Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Dehydration Stress
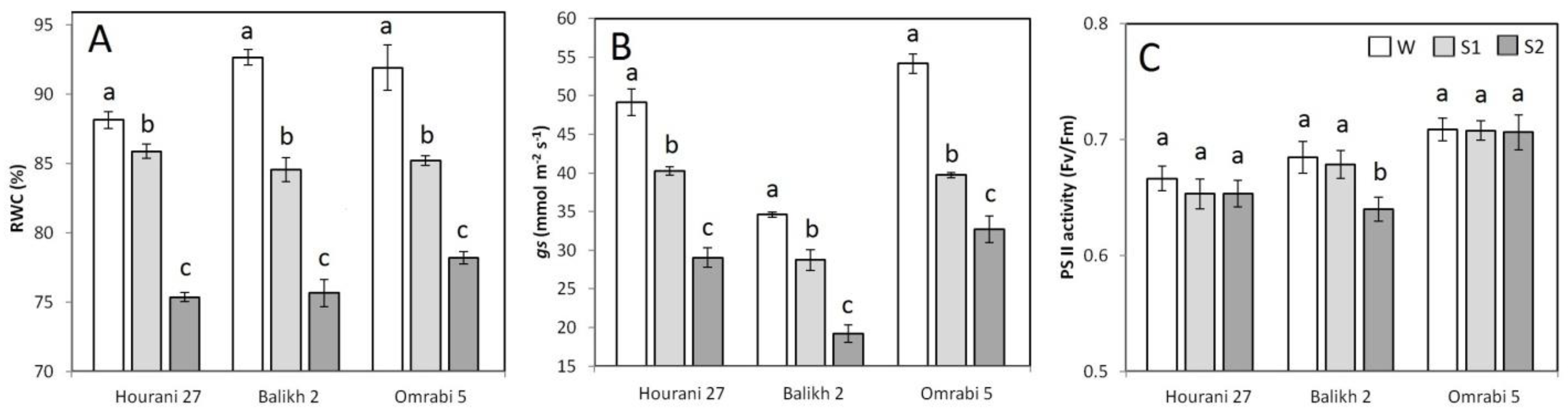
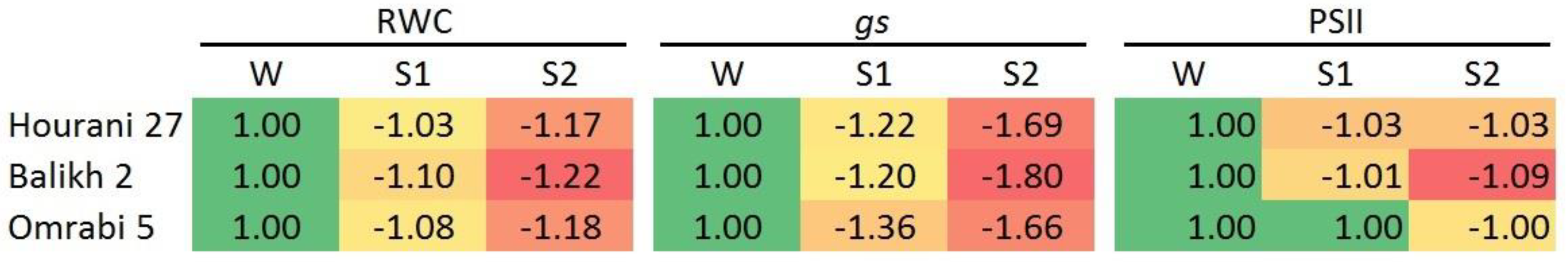
2.3. Physiological Measurements
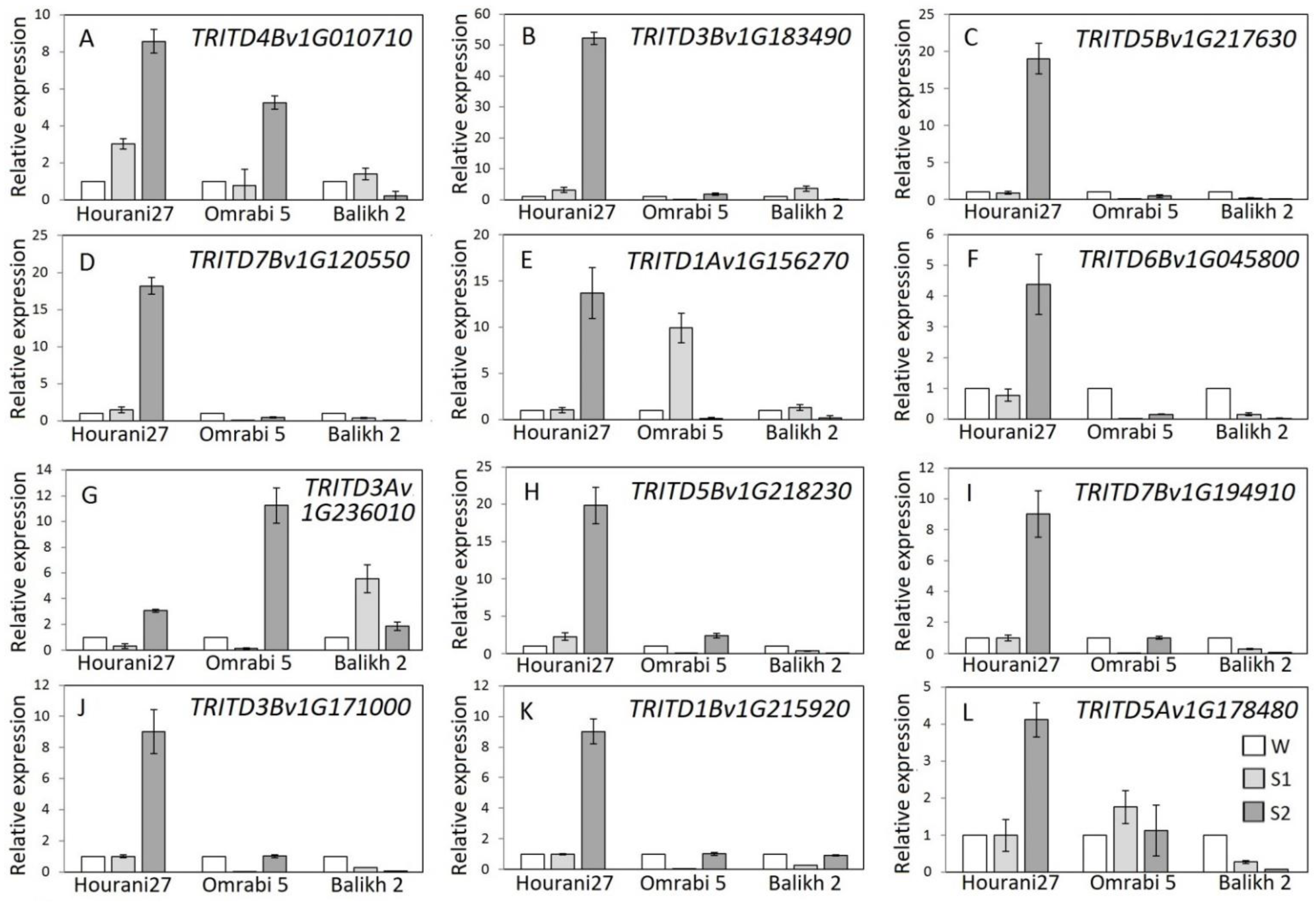
2.4. Analysis of Dehydration Responsive Memory Genes Using Quantitative Real-Time PCR
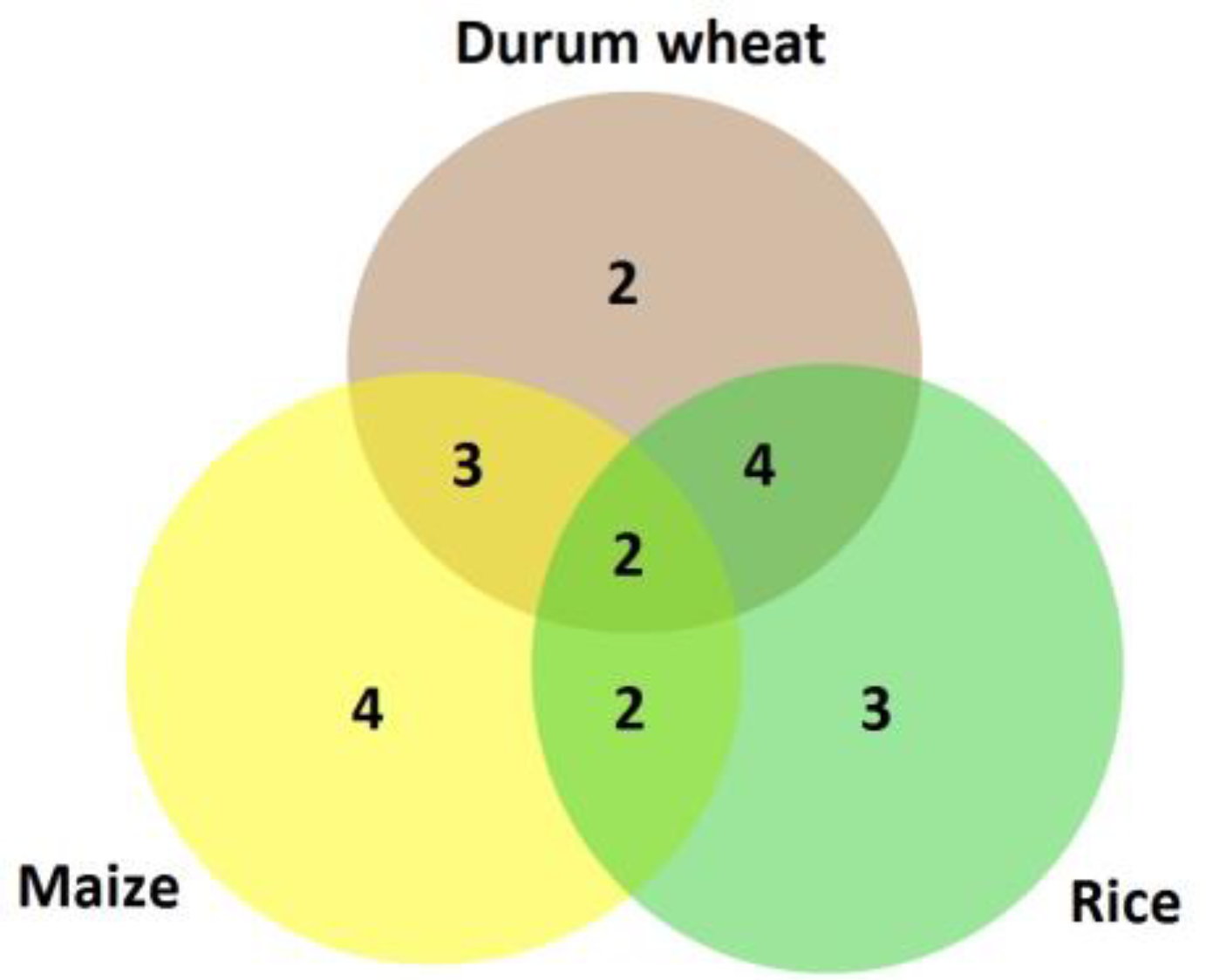
3. Results
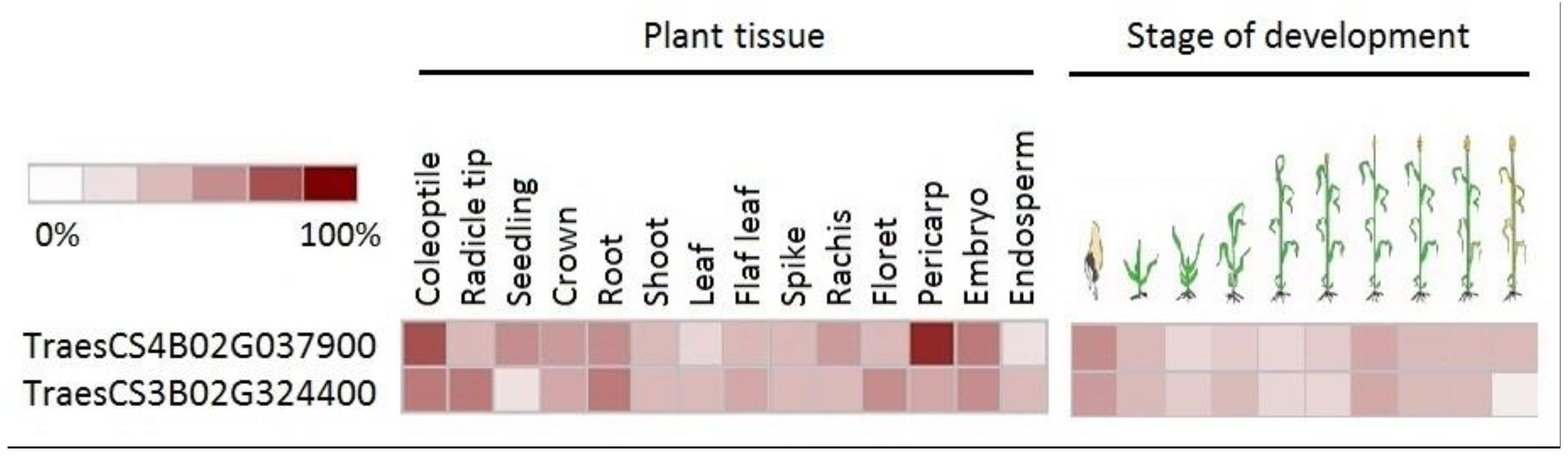
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Syouf, M.; Abu-Irmaileh, B.E.; Valkoun, J.; Bdour, S. Introgression from Durum Wheat Landraces in Wild Emmer Wheat (Triticum dicoccoides (Körn. ex Asch. et Graebner) Schweinf). Genet. Resour. Crop Evol. 2006, 53, 1165–1172. [Google Scholar] [CrossRef]
- Duwayri, M.; Migdadi, H.; Sadder, M.; Kafawin, O.; Ajlouni, M.; Amri, A.; Nachit, M. Use of SSR markers for characterizing cultivated durum wheat and its naturally occurring hybrids with wild wheat. Jordan J. Agric. Sci. 2007, 3, 398–410. [Google Scholar]
- Al-Tabbal, J.A.; Duwayri, M.A. Phenotypic variation within a Jordanian landrace of durum wheat “Safra Ma’an” (Triticum turgidum L. var. durum). J. Agric. Sci. Technol. 2013, B3, 717. [Google Scholar]
- Moaveni, P. Effect of water deficit stress on some physiological traits of wheat (Triticum aestivum). Agric. Sci. Res. J. 2011, 1, 64–68. [Google Scholar]
- Jaradat, A.A. Comparative assessment of einkorn and emmer wheat phenomes: III. Phenology. Genet. Resour. Crop Evol. 2019, 66, 1727–1760. [Google Scholar] [CrossRef]
- Wang, B.; Feng, P.; Chen, C.; Liu, D.L.; Waters, C.; Yu, Q. Designing wheat ideotypes to cope with future changing climate in South-Eastern Australia. Agric. Syst. 2019, 170, 9–18. [Google Scholar] [CrossRef]
- Duwayri, M. Comparison of wheat cultivars grown in the field under different levels of moisture. Cereal Res. Commun. 1984, 12, 27–34. [Google Scholar]
- Dura, S.; Duwayri, M.; Nachit, M.; Al-Sheyab, F. Detection of molecular markers associated with yield and yield components in durum wheat (Triticum turgidum L. var. durum) under saline conditions. Crop Pasture Sci. 2014, 64, 957–964. [Google Scholar] [CrossRef]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front. Plant Sci. 2017, 8, 1277. [Google Scholar] [CrossRef]
- Rai, A.; Mishra, U.; Singh, M.; Kumar, R.; Dubey, R.S.; Singh, N.K.; Jain, N.; Pandey, H.P. Expression Data in Response to Abiotic Stresses in Tomato at Flowering Stage; Gene Expression Omnibus, Series: GSE22304; Cornell University: Ithaca, NY, USA, 2010. [Google Scholar]
- Hazen, S.P.; Wu, Y.; Kreps, J.A. Gene expression profiling of plant responses to abiotic stress. Funct. Integr. Genom. 2003, 3, 105–111. [Google Scholar] [CrossRef]
- Lupini, A.; Preiti, G.; Badagliacca, G.; Abenavoli, M.R.; Sunseri, F.; Monti, M.; Bacchi, M. Nitrogen Use Efficiency in durum wheat under different nitrogen and water regimes in the Mediterranean Basin. Front. Plant Sci. 2021, 11, 607226. [Google Scholar] [CrossRef] [PubMed]
- Sadder, M.T.; Alsadon, A.; Wahb-Allah, M. Transcriptomic analysis of tomato lines reveals putative stress-specific biomarkers. Turk. J. Agric. For. 2014, 38, 700–715. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional ‘memory’of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef]
- Ding, Y.; Virlouvet, L.; Liu, N.; Riethoven, J.; Fromm, M.; Avramova, Z. Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol. 2014, 14, 141. [Google Scholar] [CrossRef]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef]
- Jaradat, A.; Duwayri, M. Effect of different moisture deficits on durum wheat seed germination and seedling growth. Cereal Res. Commun. 1981, 9, 55–62. [Google Scholar]
- Tadesse, W.; Nachit, M.; Abdalla, O.; Rajaram, S.; Bonjean, A. Wheat breeding at ICARDA: Achievements and prospects in the CWANA region. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., van Ginkel, M., Eds.; Lavoisier: Paris, France, 2016; Volume 3. [Google Scholar]
- Nachit, M.M. Durum breeding research to improve dryland productivity in the Mediterranean Region. In Proceedings of the The SEWANA Durum Research Network, ICARDA, Aleppo, Syria, 20–23 March 1995; pp. 1–15. [Google Scholar]
- Smart, R.E.; Bingham, G.E. Rapid Estimates of Relative Water Content. Plant Physiol. 1974, 53, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Sadder, M.T.; Al-Doss, A.A. Characterization of dehydrin AhDHN from Mediterranean saltbush (Atriplex halimus). Turk. J. Biol. 2014, 38, 469–477. [Google Scholar] [CrossRef]
- Bruce, T.J.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Wang, X.; Vignjevic, M.; Liu, F.; Jacobsen, S.; Jiang, D.; Wollenweber, B. Drought priming at vegetative growth stages improves tolerance to drought and heat stresses occurring during grain filling in spring wheat. Plant Growth Regul. 2015, 75, 677–687. [Google Scholar] [CrossRef]
- Giusti, L.; Mica, E.; Bertolini, E.; Leonardis, A.M.; De Faccioli, P.; Cattivelli, L.; Crosatti, C. microRNAs differentially modulated in response to heat and drought stress in durum wheat cultivars with contrasting water use efficiency. Funct. Integr. Genom. 2017, 17, 293–309. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2022, 27, 699–716. [Google Scholar] [CrossRef]
- Virlouvet, L.; Avenson, T.J.; Du, Q.; Zhang, C.; Liu, N.; Fromm, M.; Avramova, A.; Russo, S.E. Dehydration stress memory: Gene networks linked to physiological responses during repeated stresses of Zea mays. Front. Plant Sci. 2018, 9, 1058. [Google Scholar] [CrossRef]
- Dura, S.A.; Duwayri, M.A.; Nachit, M.M. Detection of molecular markers associated with yield and yield components in durum wheat (Triticum turgidum L. var. durum Desf.) under drought conditions. Afr. J. Agric. Res. 2013, 8, 2113–2117. [Google Scholar]
- Barutcular, C.; Toptas, I.; Turkten, H.; Yildirim, M.; Mujde, K.O.C. SPAD greenness to estimate genotypic variation in flag leaf chlorophyll in spring wheat under Mediterranean conditions. Turk. J. Field Crop. 2015, 20, 1–8. [Google Scholar] [CrossRef][Green Version]
- Xiong, D.; Chen, J.; Yu, T.; Gao, W.; Ling, X.; Li, Y.; Peng, S.; Huang, J. SPAD-based leaf nitrogen estimation is impacted by environmental factors and crop leaf characteristics. Sci. Rep. 2015, 5, 13389. [Google Scholar] [CrossRef] [PubMed]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef] [PubMed]
- Chwialkowska, K.; Nowakowska, U.; Mroziewicz, A.; Szarejko, I.; Kwasniewski, M. Water-deficiency conditions differently modulate the methylome of roots and leaves in barley (Hordeum vulgare L.). J. Exp. Bot. 2016, 67, 1109–1121. [Google Scholar] [CrossRef]
- Liu, N.; Fromm, M.; Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant 2014, 7, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Wassef, M.; Lövkvist, C.; Zielinski, D.; Aflaki, S.; Lombard, B.; Héry, T.; Loew, D.; Howard, M.; Margueron, R. A cis-acting mechanism mediates transcriptional memory at Polycomb target genes in mammals. Nat. Genet. 2021, 53, 1686–1697. [Google Scholar] [CrossRef]
- Cilliers, M. The Response of C1 and C13 Cysteine Proteases in Soybean Nodules to Drought. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2017. [Google Scholar]
- Thibaud-Nissen, F.; Wu, H.; Richmond, T.; Redman, J.C.; Johnson, C.; Green, R.; Arias, J.; Town, C.D. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006, 47, 152–162. [Google Scholar] [CrossRef]
- González-Pérez, S.; Gutiérrez, J.; García-García, F.; Osuna, D.; Dopazo, J.; Lorenzo, Ó.; Revuelta, J.L.; Arellano, J.B. Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol. 2011, 156, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, Y.; Zhou, C.; Zhang, L.; Lv, J. A wheat WRKY transcription factor TaWRKY46 enhances tolerance to osmotic stress in transgenic Arabidopsis plants. Int. J. Mol. Sci. 2020, 21, 1321. [Google Scholar] [CrossRef]
- Roy, N.; Choi, J.Y.; Lim, M.J.; Lee, S.I.; Choi, H.J.; Kim, N.S. Genetic and epigenetic diversity among dent, waxy, and sweet corns. Genes Genom. 2015, 37, 865–874. [Google Scholar] [CrossRef]
- Pandey, G.; Sharma, N.; Pankaj Sahu, P.; Prasad, M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr. Genom. 2016, 17, 490–498. [Google Scholar] [CrossRef]
- Srikant, T.; Drost, H.G. How stress facilitates phenotypic innovation through epigenetic diversity. Front. Plant Sci. 2021, 11, 606800. [Google Scholar] [CrossRef]
- Olas, J.J.; Apelt, F.; Annunziata, M.G.; John, S.; Richard, S.I.; Gupta, S.; Kragler, F.; Balazadeh, S.; Mueller-Roeber, B. Primary carbohydrate metabolism genes participate in heat-stress memory at the shoot apical meristem of Arabidopsis thaliana. Mol. Plant 2021, 14, 1508–1524. [Google Scholar] [CrossRef] [PubMed]
- Vyse, K.; Schaarschmidt, S.; Erban, A.; Kopka, J.; Zuther, E. Specific CBF transcription factors and cold-responsive genes fine-tune the early triggering response after acquisition of cold priming and memory. Physiol. Plant. 2022, 174, e13740. [Google Scholar] [CrossRef] [PubMed]
- Senapati, N.; Semenov, M.A. Assessing yield gap in high productive countries by designing wheat ideotypes. Sci. Rep. 2019, 9, 5516. [Google Scholar] [CrossRef] [PubMed]


| Durum Locus Number | Gene Description | Forward and Reveres Primers 5′-3′ |
|---|---|---|
| TRITD4Bv1G010710 | Lipoxygenase 2 | F-CTTCCATCGTCTACAAGAACTGG |
| R-CCCGTCCACCGCGTACGGGTAGTC | ||
| TRITD3Bv1G183490 | Protein kinase C-like zinc finger | F-GCGGAGCAAGTTCGCCTCCCAGACG |
| R-GCCAGCCTCGCGGTGAACTTGACGC | ||
| TRITD5Bv1G217630 | Basic helix-loop-helix (bHLH) DNA-binding | F-GTGCTGGTGCTGTTGCACAGCTGG |
| R-CGATGTCCTCGTCCATCAGCTTCGC | ||
| TRITD7Bv1G120550 | Transmembrane amino acid transporter | F-ATGTGGCTCATCATCTGCAAGCCC |
| R-ATCTATGAGTAGAACTTGTATGTC | ||
| TRITD1Av1G156270 | Late embryogenesis abundant (LEA) | F-CGTCCGAGACGGCCCAGGCCG |
| R-GCTGTCTCCCCCCATCCCCAGC | ||
| TRITD6Bv1G045800 | MYB transcription factor | F-AAGAGACCATGTTCAGAAGATAAC |
| R-TCAGCATCTTCTTATCACACTGTTAC | ||
| TRITD3Av1G236010 | Scarecrow-like protein (SCL1) | F-TCCAAGGGAAAGTCCAGATAGAATG |
| R-GAATCCAGCCATCGTCATTCTCGCC | ||
| TRITD5Bv1G218230 | Like/winged-helix DNA-binding family | F-GGAGACCAAGGCCAAGGCGGCCAAG |
| R-GACGAACTTGGCGATGGCGTACGGG | ||
| TRITD7Bv1G194910 | NAC transcription factor | F-CTAAGGGGAAGAAGACTGAGTGGG |
| R-TCCCTGTGGGTAGCTTGGCAACGG | ||
| TRITD3Bv1G171000 | WRKY transcription factor | F-GCGCAAGTACGGCCAGAAGCCCATC |
| R-GTGATCGTAGGAGTAGGTGACGAGC | ||
| TRITD1Bv1G215920 | Major facilitator superfamily | F-CGACGCTCGCCAACTGGCTGACTTC |
| R-CCAAACTCATCTGTTGCACTTCCAC | ||
| TRITD5Av1G178480 | AP2 transcription factor | F-CACGCAGTGTAAAGTTGTCGATAG |
| R-GGAGCAGAGCAGTCCCAAAC | ||
| TRITD5Av1G093080 | Actin | F-CCGAACGGGAAATTGTAAGG |
| R-TCTCTGCCCCAATGGTGATC |
| Durum Locus | Durum Response | Maize Locus | Maize Response * | Rice Locus | Rice Response ** |
|---|---|---|---|---|---|
| TRITD4Bv1G010710 | +/+, =/+, +/− | GRMZM2G102760 | +/+ | Os03g49380 | +/− |
| TRITD3Bv1G183490 | +/+, −/+, +/− | GRMZM2G106344 | +/+ | Os01g58194 | +/+ |
| TRITD5Bv1G217630 | =/+, −/= | GRMZM2G004356 | −/− | Os06g09370 | +/+ |
| TRITD7Bv1G120550 | +/+, −/= | GRMZM2G429322 | +/− | Os08g03350 | +/− |
| TRITD1Av1G156270 | =/+, +/− | GRMZM2G412436 | +/− | Os02g15250 | =/+ |
| TRITD6Bv1G045800 | =/+, −/= | GRMZM2G010920 | −/+ | OS03G25550 | +/− |
| TRITD3Av1G236010 | −/+, +/− | GRMZM2G153333 | +/= | Os07g36170 | −/+ |
| TRITD5Bv1G218230 | +/+, −/+, −/− | GRMZM2G401308 | −/= | Os07g08710 | −/+ |
| TRITD7Bv1G194910 | =/+, −/+, −/− | GRMZM2G063522 | =/+ | OS03G42630 | +/− |
| TRITD3Bv1G171000 | =/+, −/+, −/− | GRMZM2G013391 | =/+ | OS10G42850 | =/+ |
| TRITD1Bv1G215920 | =/+, −/+ | GRMZM2G028570 | =/− | Os03g24870 | +/− |
| TRITD5Av1G178480 | =/+, +/−, −/− | GRMZM2G434203 | =/− | Os04g46400 | =/− |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadder, M.T.; Musallam, A.; Allouzi, M.; Duwayri, M.A. Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.). BioTech 2022, 11, 43. https://doi.org/10.3390/biotech11030043
Sadder MT, Musallam A, Allouzi M, Duwayri MA. Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.). BioTech. 2022; 11(3):43. https://doi.org/10.3390/biotech11030043
Chicago/Turabian StyleSadder, Monther T., Anas Musallam, Majd Allouzi, and Mahmud A. Duwayri. 2022. "Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.)" BioTech 11, no. 3: 43. https://doi.org/10.3390/biotech11030043
APA StyleSadder, M. T., Musallam, A., Allouzi, M., & Duwayri, M. A. (2022). Dehydration Stress Memory Genes in Triticum turgidum L. ssp. durum (Desf.). BioTech, 11(3), 43. https://doi.org/10.3390/biotech11030043






