; Xu, Y.; Hu, Q.; et al. Epithelial SERPINB10, a novel marker of airway eosinophilia in asthma, contributes to allergic airway inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 2019, 316, L245–L254. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Ye, L.; Cai, H.; Zhu, G.; Wang, J.; Zhu, M.; Song, X.; Yang, C.; Jin, M. SERPINB10 contributes to asthma by inhibiting the apoptosis of allergenic Th2 cells. Respir Res. 2021, 22, 178. [Google Scholar] [CrossRef]
- Eden, E. Asthma and COPD in alpha-1 antitrypsin deficiency. Evidence for the Dutch hypothesis. COPD 2010, 7, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Pini, L.; Paoletti, G.; Heffler, E.; Tantucci, C.; Puggioni, F.; Asthma and Alpha1-Antitrypsin Research Group. Alpha1-antitrypsin deficiency and asthma. Curr. Opin. Allergy Clin. Immunol. 2021, 21, 46. [Google Scholar] [CrossRef]
- Benarafa, C. Regulation of Neutrophil Serine Proteases by Intracellular Serpins. Serpin Fam. 2015, 13, 59–76. [Google Scholar]
- Chen, T.; Zhou, M.; Lin, M.; Liang, S.; Yan, Y.; Wang, S.; Fang, C.; Li, D.; Ruan, Y. Research Progress on the SERPINE1 Protein and Chronic Inflammatory Diseases of the Upper Respiratory Tract: A Literature Review. Int. Arch. Allergy Immunol. 2021, 182, 1097–1102. [Google Scholar] [CrossRef]
- Bucková, D.; Izakovicová Hollá, L.; Vácha, J. Polymorphism 4G/5G in the plasminogen activator inhibitor-1 (PAI-1) gene is associated with IgE-mediated allergic diseases and asthma in the Czech population. Allergy 2002, 57, 446–448. [Google Scholar]
- Kowal, K.; Bodzenta-Lukaszyk, A.; Pampuch, A.; Szmitkowski, M.; Donati, M.B.; Iacoviello, L. Plasminogen Activator Inhibitor-1 Plasma Concentration in Allergic Asthma Patients during Allergen Challenge. Int. Arch. Allergy Immunol. 2007, 144, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.; Postma, D.S.; Bruinenberg, M.; Diemen CC van Boezen, H.M.; Koppelman, G.H.; Timens, W.; Vonk, J.M. SERPINE1-675 4G/5G polymorphism is associated with asthma severity and inhaled corticosteroid response. Eur. Respir. J. 2011, 38, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, R.; Li, S.; Kong, X.; Wang, Z.; Chen, W.; Lin, N. Genetic polymorphisms in plasminogen activator inhibitor-1 predict susceptibility to steroid-induced osteonecrosis of the femoral head in Chinese population. Diagn. Pathol. 2013, 8, 169. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, H.; Kim, S.Y.; Kim, Y.; Lee, J.-S.; Dan, K.; Seong, M.-W.; Han, D. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020, 10, 22418. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, H.; Tebbutt, S.J. Leave no one behind: Inclusion of alpha-1 antitrypsin deficiency patients in COVID-19 vaccine trials. Eur. J. Hum. Genet. 2022, 30, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, F.; Chitirala, P.; Krüger, N.; Hoffmann, M.; Zuo, W.; Lammert, F.; Smola, S.; Tov, N.; Alagem, N.; Lepper, P.M.; et al. Therapeutic Application of Alpha-1 Antitrypsin in COVID-19. Am. J. Respir. Crit. Care Med. 2021, 204, 224–227. [Google Scholar] [CrossRef]
- Yang, C.; Keshavjee, S.; Liu, M. Alpha-1 Antitrypsin for COVID-19 Treatment: Dual Role in Antiviral Infection and Anti-Inflammation. Front. Pharmacol. 2020, 11, 615398. [Google Scholar] [CrossRef]
- Ma, Z.; Paek, D.; Oh, C.K. Plasminogen activator inhibitor-1 and asthma: Role in the pathogenesis and molecular regulation. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2009, 39, 1136–1144. [Google Scholar] [CrossRef]
- Rajesh, K.; Cho, S.H.; Min, J.Y.; Kang, J.; Chan, W.; Kim, D.-Y.; Oh, S.; Torgerson, D.; del Del-Pino-Yanes, M.M.; Hu, D.; et al. PAI-1, Early Life Infections and Asthma Risk, Exacerbations, and Reduced Lung Function. J. Allergy Clin. Immunol. 2016, 137, AB178. [Google Scholar] [CrossRef]
- Cho, S.H.; Jo, A.; Casale, T.; Jeong, S.J.; Hong, S.-J.; Cho, J.K.; Holbrook, J.T.; Kumar, R.; Smith, L.J. Soy isoflavones reduce asthma exacerbation in asthmatic patients with high PAI-1–producing genotypes. J. Allergy Clin. Immunol. 2019, 144, 109–117.e4. [Google Scholar] [CrossRef]
- Zuo, Y.; Warnock, M.; Harbaugh, A.; Yalavarthi, S.; Gockman, K.; Zuo, M.; Madison, J.A.; Knight, J.S.; Kanthi, Y.; Lawrence, D.A. Plasma tissue plasminogen activator and plasminogen activator inhibitor-1 in hospitalized COVID-19 patients. Sci. Rep. 2021, 11, 1580. [Google Scholar] [CrossRef]
- Huang, P.; Zuo, Q.; Li, Y.; Oduro, P.K.; Tan, F.; Wang, Y.; Liu, X.; Li, J.; Wang, Q.; Guo, F.; et al. A Vicious Cycle: In Severe and Critically Ill COVID-19 Patients. Front. Immunol. 2022, 13, 930673. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.S. The Central Role of PAI-1 in COVID-19: Thrombosis and beyond. Am. J. Respir Cell Mol. Biol. 2021, 65, 238–240. [Google Scholar] [CrossRef]
- Lin, C.-C.; Lin, L.-J.; Wang, S.-D.; Chiang, C.-J.; Chao, Y.-P.; Lin, J.; Kao, S.-T. The Effect of Serine Protease Inhibitors on Airway Inflammation in a Chronic Allergen-Induced Asthma Mouse Model. Mediat. Inflamm. 2014, 2014, e879326. [Google Scholar] [CrossRef]
- Menou, A.; Duitman, J.; Flajolet, P.; Sallenave, J.-M.; Mailleux, A.A.; Crestani, B. Human airway trypsin-like protease, a serine protease involved in respiratory diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 312, L657–L668. [Google Scholar] [CrossRef] [PubMed]
- Miki, M.; Yasuoka, S.; Tsutsumi, R.; Nakamura, Y.; Hajime, M.; Takeuchi, Y.; Miki, K.; Kitada, S.; Maekura, R. Human airway trypsin-like protease enhances interleukin-8 synthesis in bronchial epithelial cells by activating protease-activated receptor 2. Arch. Biochem. Biophys. 2019, 664, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Müller, M.A.; Lavender, H.; Gnirss, K.; Nehlmeier, I.; Niemeyer, D.; He, Y.; Simmons, G.; Drosten, C.; et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011, 85, 13363–13372. [Google Scholar] [CrossRef] [PubMed]
- Laporte, M.; Raeymaekers, V.; Van Berwaer, R.; Vandeput, J.; Marchand-Casas, I.; Thibaut, H.-J.; Van Looveren, D.; Martens, K.; Hoffmann, M.; Maes, P.; et al. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021, 17, e1009500. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral. Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef]
- Fan, C.; Guo, L.; Gu, H.; Huo, Y.; Lin, H. Alterations in Oral–Nasal–Pharyngeal Microbiota and Salivary Proteins in Mouth-Breathing Children. Front. Microbiol. 2020, 11, 575550. [Google Scholar] [CrossRef]
- Gupta, A.; Karyakarte, R.; Joshi, S.; Das, R.; Jani, K.; Shouche, Y.; Sharma, A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2022, 24, 104880. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Kang, J.H. Effects of nasopharyngeal microbiota in respiratory infections and allergies. Clin. Exp. Pediatr. 2021, 64, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Houten CB van Boers, S.A.; Jansen, R.; Cohen, A.; Engelhard, D.; Kraaij, R.; Hiltemann, S.D.; Ju, J.; Fernández, D.; Mankoc, C.; et al. The diagnostic value of nasal microbiota and clinical parameters in a multi-parametric prediction model to differentiate bacterial versus viral infections in lower respiratory tract infections. PLoS ONE 2022, 17, e0267140. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The Lung Microbiome, Immunity and the Pathogenesis of Chronic Lung Disease. J. Immunol. Baltim. Md. 1950 2016, 196, 4839–4847. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef] [PubMed]
- Espírito Santo, C.; Caseiro, C.; Martins, M.J.; Monteiro, R.; Brandão, I. Gut Microbiota, in the Halfway between Nutrition and Lung Function. Nutrients 2021, 13, 1716. [Google Scholar] [CrossRef] [PubMed]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung–gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Brody, H. The gut microbiome. Nature 2020, 577, S5. [Google Scholar] [CrossRef]
- Harper, A.; Vijayakumar, V.; Ouwehand, A.C.; ter Haar, J.; Obis, D.; Espadaler, J.; Binda, S.; Desiraju, S.; Day, R. Viral Infections, the Microbiome, and Probiotics. Front. Cell Infect. Microbiol. 2021, 10, 596166. [Google Scholar] [CrossRef]
- Potempa, J.; Pike, R.N. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 2009, 1, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Corre, M.H.; Bachmann, V.; Kohn, T. Bacterial matrix metalloproteases and serine proteases contribute to the extra-host inactivation of enteroviruses in lake water. ISME J. 2022, 16, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Andrade, B.G.N.; Cuadrat, R.R.C.; Tonetti, F.R.; Kitazawa, H.; Villena, J. The role of respiratory microbiota in the protection against viral diseases: Respiratory commensal bacteria as next-generation probiotics for COVID-19. Biosci. Microbiota Food Health 2022, 41, 94. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Cai, Z.; Duan, X.; Zhang, H.; Cheng, H.; Han, S.; Yu, K.; Jiang, Z.; Zhang, Y.; Liu, Y.; et al. Pseudomonas aeruginosa modulates alginate biosynthesis and type VI secretion system in two critically ill COVID-19 patients. Cell Biosci. 2022, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, N.S.; Pinski, A.N.; Monsibais, A.N.; Jankeel, A.; Doratt, B.M.; Cinco, I.R.; Ibraim, I.; Messaoudi, I. Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose. Cell Rep. 2021, 36, 109637. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Qin, X.; Hou, G.; Zhang, X.; Zhang, W. Changes of Moraxella catarrhalis infection in children before and after the COVID-19 pandemic, Zhengzhou, China. J. Infect. 2023, 86, 154–225. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. An Introduction to Sequence Similarity (“Homology”) Searching. Curr. Protoc. Bioinform. 2013, 42, Unit3.1. [Google Scholar] [CrossRef]
- Hark Gan, H.; Perlow, R.A.; Roy, S.; Ko, J.; Wu, M.; Huang, J.; Yan, S.; Nicoletta, A.; Vafai, J.; Sun, D.; et al. Analysis of Protein Sequence/Structure Similarity Relationships. Biophys. J. 2002, 83, 2781–2791. [Google Scholar] [CrossRef]
- Singh, S.; O’Reilly, S.; Gewaid, H.; Bowie, A.G.; Gautier, V.; Worrall, D.M. Reactive Centre Loop Mutagenesis of SerpinB3 to Target TMPRSS2 and Furin: Inhibition of SARS-CoV-2 Cell Entry and Replication. Int. J. Mol. Sci. 2022, 23, 12522. [Google Scholar] [CrossRef]
- He, M.; Gao, J.; Wu, J.; Zhou, Y.; Fu, H.; Ke, S.; Yang, H.; Chen, C.; Huang, L. Host Gender and Androgen Levels Regulate Gut Bacterial Taxa in Pigs Leading to Sex-Biased Serum Metabolite Profiles. Front. Microbiol. 2019, 10, 1359. [Google Scholar] [CrossRef]
- Baratchian, M.; McManus, J.M.; Berk, M.; Nakamura, F.; Mukhopadhyay, S.; Xu, W.; Erzurum, S.; Drazba, J.; Peterson, J.; Klein, E.A.; et al. Sex, androgens and regulation of pulmonary AR, TMPRSS2 and ACE2. BioRxiv, 2020; preprint. [Google Scholar]
- Baratchian, M.; McManus, J.M.; Berk, M.P.; Nakamura, F.; Mukhopadhyay, S.; Xu, W.; Erzurum, S.; Drazba, J.; Peterson, J.; Klein, E.A.; et al. Androgen regulation of pulmonary AR, TMPRSS2 and ACE2 with implications for sex-discordant COVID-19 outcomes. Sci. Rep. 2021, 11, 11130. [Google Scholar] [CrossRef] [PubMed]
- Colldén, H.; Landin, A.; Wallenius, V.; Elebring, E.; Fändriks, L.; Nilsson, M.E.; Ryberg, H.; Poutanen, M.; Sjögren, K.; Vandenput, L.; et al. The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E1182–E1192. [Google Scholar] [CrossRef] [PubMed]
- Aman, F.; Masood, S. How Nutrition can help to fight against COVID-19 Pandemic. Pak. J. Med. Sci. 2020, 36, S121–S123. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Nutrition and immunity: Lessons for COVID-19. Eur. J. Clin. Nutr. 2021, 75, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Kahleova, H.; Barnard, N.D. Can a plant-based diet help mitigate COVID-19? Eur. J. Clin. Nutr. 2022, 76, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Rebholz, C.M.; Hegde, S.; LaFiura, C.; Raghavan, M.; Lloyd, J.F.; Cheng, S.; Seidelmann, S.B. Plant-based diets, pescatarian diets and COVID-19 severity: A population-based case–control study in six countries. BMJ Nutr. Prev. Health 2021, 4, 257. [Google Scholar] [CrossRef] [PubMed]
- Dattner, I.; Goldberg, Y.; Katriel, G.; Yaari, R.; Gal, N.; Miron, Y.; Ziv, A.; Sheffer, R.; Hamo, Y.; Huppert, A. The role of children in the spread of COVID-19: Using household data from Bnei Brak, Israel, to estimate the relative susceptibility and infectivity of children. PLOS Comput. Biol. 2021, 17, e1008559. [Google Scholar] [CrossRef]
- Paul, L.A.; Daneman, N.; Schwartz, K.L.; Science, M.; Brown, K.A.; Whelan, M.; Chan, E.; Buchan, S.A. Association of Age and Pediatric Household Transmission of SARS-CoV-2 Infection. JAMA Pediatr. 2021, 175, 1151–1158. [Google Scholar] [CrossRef]
- Tracy, M.; Cogen, J.; Hoffman, L.R. The Pediatric Microbiome and the Lung. Curr. Opin. Pediatr. 2015, 27, 348–355. [Google Scholar] [CrossRef]
- Liu, T.; Lin, C.-H.; Chen, Y.-L.; Jeng, S.-L.; Tsai, H.-J.; Ho, C.-L.; Kuo, W.-S.; Hsieh, M.-H.; Chen, P.-C.; Wu, L.S.-H.; et al. Nasal Microbiome Change During and After Exacerbation in Asthmatic Children. Front. Microbiol. 2022, 12, 833726. [Google Scholar] [CrossRef]
- Anand, S.; Mande, S.S. Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef] [PubMed]
- Strain, R.; Stanton, C.; Ross, R.P. Effect of diet on pathogen performance in the microbiome. Microbiome Res. Rep. 2022, 1, 13. [Google Scholar] [CrossRef]
- Clark, S.E. Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr. Opin. Immunol. 2020, 66, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lei, H.; Zhang, K.; Ke, F.; Song, C. Diversification of animal gut microbes and NRPS gene clusters in some carnivores, herbivores and omnivores. Biotechnol. Biotechnol. Equip. 2020, 34, 1280–1287. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Teng, J.L.L.; Chiu, T.H.; Chan, E.; Tsang, A.K.L.; Panagiotou, G.; Zhai, S.-L.; Woo, P.C.Y. Differential Microbial Communities of Omnivorous and Herbivorous Cattle in Southern China. Comput. Struct. Biotechnol. J. 2018, 16, 54–60. [Google Scholar] [CrossRef]
- Senghor, B.; Sokhna, C.; Ruimy, R.; Lagier, J.-C. Gut microbiota diversity according to dietary habits and geographical provenance. Hum. Microbiome J. 2018, 7–8, 1–9. [Google Scholar] [CrossRef]
- Nishida, A.H.; Ochman, H. Rates of Gut Microbiome Divergence in Mammals. Mol. Ecol. 2018, 27, 1884. [Google Scholar] [CrossRef]
- Zoelzer, F.; Burger, A.L.; Dierkes, P.W. Unraveling differences in fecal microbiota stability in mammals: From high variable carnivores and consistently stable herbivores. Anim. Microbiome 2021, 3, 77. [Google Scholar] [CrossRef]
- Finlay, B.B.; Amato, K.R.; Azad, M.; Blaser, M.J.; Bosch, T.C.G.; Chu, H.; Dominguez-Bello, M.G.; Ehrlich, S.D.; Elinav, E.; Geva-Zatorsky, N.; et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc. Natl. Acad. Sci. USA 2021, 118, e2010217118. [Google Scholar] [CrossRef]
- Xu, X.-W.; Wu, X.-X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-CoV-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef]
- Yeo, W.S.; Ng, Q.X. Passive inhaled mRNA vaccination for SARS-Cov-2. Med. Hypotheses 2021, 146, 110417. [Google Scholar] [CrossRef] [PubMed]
- Sörensen, M.; Kantorek, J.; Byrnes, L.; Boutin, S.; Mall, M.A.; Lasitschka, F.; Zabeck, H.; Nguyen, D.; Dalpke, A.H. Pseudomonas aeruginosa Modulates the Antiviral Response of Bronchial Epithelial Cells. Front. Immunol. 2020, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Ruffin, M.; Bigot, J.; Calmel, C.; Mercier, J.; Pizzorno, A.; Rosa-Calatrava, M.; Corvol, H.; Balloy, V.; Terrier, O.; Guillot, L. 410: Pseudomonas aeruginosa modulates SARS-CoV-2 infectivity in CF airway epithelial cells by increasing expression of the host protease TMPRSS2. J. Cyst. Fibros 2021, 20, S193. [Google Scholar] [CrossRef]
- Ruffin, M.; Bigot, J.; Calmel, C.; Mercier, J.; Givelet, M.; Oliva, J.; Pizzorno, A.; Rosa-Calatrava, M.; Corvol, H.; Balloy, V.; et al. Flagellin from Pseudomonas aeruginosa Modulates SARS-CoV-2 Infectivity in Cystic Fibrosis Airway Epithelial Cells by Increasing TMPRSS2 Expression. Front. Immunol. 2021, 12, 714027. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.A.; Hampton, T.H.; Ashare, A. SARS-CoV-2 (COVID-19) and cystic fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 319, L408–L415. [Google Scholar] [CrossRef] [PubMed]
- Camiolo, M.; Gauthier, M.; Kaminski, N.; Ray, A.; Wenzel, S.E. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J. Allergy Clin. Immunol. 2020, 146, 315–324.e7. [Google Scholar] [CrossRef]
- Zhang, Y.-H.; Zhang, Y.; Dong, X.-F.; Hao, Q.-Q.; Zhou, X.-M.; Yu, Q.-T.; Li, S.-Y.; Chen, X.; Tengbeh, A.F.; Dong, B.; et al. ACE2 and Ang-(1-7) protect endothelial cell function and prevent early atherosclerosis by inhibiting inflammatory response. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. Al 2015, 64, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Salka, K.; Abutaleb, K.; Chorvinsky, E.; Thiruvengadam, G.; Arroyo, M.; Gomez, J.L.; Gutierrez, M.J.; Pillai, D.K.; Jaiswal, J.K.; Nino, G. IFN Stimulates ACE2 Expression in Pediatric Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2021, 64, 515–518. [Google Scholar] [CrossRef]
- Katze, M.G.; He, Y.; Gale, M. Viruses and interferon: A fight for supremacy. Nat. Rev. Immunol. 2002, 2, 675–687. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. CR 2019, 38, 173. [Google Scholar] [CrossRef]
- de Paula Gonzaga, A.L.A.C.; Palmeira, V.A.; Ribeiro, T.F.S.; Costa, L.B.; de Sá Rodrigues, K.E.; Simões-E-Silva, A.C. ACE2/Angiotensin-(1-7)/Mas Receptor Axis in Human Cancer: Potential Role for Pediatric Tumors. Curr. Drug Targets 2020, 21, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, C.P.; Nguyen, J.; Yamaguchi, Y.; Werts, A.D.; Lu, P.; Ladd, M.R.; Fulton, W.B.; Kovler, M.L.; Wang, S.; Prindle, T., Jr.; et al. A Dynamic Variation of Pulmonary ACE2 Is Required to Modulate Neutrophilic Inflammation in Response to Pseudomonas aeruginosa Lung Infection in Mice. J. Immunol. 2019, 203, 3000–3012. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Yue, X.; Lazartigues, E. ACE2 mouse models: A toolbox for cardiovascular and pulmonary research. Nat. Commun. 2020, 11, 5165. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, A.; Haarmann, H.; Zahradnik, S.; Frenzel, K.; Schreiber, F.; Klassert, T.E.; Heyl, K.A.; Endres, A.-S.; Schmidtke, M.; Hofmann, J.; et al. Moraxella catarrhalis decreases antiviral innate immune responses by down-regulation of TLR3 via inhibition of p53 in human bronchial epithelial cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016, 30, 2426–2434. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Cazzolla, A.P.; Arena, C.; Sovereto, D.; Caloro, G.A.; Dioguardi, A.; Crincoli, V.; Laino, L.; Troiano, G.; Lo Muzio, L. Innate Immunity in Children and the Role of ACE2 Expression in SARS-CoV-2 Infection. Pediatr. Rep. 2021, 13, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.; Purohit, P.; Misra, R.; Pareek, P.; Goel, A.; Khattri, S.; Pant, K.K.; Misra, S.; Sharma, P. Diseases and Molecular Diagnostics: A Step Closer to Precision Medicine. Indian J. Clin. Biochem. 2017, 32, 374–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ha, D.; Yoshitake, R.; Chen, S. White button mushroom interrupts tissue AR-mediated TMPRSS2 expression and attenuates pro-inflammatory cytokines in C57BL/6 mice. Npj Sci. Food 2021, 5, 20. [Google Scholar] [CrossRef]
- Fraser, B.J.; Beldar, S.; Seitova, A.; Hutchinson, A.; Mannar, D.; Li, Y.; Kwon, D.; Tan, R.; Wilson, R.P.; Leopold, K.; et al. Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 2022, 18, 963–971. [Google Scholar] [CrossRef]
- Pelay-Gimeno, M.; Glas, A.; Koch, O.; Grossmann, T.N. Structure-Based Design of Inhibitors of Protein–Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem. Int. Ed. Engl. 2015, 54, 8896–8927. [Google Scholar] [CrossRef]
- Ruiz-Gómez, G.; Tyndall, J.D.A.; Pfeiffer, B.; Abbenante, G.; Fairlie, D.P. Update 1 of: Over one hundred peptide-activated G protein-coupled receptors recognize ligands with turn structure. Chem. Rev. 2010, 110, PR1-41. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Prior, P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- Pawar, N.R.; Buzza, M.S.; Antalis, T.M. Membrane-Anchored Serine Proteases and Protease-Activated Receptor-2-Mediated Signaling: Co-Conspirators in Cancer Progression. Cancer Res. 2019, 79, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Butowt, R.; von Bartheld, C.S. Anosmia in COVID-19: Underlying Mechanisms and Assessment of an Olfactory Route to Brain Infection. Neuroscientist 2021, 27, 582–603. [Google Scholar] [CrossRef] [PubMed]
- Abdel Hameid, R.; Cormet-Boyaka, E.; Kuebler, W.M.; Uddin, M.; Berdiev, B.K. SARS-CoV-2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am. J. Physiol. Lung Cell Mol. Physiol. 2021, 320, L430–L435. [Google Scholar] [CrossRef] [PubMed]
- Szewczykowski, C.; Mardin, C.; Lucio, M.; Wallukat, G.; Hoffmanns, J.; Schröder, T.; Raith, F.; Rogge, L.; Heltmann, F.; Moritz, M.; et al. Long COVID: Association of Functional Autoantibodies against G-Protein-Coupled Receptors with an Impaired Retinal Microcirculation. Int. J. Mol. Sci. 2022, 23, 7209. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.M.C.; Coimbra, J.T.S.; Ramos, M.J.; Fernandes, P.A. Transmembrane Protease Serine 2 Proteolytic Cleavage of the SARS-CoV-2 Spike Protein: A Mechanistic Quantum Mechanics/Molecular Mechanics Study to Inspire the Design of New Drugs to Fight the COVID-19 Pandemic. J. Chem. Inf. Model. 2022, 62, 2510–2521. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Lushington, G.H.; McGee, J.; Chaguturu, R. Assay Technologies for Proteases. In Enzyme Technologies; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 1–53. [Google Scholar]
- Patel, S. A critical review on serine protease: Key immune manipulator and pathology mediator. Allergol. Immunopathol. 2017, 45, 579–591. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Krzemińska, A.; Moliner, V.; Swiderek, K. Dynamic and Electrostatic Effects on the Reaction Catalyzed by HIV-1 Protease. J. Am. Chem. Soc. 2016, 138, 16283–16298. [Google Scholar] [CrossRef]
- Hyland, L.J.; Tomaszek, T.A.; Roberts, G.D.; Carr, S.A.; Magaard, V.W.; Bryan, H.L.; Fakhoury, S.A.; Moore, M.L.; Minnich, M.D.; Culp, J.S. Human immunodeficiency virus-1 protease. 1. Initial velocity studies and kinetic characterization of reaction intermediates by 18O isotope exchange. Biochemistry 1991, 30, 8441–8453. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zheng, X.; Zhou, B.; Li, J.; Chen, M.; Deng, R.; Wong, G.; Lavillette, D.; Meng, G. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proc. Natl. Acad. Sci. USA 2022, 119, e2111199119. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Li, Q.; Wang, Y.; Shi, Y. Syncytia formation during SARS-CoV-2 lung infection: A disastrous unity to eliminate lymphocytes. Cell Death Differ. 2021, 28, 2019–2021. [Google Scholar] [CrossRef]
- Magesh, S.; John, D.; Li, W.T.; Li, Y.; Mattingly-app, A.; Jain, S.; Chang, E.Y.; Ongkeko, W.M. Disparities in COVID-19 Outcomes by Race, Ethnicity, and Socioeconomic Status: A Systematic Review and Meta-analysis. JAMA Netw Open 2021, 4, e2134147. [Google Scholar] [CrossRef] [PubMed]
- McQuillen, K. Protein metabolism of bacteria. In Der Stickstoffumsatz/Nitrogen Metabolism; Allen, E.K., Allen, O.N., Böttger, I., Caspersson, T., Dillemann, G., Engel, H., Fischer, H., Guggenheim, M., Haas, P., et al., Eds.; Springer: Berlin/Heidelberg, Germany, 1958; pp. 659–673. [Google Scholar]
- Marshall, N.C.; Finlay, B.B.; Overall, C.M. Sharpening Host Defenses during Infection: Proteases Cut to the Chase. Mol. Cell Proteom. MCP 2017, 16, S161–S171. [Google Scholar] [CrossRef] [PubMed]
- Potempa, M.; Potempa, J. Protease-dependent mechanisms of complement evasion by bacterial pathogens. Biol. Chem. 2012, 393, 873–888. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Ouertani, A.; Chaabouni, I.; Mosbah, A.; Long, J.; Barakat, M.; Mansuelle, P.; Mghirbi, O.; Najjari, A.; Ouzari, H.-I.; Masmoudi, A.S.; et al. Two New Secreted Proteases Generate a Casein-Derived Antimicrobial Peptide in Bacillus cereus Food Born Isolate Leading to Bacterial Competition in Milk. Front. Microbiol. 2018, 9, 1148. [Google Scholar] [CrossRef]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef]
- Roach, K. Pseudomonas sinus Infection Tricky to Eradicate. Ariz Dly Star. 2017. Available online: https://tucson.com/lifestyles/Pseudomonas-sinus-infection-tricky-to-eradicate/article_f470ccda-25f5-5bac-abc2-86f2528e1c87.html (accessed on 12 March 2023).
- Fothergill, J.L.; Neill, D.R.; Loman, N.; Winstanley, C.; Kadioglu, A. Pseudomonas aeruginosa adaptation in the nasopharyngeal reservoir leads to migration and persistence in the lungs. Nat. Commun. 2014, 5, 4780. [Google Scholar] [CrossRef]
- Kusaka, Y.; Ogawa, T.; Yamada, T.; Minami, K.; Umegaki, O.; Ukimura, A.; Kusaka, Y.; Ogawa, T.; Yamada, T.; Minami, K.; et al. Young Healthy Patient with Severe COVID-19 and Fulminant Community-Acquired Pseudomonas aeruginosa Pneumonia: A Case Report. Cureus 2022, 14, e32617. [Google Scholar] [CrossRef]
- Qu, J.; Cai, Z.; Liu, Y.; Duan, X.; Han, S.; Liu, J.; Zhu, Y.; Jiang, Z.; Zhang, Y.; Zhuo, C.; et al. Persistent Bacterial Coinfection of a COVID-19 Patient Caused by a Genetically Adapted Pseudomonas aeruginosa Chronic Colonizer. Front. Cell Infect. Microbiol. 2021, 11, 641920. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Pang, Y.T.; Miao, Y.; Wang, Y.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics in NAMD. J. Chem. Theory Comput. 2017, 13, 9–19. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Wang, K.; Jia, Z.; Bao, L.; Wang, L.; Cao, L.; Chi, H.; Hu, Y.; Li, Q.; Zhou, Y.; Jiang, Y. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 2022, 603, 919–925. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Delano, W. Pymol [Software]. Schrodinger, Inc.: New York, NY, USA, 2022. Available online: https://pymol.org/2/ (accessed on 1 January 2022).
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Im, W.; Feig, M.; Brooks, C.L. An Implicit Membrane Generalized Born Theory for the Study of Structure, Stability, and Interactions of Membrane Proteins. Biophys. J. 2003, 85, 2900–2918. [Google Scholar] [CrossRef]
- Onufriev, A.; Case, D.A.; Bashford, D. Effective Born radii in the generalized Born approximation: The importance of being perfect. J. Comput. Chem. 2002, 23, 1297–1304. [Google Scholar] [CrossRef]
- Koehl, P. Electrostatics calculations: Latest methodological advances. Curr. Opin. Struct. Biol. 2006, 16, 142–151. [Google Scholar] [CrossRef]
- Chipot, C.; Pohorille, A. Free Energy Calculations: Theory and Applications in Chemistry and Biology; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]

| PAI-1/SERPINe1 | A1AC/SERPINa3 | HAT/TMPRSS11D | ||||
|---|---|---|---|---|---|---|
| % Identity | % Coverage | % Identity | % Coverage | % Identity | % Coverage | |
| Staphylococcus | 28.76% | 90% | 25.87% | 87% | 30.30% | 31% |
| (MCM1356220.1) | (MCM1356220.1) | (PAL08087.1) | ||||
| Pseudomonas | 27.34% | 93% | 32.86% | 16% | 36.21% | 54% |
| (MAK72596.1) | (PNB35482.1) | (MCL6711730.1) | ||||
| Dolosigranulum | n/a | n/a | n/a | n/a | n/a | n/a |
| (none) | (none) | (none) | ||||
| Corynebacterium | 32.27% | 60% | 29.27% | 57% | 27.73% | 51% |
| (EEG25322.1) | (WP_232022389.1) | (WP_003858612.1) | ||||
| Moraxella | 28.41% | 85% | 28.17% | 97% | 37.82% | 56% |
| (MBC7753780.1) | (WP_219332546.1) | (MBC7754020.1) | ||||
| Phylum Family | Mean | (St. Dev.) | Count | Maximum | Minimum |
|---|---|---|---|---|---|
| Moraxella sp. | 63.52 | (25.97) | 40 | 97.18 | 13.47 |
| Gammaproteobacteria sp. | 4.02 | (10.48) | 13 | 61.38 | 0.00 |
| Pseudomonas aeruginosa | 15.89 | (22.05) | 29 | 81.37 | 0.00 |
| Microbacteriacae sp. | 2.28 | (4.45) | 15 | 21.30 | 0.00 |
| Candidatus Gracilibacteria | 1.70 | (4.69) | 10 | 26.13 | 0.00 |
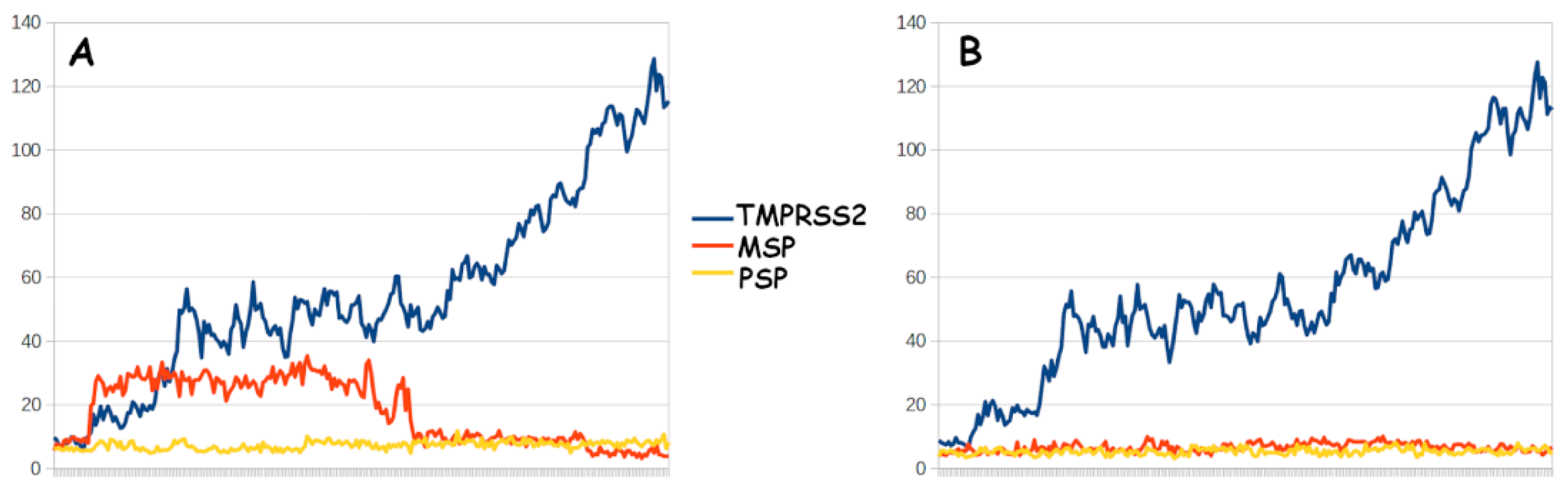
| TMPRSS2 | MSP | PSP | ||||
|---|---|---|---|---|---|---|
| Conf. 1 | Conf. 2 | Conf. 1 | Conf. 2 | Conf. 1 | Conf. 2 | |
| Binding energy | −77.45 | −30.98 | 5.33 | −19.25 | −55.5 | 28.78 |
| Std. err. | 4.38 | 4.77 | 4.46 | 4.36 | 3.26 | 3.42 |
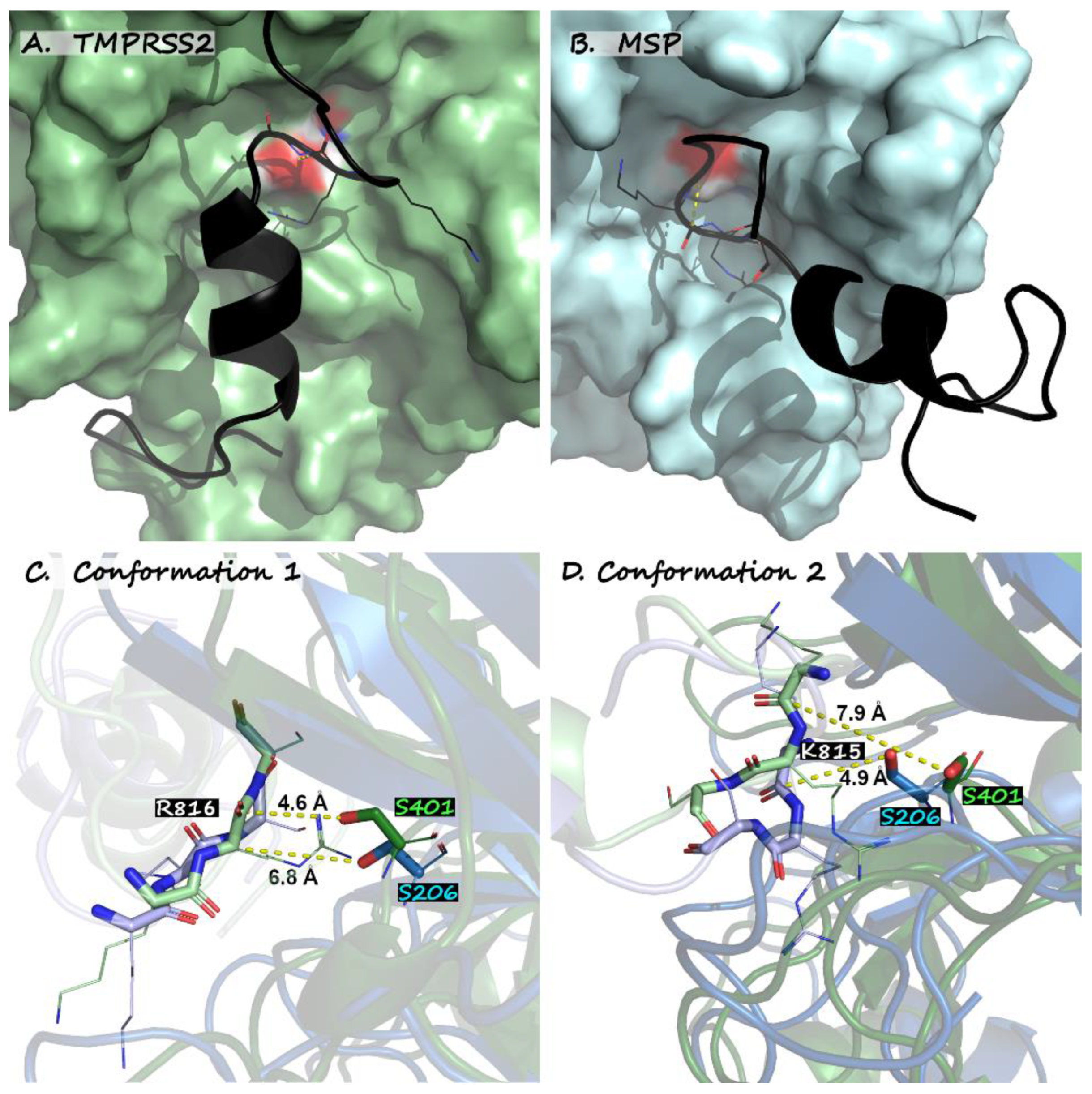
| Catalytic approach | 4.94 Å to R815 (conf. 1) | 5.11 Å to K814 (conf. 2) | 4.93 Å to K814 (conf. 1) | |||
| Std. err. | 0.04 Å | 0.06 Å | 0.04 Å | |||
| Minimum distance | 4.42 Å | 4.43 Å | 4.39 Å | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lushington, G.H.; Linde, A.; Melgarejo, T. Bacterial Proteases as Potentially Exploitable Modulators of SARS-CoV-2 Infection: Logic from the Literature, Informatics, and Inspiration from the Dog. BioTech 2023, 12, 61. https://doi.org/10.3390/biotech12040061
Lushington GH, Linde A, Melgarejo T. Bacterial Proteases as Potentially Exploitable Modulators of SARS-CoV-2 Infection: Logic from the Literature, Informatics, and Inspiration from the Dog. BioTech. 2023; 12(4):61. https://doi.org/10.3390/biotech12040061
Chicago/Turabian StyleLushington, Gerald H., Annika Linde, and Tonatiuh Melgarejo. 2023. "Bacterial Proteases as Potentially Exploitable Modulators of SARS-CoV-2 Infection: Logic from the Literature, Informatics, and Inspiration from the Dog" BioTech 12, no. 4: 61. https://doi.org/10.3390/biotech12040061
APA StyleLushington, G. H., Linde, A., & Melgarejo, T. (2023). Bacterial Proteases as Potentially Exploitable Modulators of SARS-CoV-2 Infection: Logic from the Literature, Informatics, and Inspiration from the Dog. BioTech, 12(4), 61. https://doi.org/10.3390/biotech12040061







