Detection and Quantification of Immunoregulatory miRNAs in Human Milk and Infant Milk Formula
Abstract
:1. Introduction
2. Materials and Methods
2.1. Type of Study
2.2. Sample
2.3. Oligonucleotide Selection and Design
2.4. RNA Extraction
2.5. cDNA Synthesis
2.6. miRNA Quantification by qPCR
2.7. Bioinformatics and Statistical Analyses
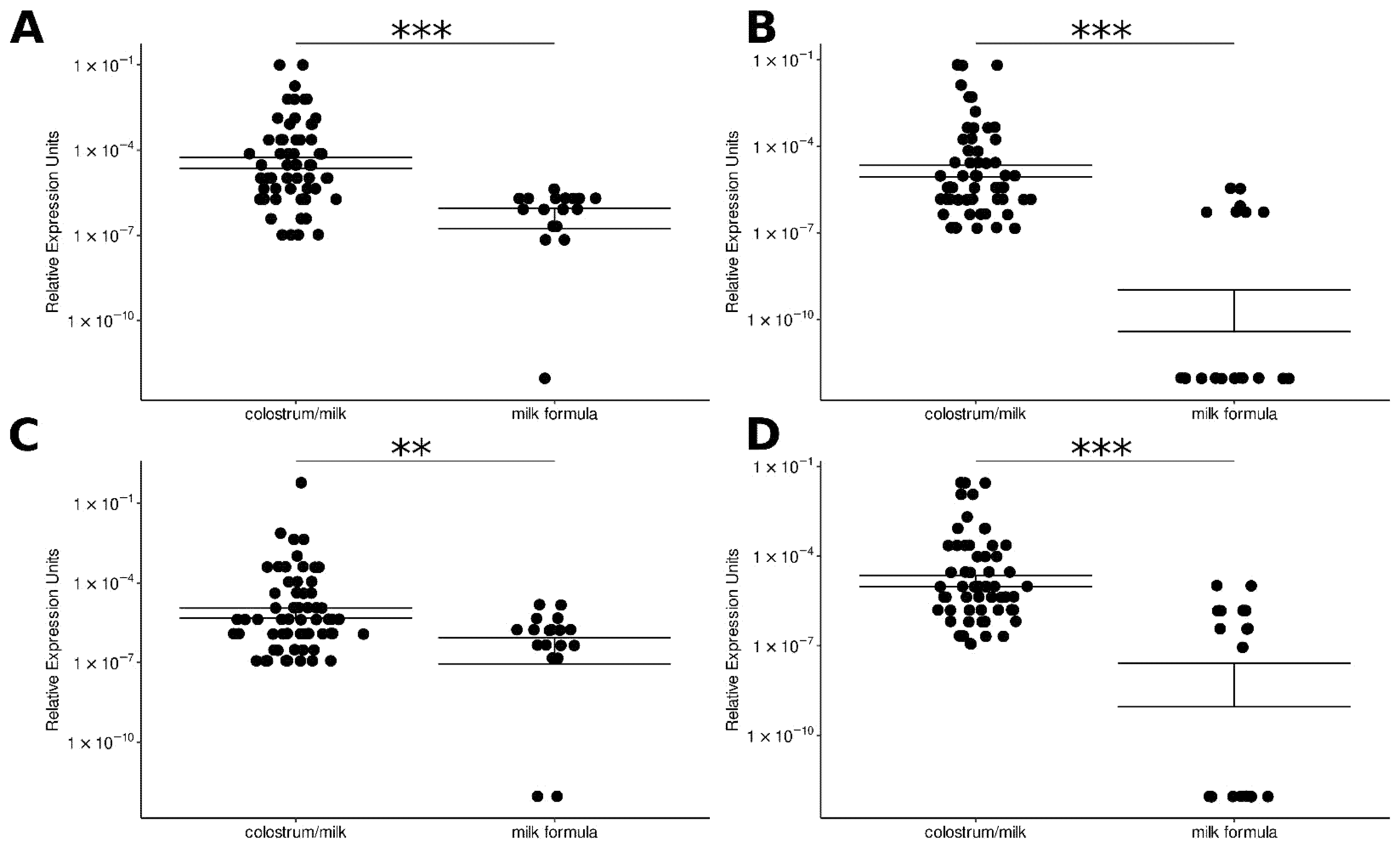
3. Results
3.1. miRNA Relative Expression Is Comparable among the Studied Human Milk Samples
3.2. Selected miRNAs Were Identified in Infant Milk Formulae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vista de Situación de las Prácticas de Lactancia Materna y Alimentación Complementaria en México: Resultados de la Ensanut 2018-19. Available online: https://www.saludpublica.mx/index.php/spm/article/view/11567/11972 (accessed on 6 December 2021).
- De Cosío-Martínez, T.G.; Hernández-Cordero, S.; Rivera-Dommarco, J.; Hernández-Ávila, M. Recomendaciones para una política nacional de promoción de la lactancia materna en México: Postura de la Academia Nacional de Medicina. TT—Recommendations for a multisectorial national policy to promote breastfeeding in Mexico: Position of the Nationa. Salud Publica Mex. 2017, 59, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Jung, Y.; Uhm, H.; Song, M.; Son, S.; Goo, J.; Jeong, C.; Song, J.J.; Kim, V.N.; Hohng, S. Quantification of purified endogenous miRNAs with high sensitivity and specificity. Nat. Commun. 2020, 11, 6033. [Google Scholar] [CrossRef]
- Bauman, D.E.; Mather, I.H.; Wall, R.J.; Lock, A.L. Major advances associated with the biosynthesis of milk. J. Dairy Sci. 2006, 89, 1235–1243. [Google Scholar] [CrossRef]
- Mosca, F.; Giannì, M.L. Human milk: Composition and health benefits. Pediatr. Med. Chir. 2017, 39, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating micrornas in breast milk and their potential impact on the infant. Nutrients 2020, 12, 3066. [Google Scholar] [CrossRef]
- Reif, S.; Elbaum Shiff, Y.; Golan-Gerstl, R. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J. Transl. Med. 2019, 17, 325. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010, 184, 6773–6781. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Floris, I.; Billard, H.; Boquien, C.Y.; Joram-Gauvard, E.; Simon, L.; Legrand, A.; Boscher, C.; Rozé, J.C.; Bolaños-Jiménez, F.; Kaeffer, B. MiRNA analysis by quantitative PCR in preterm human breast milk reveals daily fluctuations of hsa-miR-16-5p. PLoS ONE 2015, 10, e0140488. [Google Scholar] [CrossRef] [Green Version]
- Leiferman, A.; Shu, J.; Upadhyaya, B.; Cui, J.; Zempleni, J. Storage of Extracellular Vesicles in Human Milk, and MicroRNA Profiles in Human Milk Exosomes and Infant Formulas. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 235–238. [Google Scholar] [CrossRef]
- García-González, I.; Corona-Cervantes, K.; Hernández-Quiroz, F.; Villalobos-Flores, L.E.; Galván-Rodríguez, F.; Romano, M.C.; Miranda-Brito, C.; Piña-Escobedo, A.; Borquez-Arreortúa, F.G.; Rangel-Calvillo, M.N.; et al. The Effect of Holder Pasteurization on the Diversity of the Human Milk Bacterial Microbiota Using High-Throughput DNA Sequencing. J. Hum. Lact. 2021. [Google Scholar] [CrossRef]
- Kramer, M.F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011, 95, 15.10.1–15.10.15. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 December 2021).
- RStudio Team. RStudio: Integrated Development Environment for R. 2021. Available online: https://www.rstudio.com (accessed on 1 December 2021).
- Wickham, H.; Hester, J. Readr: Read Rectangular Text Data. 2020. Available online: https://readr.tidyverse.org/ (accessed on 1 December 2021).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. 2021. Available online: https://dplyr.tidyverse.org// (accessed on 1 December 2021).
- Wickham, H.; Seidel, D. Scales: Scale Functions for Visualization. 2020. Available online: https://scales.r-lib.org/ (accessed on 1 December 2021).
- Wickham, H. Tidyr: Tidy Messy Data. 2021. Available online: https://tidyr.tidyverse.org/ (accessed on 1 December 2021).
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lüdecke, D. sjPlot: Data Visualization for Statistics in Social Science. 2021. Available online: https://strengejacke.github.io/sjPlot/ (accessed on 1 December 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Thomas, J.P.; Ölbei, M.; Brooks-Warburton, J.; Korcsmaros, T.; Modos, D. Analysing miRNA-Target Gene Networks in Inflammatory Bowel Disease and Other Complex Diseases Using Transcriptomic Data. Genes 2022, 2022, 370. [Google Scholar] [CrossRef]
- Soroosh, A.; Koutsioumpa, M.; Pothoulakis, C.; Iliopoulos, D. Functional role and therapeutic targeting of microRNAs in inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G256. [Google Scholar] [CrossRef]
- Cao, B.; Zhou, X.; Ma, J.; Zhou, W.; Yang, W.; Fan, D.; Hong, L. Role of MiRNAs in Inflammatory Bowel Disease. Dig. Dis. Sci. 2017, 62, 1426–1438. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef] [PubMed]
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-Coding RNAs in Human Breast Milk: A Systematic Review. Front. Immunol. 2021, 12, 725323. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef]
- Shah, K.B.; Chernausek, S.D.; Garman, L.D.; Pezant, N.P.; Plows, J.F.; Kharoud, H.K.; Demerath, E.W.; Fields, D.A. Human Milk Exosomal MicroRNA: Associations with Maternal Overweight/Obesity and Infant Body Composition at 1 Month of Life. Nutrients 2021, 13, 1091. [Google Scholar] [CrossRef]
- Shiff, Y.E.; Reif, S.; Marom, R.; Shiff, K.; Reifen, R.; Golan-Gerstl, R. MiRNA-320a is less expressed and miRNA-148a more expressed in preterm human milk compared to term human milk. J. Funct. Foods 2019, 57, 68–74. [Google Scholar] [CrossRef]
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human Breast Milk miRNA, Maternal Probiotic Supplementation and Atopic Dermatitis in Offspring. PLoS ONE 2015, 10, e0143496. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef]
- Tomé-Carneiro, J.; Fernández-Alonso, N.; Tomás-Zapico, C.; Visioli, F.; Iglesias-Gutierrez, E.; Dávalos, A. Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help. Pharmacol. Res. 2018, 132, 21–32. [Google Scholar] [CrossRef]
- Kakimoto, Y.; Matsushima, Y.; Tanaka, M.; Hayashi, H.; Wang, T.; Yokoyama, K.; Ochiai, E.; Osawa, M. MicroRNA profiling of gastric content from breast-fed and formula-fed infants to estimate last feeding: A pilot study. Abbreviations BM Breast milk FM Formula milk GC Gastric content GCB GC of breast-fed infant GCF GC of formula-fed infant GJ Gastric juice. Int. J. Leg. Med. 2020, 134, 903–909. [Google Scholar] [CrossRef]

| Anthropometric Data | Mean | SD | |
| Age (years) | 22.93 | ±4.95 | |
| Height (m) α | 1.57 | ±0.07 | |
| Weight (Kg) β | 56.48 | ±10.01 | |
| BMI γ | 23.22 | ±3.74 | |
| Type of delivery | n | % | |
| Elective caesarean | 4 | 6.67 | |
| Non elective caesarean | 13 | 21.67 | |
| Vaginal | 43 | 71.66 | |
| Parity | n | % | |
| Primiparous δ | 16 | 27.12 | |
| Multiparous δ | 43 | 72.88 | |
| Milk extraction | n | % | |
| Manual ε | 34 | 82.93 | |
| Pump ε | 7 | 17.07 | |
| Socioeconomic data | n | % | |
| Residence | Mexico City (19°25′10″ N 99°08′44″ O) | 15 | 25.00 |
| Estado de México (19°21′15″ N 99°37′51″ O) | 36 | 60.00 | |
| Guerrero State (17°36′47″ N 99°57′00″ O) | 2 | 3.33 | |
| Hidalgo State (20°28′42″ N 98°51′49″ O) | 1 | 1.67 | |
| Oaxaca State (16°53′53″ N 96°24′51″ O) | 3 | 5.00 | |
| Puebla State (19°00′13″ N 97°53′18″ O) | 2 | 3.33 | |
| Veracruz State (19°26′05″ N 96°22′59″ O) | 1 | 1.67 | |
| Activity | Housewife δ | 52 | 88.14 |
| Student δ | 2 | 3.39 | |
| General employee δ | 5 | 8.47 | |
| Education | Primary school (6 years) | 20 | 33.33% |
| Secondary school (3 years) | 36 | 60.00% | |
| High school (3 years) | 1 | 1.67% | |
| University school (4–5 years) | 3 | 5.00% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vélez-Ixta, J.M.; Benítez-Guerrero, T.; Aguilera-Hernández, A.; Martínez-Corona, H.; Corona-Cervantes, K.; Juárez-Castelán, C.J.; Rangel-Calvillo, M.N.; García-Mena, J. Detection and Quantification of Immunoregulatory miRNAs in Human Milk and Infant Milk Formula. BioTech 2022, 11, 11. https://doi.org/10.3390/biotech11020011
Vélez-Ixta JM, Benítez-Guerrero T, Aguilera-Hernández A, Martínez-Corona H, Corona-Cervantes K, Juárez-Castelán CJ, Rangel-Calvillo MN, García-Mena J. Detection and Quantification of Immunoregulatory miRNAs in Human Milk and Infant Milk Formula. BioTech. 2022; 11(2):11. https://doi.org/10.3390/biotech11020011
Chicago/Turabian StyleVélez-Ixta, Juan Manuel, Tizziani Benítez-Guerrero, Arlene Aguilera-Hernández, Helga Martínez-Corona, Karina Corona-Cervantes, Carmen Josefina Juárez-Castelán, Martín Noé Rangel-Calvillo, and Jaime García-Mena. 2022. "Detection and Quantification of Immunoregulatory miRNAs in Human Milk and Infant Milk Formula" BioTech 11, no. 2: 11. https://doi.org/10.3390/biotech11020011
APA StyleVélez-Ixta, J. M., Benítez-Guerrero, T., Aguilera-Hernández, A., Martínez-Corona, H., Corona-Cervantes, K., Juárez-Castelán, C. J., Rangel-Calvillo, M. N., & García-Mena, J. (2022). Detection and Quantification of Immunoregulatory miRNAs in Human Milk and Infant Milk Formula. BioTech, 11(2), 11. https://doi.org/10.3390/biotech11020011







