Abstract
The 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) is the central enzyme of the shikimate pathway to synthesize the three aromatic amino acids in fungi, plants, and prokaryotes. This enzyme is the target of the herbicide glyphosate. In most plants and prokaryotes, the EPSPS protein is constituted by a single domain family, the EPSP synthase (PF00275) domain, whereas in fungi, the protein is formed by a multi-domain structure from combinations of 22 EPSPS-associated domains. The most common multi-domain EPSPS structure in fungi involves five EPSPS-associated domains of the shikimate pathway. In this article, we analyze 390 EPSPS proteins of fungi to determine the extent of the EPSPS-associated domains. Based on the current classification of the EPSPS protein, most fungal species are intrinsically sensitive to glyphosate. However, complex domain architectures may have multiple responses to the herbicide. Further empirical studies are needed to determine the effect of glyphosate on fungi, taking into account the diversity of multi-domain architectures of the EPSPS. This research opens the door to novel biotechnological applications for microbial degradation of glyphosate.
1. Introduction
Glyphosate-based products are the herbicide most used against weeds worldwide. The herbicide targets an almost universal enzyme in plants, the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), also known as aroA [1,2]. The EPSPS is a key enzyme in the shikimate pathway for the synthesis of Tyrosine, phenylalanine, and tryptophan [3]. Because the epsps gene is not found in animals, the use of glyphosate is supposed to be safe for human health. However, the epsps gene is also present in most fungal and bacterial species. Thus, glyphosate may have an effect on microbial communities of free-living and host-associated microorganisms [4,5,6,7]. Recent studies of the EPSPS-glyphosate relationship have provided clues on the potential effect of the herbicide on the microbiota [8]. A comprehensive analysis of the potential sensitivity of the EPSPS protein has shown the potential impact of the herbicide on several species of plant, fungi, and bacteria [4,5] (e.g., the herbicide has the potential to affect half of the human gut microbiota [4]). The current classification of the EPSPS enzyme includes four EPSPS classes (class I: potentially sensitive and class II–IV: potentially resistant) based on amino acid markers in the EPSPS single-domain protein characteristic of plants and bacteria. However, the multi-domain EPSPS structure in fungi may lead to a complex response to the herbicide that has been largely overlooked.
Here, we analyze the evolution of the EPSPS domain in fungi and the distribution of additional EPSPS-associated domains. The EPSPS enzyme, at least in its single-domain structure characteristic of plants and prokaryotes, closes after its interaction with the two substrates, shikimate 3-phosphate (S3P) and phosphoenol pyruvate (PEP) [9]. Most of the EPSPS protein structures in the Protein Data Bank are in the closed form [10], and there are no representatives of any multi-domain EPSPS structure characteristic of fungi. The target-site sensitivity to glyphosate, also known as intrinsic sensitivity, was estimated based on the presence of amino acid markers in the EPSPS active site [4]. In addition, there are non-target-site factors (e.g., levels of gene expression of the epsps gene) that highly contribute to modulating the response of organisms to the herbicide [11,12,13]. The intrinsic sensitivity of the EPSPS to the herbicide has been largely studied in bacteria [4,5,14], and the results are in agreement with empirical microbiome studies [15,16,17,18,19]. Although more than 90% of fungal species have been classified as potentially sensitive to glyphosate (n = 789; 726 sensitive, 6 resistant, and 57 unclassified) [4], the response of a fungal multi-domain EPSPS to the herbicide glyphosate is yet unclear. The results of our survey of EPSPS-associated domains in fungi will help determine the effect of glyphosate on fungal species. Moreover, finding intrinsically resistant fungal strains is relevant in the development of agro-biotechnological applications to identify novel strategies for microbial degradation of glyphosate.
2. Materials and Methods
2.1. Dataset
EPSPS-associated domains present in EPSPS proteins were obtained from the PFAM (http://pfam.xfam.org, accessed on 8 July 2022), a comprehensive database of protein domains [20]. The dataset of EPSPS proteins was gathered from https://ppuigbo.me/programs/EPSPSClass, accessed on 20 July 2022 [4], and included 1175 EPSPS proteins with multi-domain structure prokaryotes and eukaryotes. The dataset included a subset of 390 out of 422 fungal proteins with multi-domain EPSPS structure (Supplementary Table S1).
2.2. Bipartite Network
A bipartite network of protein domains in fungal species was built with the program Cytoscape [21]. This network was used to visualize the presence and absence of EPSPS-associated domains and the distribution of the different architectures of the multidomain EPSPS protein in fungi.
2.3. Phylogenetics Analysis
EPSPS domains, from the dataset of EPSPS protein sequences of fungi, were aligned with the programs MUSCLE [22] and curated with Gblocks [23]. The program FastTree2 [24] was used to build a phylogenetic tree of the EPSPS domain. We utilized Dollon parsimony with the program Count [25] to analyze the evolution of the EPSPS-associated domains in fungi.
2.4. Potential Sensitivity to Glyphosate
The potential sensitivity to glyphosate was estimated using the EPSPSClass web server (https://ppuigbo.me/programs/EPSPSClass, accessed on 20 July 2022) [4]. EPSPS proteins are currently divided into four main classes (class I, sensitive; class II–IV, resistant).
3. Results and Discussion
3.1. Functional Characterization of EPSPS-Associated Domains
EPSPS proteins were defined by the EPSPS domain, which is approximately 1350 nucleotides long (450 amino acids) [4]. However, there were variations in length of the EPSPS protein, depending on the total number of EPSPS-associated domains and ranges between 163 (A0A101J2R9_9PORP) and 3206 (A0A094CHT8_9PEZI) amino acids. A multi-domain EPSPS structure was observed in most fungi, but it was rarely observed in plants and bacteria. Usually, multi-domain EPSPS genes of bacteria and plants are formed by two domains, whereas the fungal EPSPS is a larger sequence composed of more than five EPSPS-associated domains. Although most of the EPSPS-associated domains are involved in the shikimate pathways for the synthesis of the aromatic amino acids, promiscuous domains were also present (Table 1 and Supplementary Table S2).

Table 1.
Most common functions of the EPSPS-associated domains.
The EPSPS-associated domains can be classified into four main functional categories: shikimate (involved in shikimate pathway proteins), enzymes (proteins with catalytic function), expression (domains involved in gene expression), and structural function (proteins that do not have a catalytic function, such as binding sites, histones, and helix-turn-helix domains). The distribution of the EPSPS-associated domains in a dataset of 1175 multi-domain proteins showed certain dominance of the domains EPSP_synthase (as a marker of the EPSPS proteins), Shikimate kinase (SKI), 3-dehydroquinate synthase (DHQ_synthase), 3-dehydroquinate dehydratase (DHquinase_I), and Shikimate dehydrogenase substrate binding domain (Shikimate_dh_N; Table 1 and Supplementary Table S2). In some proteins, the multi-domain structure of the EPSPS included more than one hit to the EPSPS domain in pfam (e.g., S8DP49_FOMPI and A0A067M4R2_9AGAM).
3.2. Distribution of the EPSPS-Associated Domains in Fungi
Most of the EPSPS-associated in fungi were involved in the shikimate pathway (e.g., SKI, DHQ_synthase, DHquinase_I) and in the synthesis of aromatic amino acids (e.g., Shikimate_dh_N, PDH). There were also some promiscuous domains (e.g., HTH_3) associated with the EPSPS in some fungal species. Infrequent, but amply distributed, EPSPS-associated domains in fungi were involved in DNA modification and gene expression. A total of 22 domains were present in diverse domain architectures of the EPSPS protein, across 390 fungal species (Supplementary Table S3). However, in fungi, the most common multi-domain structure of the EPSPS consisted of five EPSPS-associated domains (Figure 1), mostly involved in the shikimate pathway, such as SKI (n = 374); DHQ_synthase (n = 374); DHquinase_I (n = 367); Shikimate_dh_N (n = 366); and Shikimate/quinate 5-dehydrogenase (Shikimate_DH; n = 136). In fungi, 16 out of 22 EPSPS-associated domains were only present in less than three proteins (Supplementary Table S3).
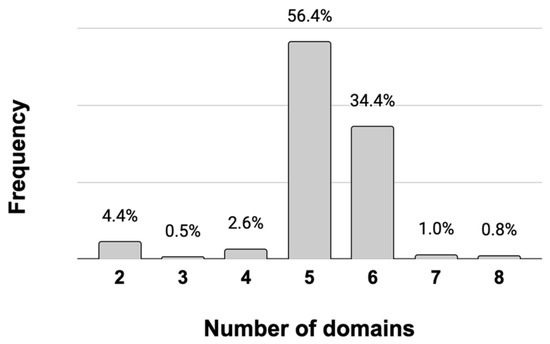
Figure 1.
Frequency of multi-domain structures of the EPSPS in fungi. There are 390 out of 420 EPSPS proteins with multi-domain structure in fungi. The majority of these proteins have five or six domains.
We have analyzed the distribution of the EPSPS-associated domains in different taxonomic groups of fungi (ascomycota, basidiomycota, mucoromycota, chytridiomycota, blastocladiomycota, zoopagomycota) (Figure 2). Ascomycota was the most variable phylum in terms of EPSPS-associated domains and contained several infrequent domains. Moreover, the least number of Shikimate_DH domains were present in ascomycota (e.g., this domain was least prevalent in Eurotiomycetes, Dothideomycetes, and Leotiomycetes). Most multi-domain architectures (n = 220) contained structures with five domains involved in the shikimate pathway, and approximately 1/3 of the protein sequences (n = 134) had all six domains of the shikimate pathway. Thus, the overall trend in fungal EPSPS proteins, as shown in the bipartite network, was an association of the EPSPS domain with other domains of the shikimate pathway.
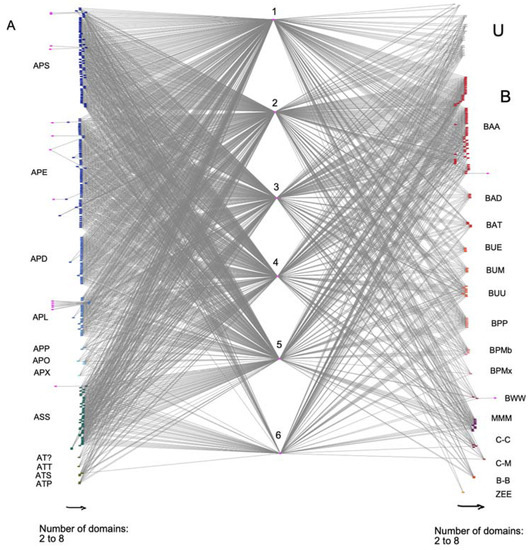
Figure 2.
A bipartite network of EPSPS-associated domains in fungal species. Letters stand for different phylums and classes of fungi. On the left side, there are fungi of the phylum ascomycota (A), and on the right side, there are fungi of phylum Basidiomycota (B), Mucoromycota (M), Chytridiomycota (C), Blastocladiomycota (Bl), Zoopagomygota (Z), and Unknown taxa (U). Numbers correspond to fungal multi-domain: (1) DH synthase, (2) EPSP synthase, (3) SKI, (4) DHquinase_1, (5) Shikimate_dh_N, and (6) Shikimate_DH. Arrows on the bottom of the figure note the number of domains in the multi-domain, which ranges from two to eight domains. Subphylum and class starting from the upper left are Pezizomycotina: Sordariomycetes (APS), Eurotiomycetes (APE), Dothideomycetes (APD), Leotiomycetes (APL), Pezizomycetes (APP), Orbiliomycetes (APO), and Xylonomycetes (APX); Saccharomycotina: Saccharomycetes (ASS); Taphrinomycotina: incertae sedis (AT?), Taphrinomycetes (ATT), Schizosaccharomycetes (ATS), and Pneumocystidomycetes (ATP); Agariomycotiina: Agaricomycetes (BAA), Dacrymycetes (BAD), and Tremellomycetes (BAT); Ustilaginomycotina: Exobasidiomycetes (BUE), Malasseziomycetes (BUM), and Ustilaginomycetes (BUU); Pucciniomycotina: Pucciniomycetes (BPP), Microbotryomycetes (BPMb), and Mixiomycetes (BPMx); Wallemiomycotina: Wallemiomycetes (BWW); Mucoromycotina: Mucoromycetes (MMM); Chytridiomycota: Chytridiomycetes (C-C) and Monoblepharidomycetes (C-M); Blastocladiomycota: Blastocladiomycetes (BI-B) and Entomophthoromycotina: Entomophthoromycetes (ZEE).
3.3. Phylogenetics Analysis of the EPSPS Protein in Fungi
The EPSPS multi-domain structure in fungi was heterogeneous across the phylogenetic tree. However, most of the species had five or six domains in the EPSPS protein (Figure 3). Our analysis indicated that a protein sequence with all six most abundant domains (i.e., a six-domain multi-domain structure) was the original EPSPS sequence in fungi (Table 1). Thus, the majority of sequences with five multi-domain structures raised by loss of the Shikimate_DH independently in different branches of the evolutionary tree. Moreover, many domains have been independently lost at early and late stages in the evolution of fungi. Notice that in some sequences, the Shikimate_DH (located at the C-term of the EPSPS protein) was disrupted. We speculate that in some cases, this domain was lost in a crossover event without affecting the functionality of the shikimate pathway. Moreover, we do not know if the domain function was preserved in a different protein. On the other hand, the Dollon parsimony analysis of the fungal phylogeny (Figure 3) indicated that infrequent domains were late inclusions into the multi-domain structure.
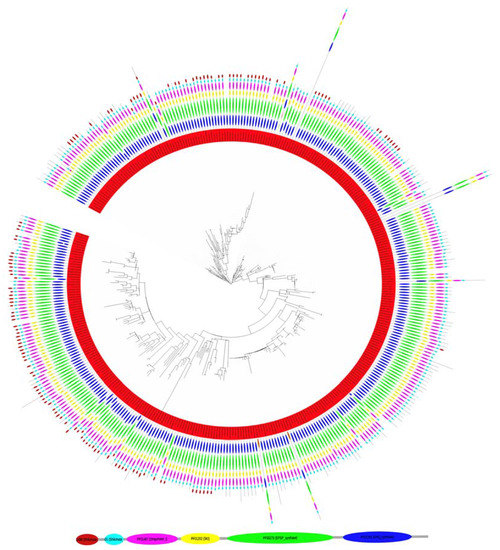
Figure 3.
Phylogenetic conservation of the multi-domain structure of the EPSPS protein in 390 fungi. The ancestral domain structure includes the EPSPS (PF00275; green) and EPSPS-associated domains Shikimate_DH (PF01488l; maroon), Shikimate_dH_N (PF08501; light blue), DHQuinase_I (PF01487; pink), SKI (PF01202, yellow), and 3-dehydroquinate synthase (PF01761; blue). Figure modified from [4] with permission. A detailed view of the phylogenetic tree is freely accessible at https://itol.embl.de/tree/13023210641470501567596434, accessed on 20 December 2021.
3.4. Potential Sensitivity to Glyphosate in Fungi
Here, we analyzed the frequency and evolution of EPSPS-associated domains to determine variations in the intrinsic sensitivity of the EPSPS protein to glyphosate. In bacteria and plants, the EPSPS protein sequence has a single domain, whereas fungal EPSPS proteins contain several domains [4]. Therefore, the EPSPS protein folding in fungi may result in a different interaction with the herbicide compared to the plant and bacteria EPSPS [8]. These potential effects of the multi-domain structure of the EPSPS have been mostly neglected. Moreover, additional non-target mechanisms of resistance (e.g., efflux pumps, vacuolar sequestration, and metabolization of glyphosate) or sensitivity (e.g., toxic effect on the mitochondria) to glyphosate modulate the intrinsic sensitivity status in the EPSPS protein [8,27] and may have a differential effect on fungal species. Several experimental and field studies have shown a negative effect of glyphosate on fungal communities in soil [28] and underground host-associated interactions [29]. Other fungi have developed non-target site resistance mechanisms (e.g., Purpureocillium lilacinum is able to degrade glyphosate and use glyphosate as a nutritional source [30]). The EPSPS of P. lilacinum (PWI66746.1) is sensitive to glyphosate. Moreover, it has been suggested that the carbon-phosphorus bond in glyphosate is the major metabolic degradation mechanism utilized by fungi [31].
The EPSPS is a two-substrate enzyme with an open (without ligand) and closed (with ligand) conformation. Glyphosate’s mode of action is competitive against the PEP and noncompetitive against the S3P [9]. However, the dual conformation of the EPSPS has been mostly studied in single-domain proteins of plants and bacteria; thus, its effect in a multi-domain structure is quite uncertain [8]. Our results showed that 354 (90.8%) fungi were potentially sensitive to glyphosate, 5 (1.3%) were resistant, and 31 (7.9%) were unknown (i.e., unclassified EPSPS proteins based on the current classification system). Interestingly, all EPSPS resistant species were class III, a not yet fully understood mechanism of resistance to glyphosate only present in a very small fraction of species [4]. However, the general trend changed depending on the number of domains (Figure 4). Multi-domain structures of the EPSPS protein with less than five domains had a significantly larger amount of unclassified sequences (Figure 4). Thus, further experimental evidence and new models are needed to determine the sensitivity of fungal organisms to glyphosate.
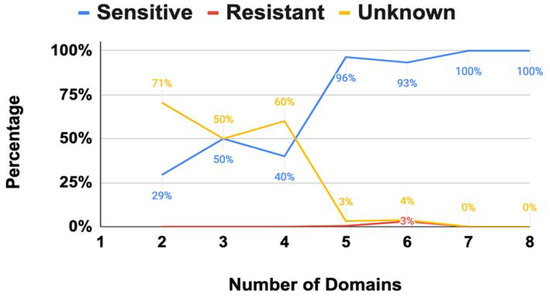
Figure 4.
Distribution of sensitive, resistant, and unknown EPSPS sequences to glyphosate.
4. Conclusions
In fungi, the most common multi-domain structure of the EPSPS ranges from two to eight domains. The ancestral state of the EPSPS protein included six domains (DHquinase_I DHQ_synthase, EPSPS, SKI, Shikimate_DH, and Shikimate_dH_N), as shown in the phylogenetic analysis. The wide diversity of EPSPS multi-domain structure in fungi is the product of several independent rearrangements of domains throughout evolution. Analyses of the EPSPS enzyme showed that most fungi are potentially sensitive to glyphosate. However, the total number of EPSPS-associated domains have an effect on the potential sensitivity status. Future analyses will be necessary to determine how different EPSPS multi-domain architectures affect the sensitivity of the EPSPS enzyme to glyphosate. These studies may have a substantial contribution to the development of novel biotechnological applications for microbial degradation of glyphosate.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biotech11030028/s1. Table S1: List of EPSPS proteins with more than one domain in fungi analyzed; Table S2: List of EPSPS-associated domains; Table S3: List of EPSPS-associated domains in fungi.
Author Contributions
Conceptualization, P.P.; formal analysis, T.T.; investigation, T.T. and P.P.; methodology, P.P.; resources, P.P. and T.T.; draft preparation, T.T. and P.P.; editing, P.P.; visualization, T.T.; supervision, P.P. All authors have read and agreed to the published version of the manuscript.
Funding
Funds from the Turku Collegium for Science, Medicine, and Technology (PP) were used to support this research. PP research is currently funded by a TQV grant PTQ2018-009846.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
A dataset of pre-computed EPSPS proteins is freely available from the EPSPSClass web server at http://ppuigbo.me/programs/EPSPSClass/, accessed on 20 July 2022.
Acknowledgments
We really appreciate the insightful comments from both anonymous reviewers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bentley, R.; Haslam, E. The Shikimate Pathway—A Metabolic Tree with Many Branche. Crit. Rev. Biochem. Mol. Biol. 1990, 25, 307–384. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Dacks, J.B.; Campbell, S.A.; Blanchard, J.L.; Foster, P.G.; McLeod, R.; Roberts, C.W. Evolutionary Origins of the Eukaryotic Shikimate Pathway: Gene Fusions, Horizontal Gene Transfer, and Endosymbiotic Replacements. Eukaryot. Cell 2006, 5, 1517–1531. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Leino, L.; Tall, T.; Helander, M.; Saloniemi, I.; Saikkonen, K.; Ruuskanen, S.; Puigbò, P. Classification of the glyphosate target enzyme (5-enolpyruvylshikimate-3-phosphate synthase) for assessing sensitivity of organisms to the herbicide. J. Hazard. Mater. 2020, 408, 124556. [Google Scholar] [CrossRef] [PubMed]
- Rainio, M.J.; Ruuskanen, S.; Helander, M.; Saikkonen, K.; Saloniemi, I.; Puigbò, P. Adaptation of bacteria to glyphosate: A microevolutionary perspective of the enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Environ. Microbiol. Rep. 2021, 13, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Balbuena, M.S.; Tison, L.; Hahn, M.-L.; Greggers, U.; Menzel, R.; Farina, W.M. Effects of sublethal doses of glyphosate on honeybee navigation. J. Exp. Biol. 2015, 218 Pt 17, 2799–2805. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Blumberg, B.; Antoniou, M.N.; Benbrook, C.M.; Carroll, L.; Colborn, T.; Everett, L.G.; Hansen, M.; Landrigan, P.J.; Lanphear, B.P.; et al. Is it time to reassess current safety standards for glyphosate-based herbicides? J. Epidemiol. Community Health 2017, 71, 613–618. [Google Scholar] [CrossRef]
- Mathew, S.A.; Muola, A.; Saikkonen, K.; Saloniemi, I.; Helander, M.; Puigbò, P. Quantification of the Potential Impact of Glyphosate-Based Products on Microbiomes. J. Vis. Exp. 2022, 179, e63109. [Google Scholar] [CrossRef]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.S.; Kabsch, W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Ruszkowski, M.; Forlani, G. Deciphering the structure of Arabidopsis thaliana 5-enol-pyruvyl-shikimate-3-phosphate synthase: An essential step toward the discovery of novel inhibitors to supersede glyphosate. Comput. Struct. Biotechnol. J. 2022, 20, 1494–1505. [Google Scholar] [CrossRef]
- Burchfield, S.L.; Bailey, D.C.; Todt, C.E.; Denney, R.D.; Negga, R.; Fitsanakis, V.A. Acute exposure to a glyphosate-containing herbicide formulation inhibits Complex II and increases hydrogen peroxide in the model organism Caenorhabditis elegans. Environ. Toxicol. Pharmacol. 2018, 66, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Nerozzi, C.; Recuero, S.; Galeati, G.; Bucci, D.; Spinaci, M.; Yeste, M. Effects of Roundup and its main component, glyphosate, upon mammalian sperm function and survival. Sci. Rep. 2020, 10, s41598-020. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.P.; Le Manac’h, S.G.; Hénault-Ethier, L.; Labrecque, M.; Lucotte, M.; Juneau, P. Glyphosate-Dependent Inhibition of Photosynthesis in Willow. Front. Plant Sci. 2017, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Van Bruggen, A.H.C.; He, M.M.; Shin, K.; Mai, V.; Jeong, K.C.; Finckh, M.R.; Morris, J.G., Jr. Environmental and health effects of the herbicide glyphosate. Sci. Total Environ. 2018, 616–617, 255–268. [Google Scholar] [CrossRef]
- Ruuskanen, S.; Rainio, M.J.; Gómez-Gallego, C.; Selenius, O.; Salminen, S.; Collado, M.C.; Saikkonen, K.; Saloniemi, I.; Helander, M. Glyphosate-based herbicides influence antioxidants, reproductive hormones and gut microbiome but not reproduction: A long-term experiment in an avian model. Environ. Pollut. 2020, 266, 115108. [Google Scholar] [CrossRef]
- Motta, E.V.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed]
- van Bruggen, A.H.C.; Finckh, M.R.; He, M.; Ritsema, C.J.; Harkes, P.; Knuth, D.; Geissen, V. Indirect effects of the herbicide glyphosate on plant, animal and human health through its effects on microbial communities. Front. Environ. Sci. 2021, 9, 589618. [Google Scholar] [CrossRef]
- Gómez-Gallego, C.; Rainio, M.J.; Collado, M.C.; Mantziari, A.; Salminen, S.; Saikkonen, K.; Helander, M. Glyphosate-based herbicide affects the composition of microbes associated with Colorado potato beetle (Leptinotarsa decemlineata). FEMS Microbiol. Lett. 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Mesnage, R.; Teixeira, M.; Mandrioli, D.; Falcioni, L.; Ducarmon, Q.R.; Zwittink, R.D.; Mazzacuva, F.; Caldwell, A.; Halket, J.; Amiel, C.; et al. Use of Shotgun Metagenomics and Metabolomics to Evaluate the Impact of Glyphosate or Roundup MON 52276 on the Gut Microbiota and Serum Metabolome of Sprague-Dawley Rats. Environ. Health Perspect. 2021, 129, 017005. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Csűrös, M. Ancestral Reconstruction by Asymmetric Wagner Parsimony over Continuous Characters and Squared Parsimony over Distributions. In Proceedings of the International Workshop on Comparative Genomics, Paris, France, 13–15 October 2008; pp. 72–86. [Google Scholar] [CrossRef]
- Tall, T.; Puigbò, P. The Glyphosate Target Enzyme 5-Enolpyruvyl Shikimate 3-Phosphate Synthase (EPSPS) Contains Several EPSPS-Associated Domains in Fungi. Proceedings 2020, 76, 6. [Google Scholar] [CrossRef]
- Duke, S.O. Enhanced metabolic degradation: The last evolved glyphosate resistance mechanism of weeds? Plant Physiol. 2019, 181, 1401–1403. [Google Scholar] [CrossRef]
- Vázquez, M.B.; Moreno, M.V.; Amodeo, M.R.; Bianchinotti, M.V. Effects of glyphosate on soil fungal communities: A field study. Rev. Argent. Microbiol. 2021, 53, 349–358. [Google Scholar] [CrossRef]
- Zaller, J.G.; Heigl, F.; Ruess, L.; Grabmaier, A. Glyphosate herbicide affects belowground interactions between earthworms and symbiotic mycorrhizal fungi in a model ecosystem. Sci. Rep. 2014, 4, srep05634. [Google Scholar] [CrossRef]
- Spinelli, V.; Ceci, A.; Bosco, C.D.; Gentili, A.; Persiani, A.M. Glyphosate-Eating Fungi: Study on Fungal Saprotrophic Strains’ Ability to Tolerate and Utilise Glyphosate as a Nutritional Source and on the Ability of Purpureocillium lilacinum to Degrade It. Microorganisms 2021, 9, 2179. [Google Scholar] [CrossRef]
- Adelowo, F.E.; Olu-Arotiowa, O.A.; Amuda, O.S. Biodegradation of Glyphosate by Fungi Species. Adv. Biosci. Bioeng. 2014, 2, 359–381. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).