PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Division of Data into Training and Testing Datasets
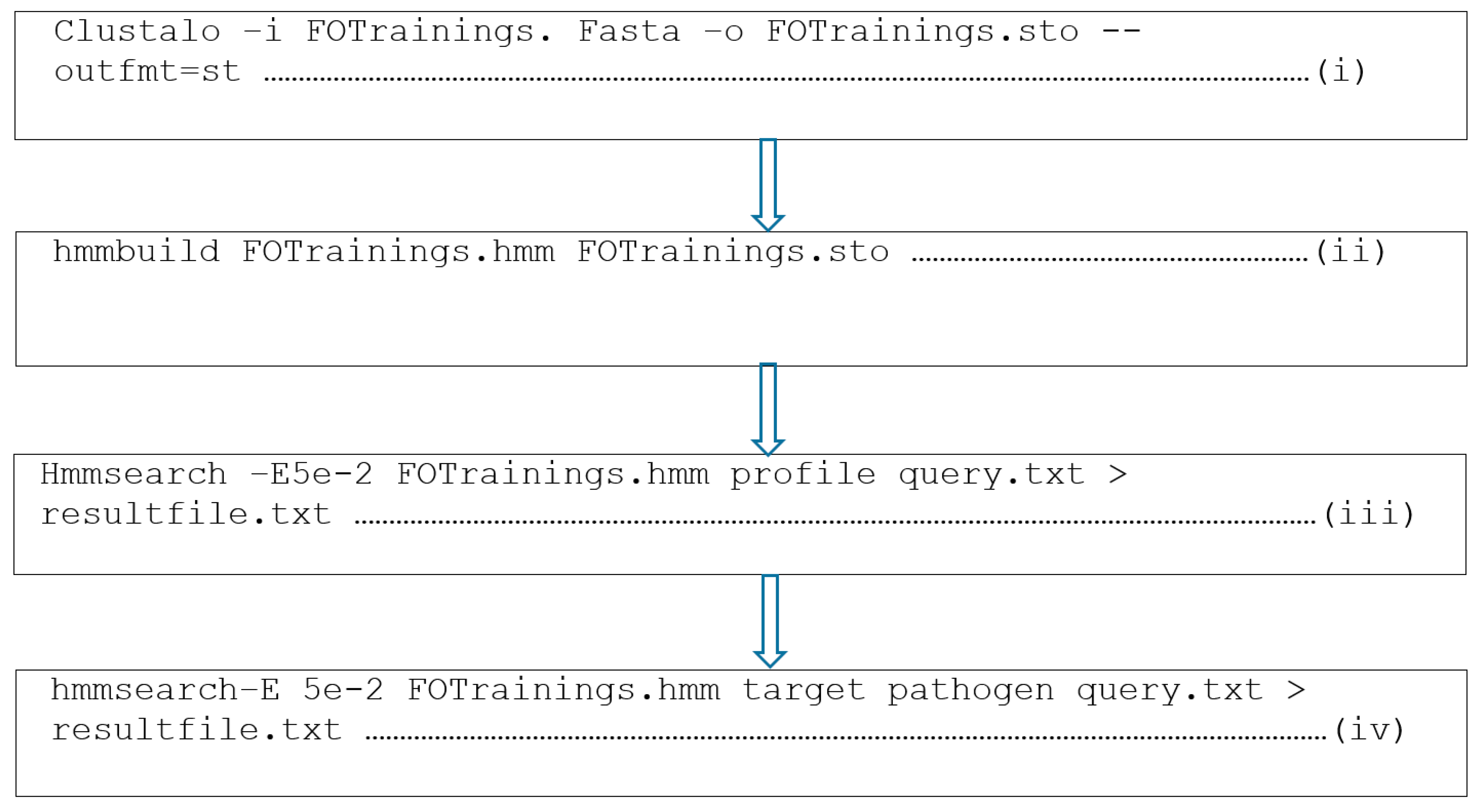
2.3. Construction of AMPs Profiles
2.4. Independent Profile Testing
2.5. Performance Measurement of Each Profile
2.6. Novel Putative Anti-Fusarium oxysporum AMPs Identification
2.7. Identification of Receptors
2.8. Evaluation of the Protein Receptor Model
2.9. Physicochemical Properties of the Putative Anti-Fusarium oxysporum AMPs and the Fusarium oxysporum Fpr1 Protein
2.10. Structure Predictions of the Putative Anti-Fusarium oxysporum AMPs and Fusarium oxysporum Proteins
2.11. Interaction Analysis of the Putative Anti-Fusarium oxysporum AMPs and Fusarium oxysporum Protein
3. Results
3.1. Data Collection
3.2. Profile Construction
3.3. Testing and Performance Measurement of the Profile
3.4. Proteome Sequence Database Query and Discovery of Anti-Fusarium oxysporum AMPs
3.5. Receptor Identification
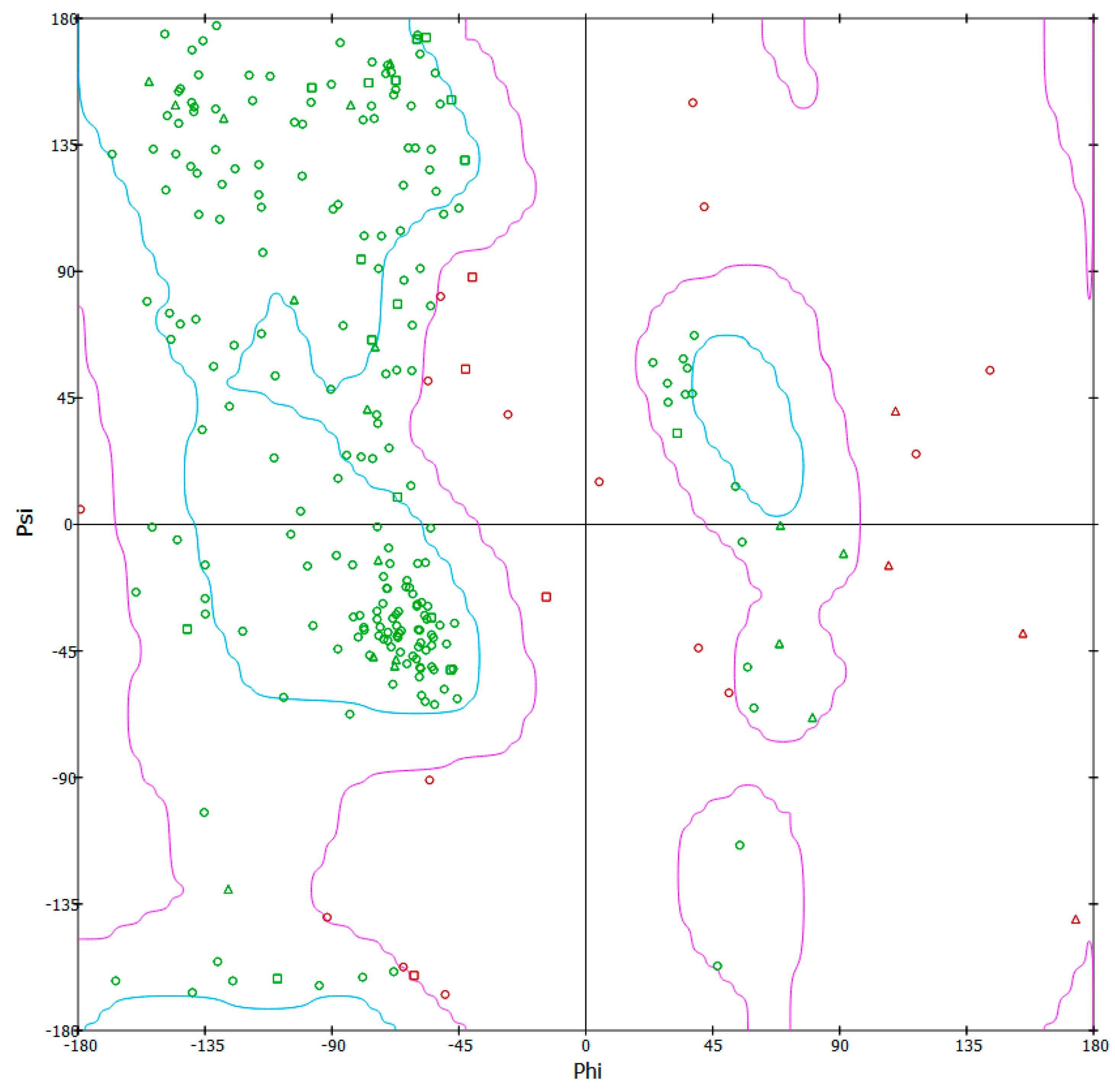
3.6. 3-D Model Structure Validation
3.7. Physicochemical Analysis of the Anti-Fusarium oxysporum AMPs and Fusarium oxysporum PR-1-Like Protein
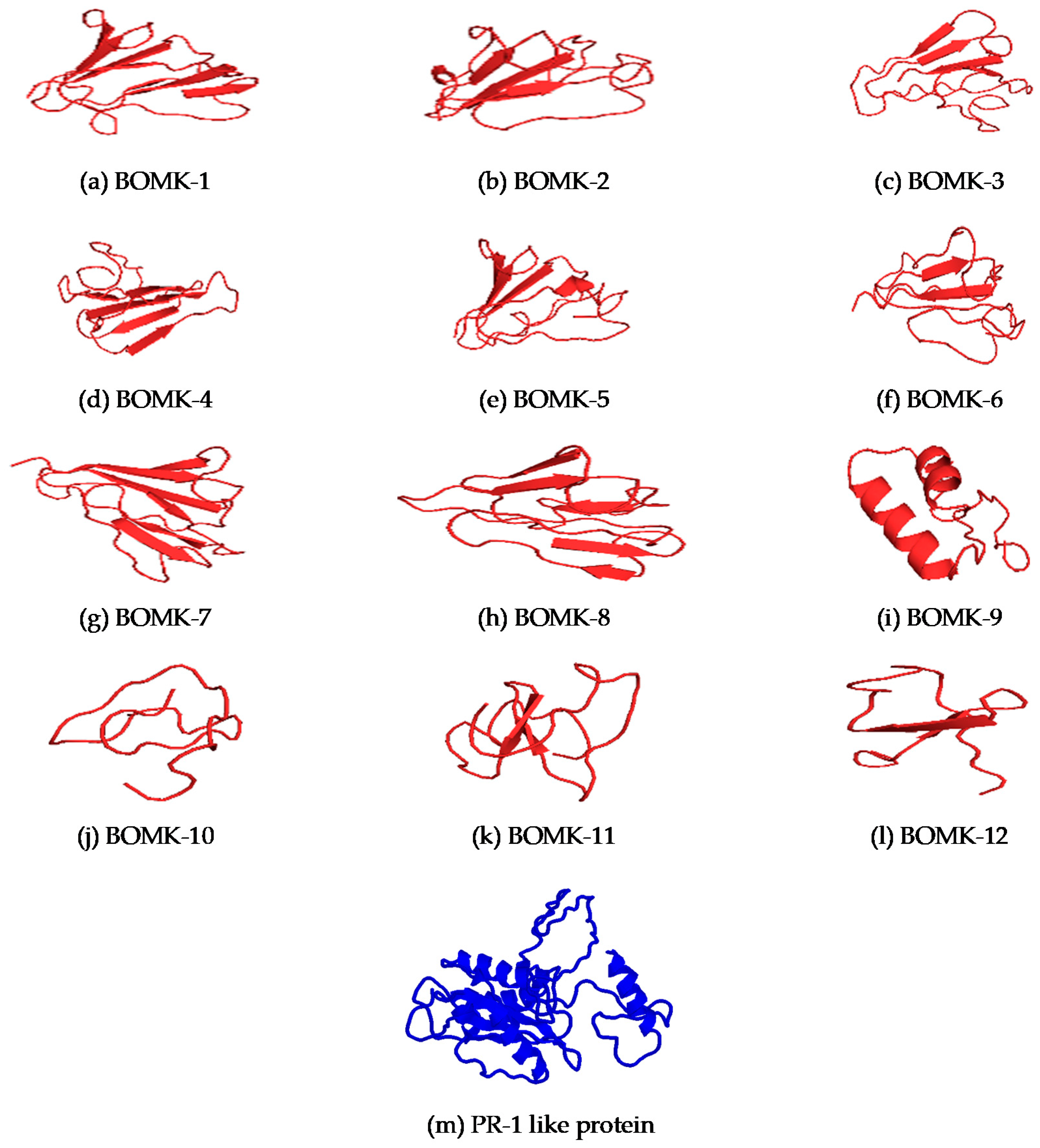
3.8. Structure Prediction and Docking
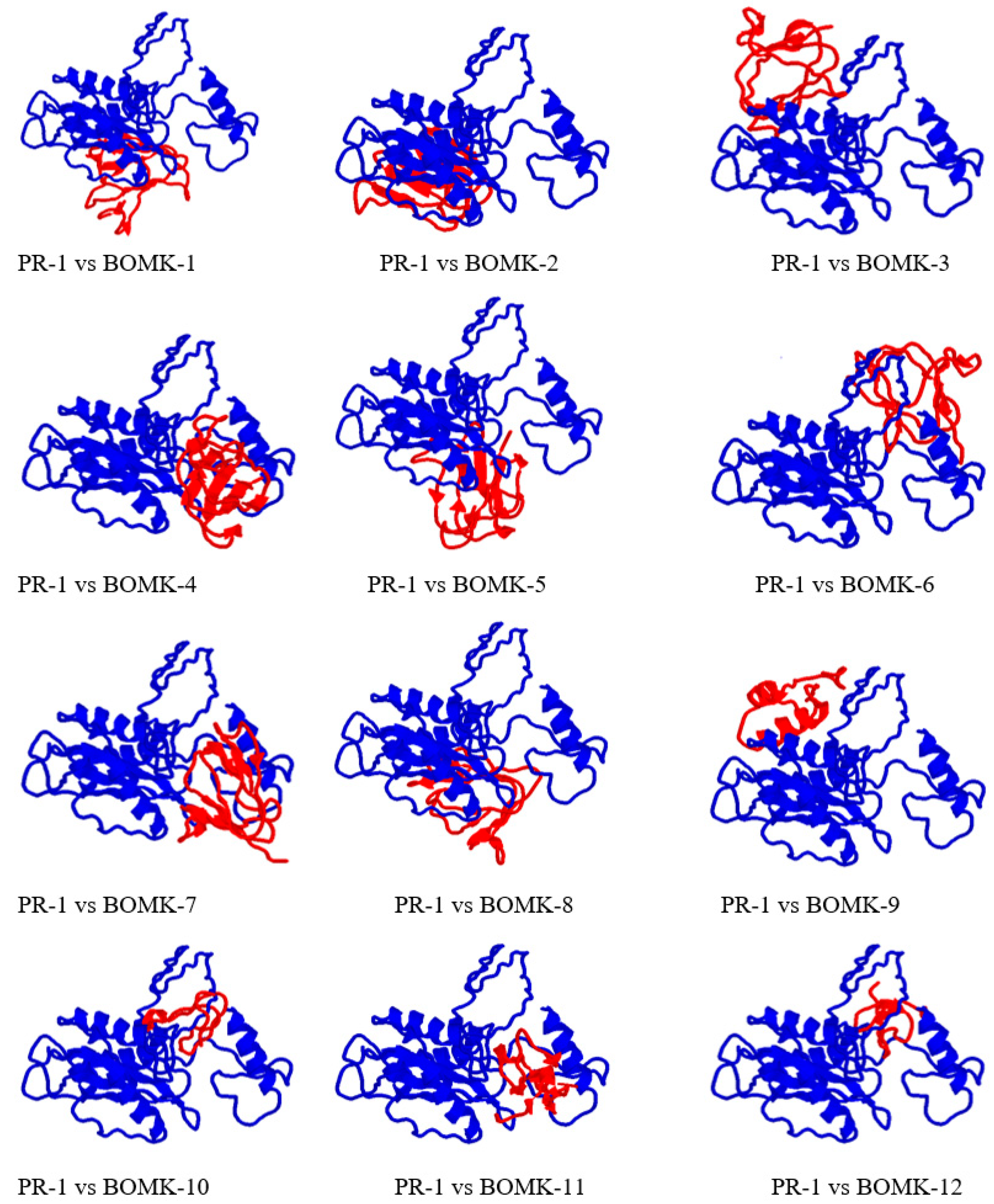
3.9. Protein-Peptide Interaction between Anti-Fusarium oxysporum and Fusarium oxysporum Fpr1
4. Discussion
4.1. Data Retrieval and Profile Construction of the Anti-Fusarium oxysporum AMPs
4.2. Testing of the Profiles
4.3. Proteome Sequence Database Query and Discovery of Anti-Fusarium oxysporum AMPs
4.4. Receptor Identification
4.5. Physicochemical Analysis
4.6. Structure Prediction and Docking Interaction Analysis of the Putative Anti-Fusarium oxysporum and Fusarium oxysporum PR-1-Like Protein
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.T.; Gordon, T.R. Management of Fusarium wilt of strawberry. Crop. Prot. 2015, 73, 67–72. [Google Scholar] [CrossRef]
- Ploetz, R.C.; Herbert, J.; Sebasigari, K.; Hernandez, J.H.; Pegg, K.G.; Ventura, J.A.; Mayato, L.S. Importance of Fusarium Wilt in Different Banana Growing Regions; APS Press: St. Paul, MN, USA, 1990; pp. 9–26. [Google Scholar]
- Jacobson, D.; Gordon, T. Fusarium oxysporum f. sp. melonis: A case study of diversity within a forma specialis. Phytopathologyogy 1991, 81, 1064–1067. [Google Scholar]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathologyogy 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Ordoñez, N.; García-Bastidas, F.; Laghari, H.B.; Akkary, M.Y.; Harfouche, E.N.; Al Awar, B.N.; Kema, G.H.J. First Report of Fusarium oxysporum f. sp. cubense Tropical Race 4 Causing Panama Disease in Cavendish Bananas in Pakistan and Lebanon. Plant Dis. 2016, 100, 209. [Google Scholar] [CrossRef]
- Lu, S.; Edwards, M.C. Molecular Characterization and Functional Analysis of PR-1-Like Proteins Identified from the Wheat Head Blight Fungus Fusarium graminearum. Phytopathology 2018, 108, 510–520. [Google Scholar] [CrossRef]
- Peng, Y.; Van Wersch, R.; Zhang, Y. Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef]
- Baysal, O.; Karaaslan, C.; Siragusa, M.; Alessandro, R.; Carimi, F.; De Pasquale, F.; Da Silva, J.A.T. Molecular markers reflect differentiation of Fusarium oxysporum forma speciales on tomato and forma on eggplant. Biochem. Syst. Ecol. 2013, 47, 139–147. [Google Scholar] [CrossRef]
- Gopinath, V.; Velusamy, P. Extracellular biosynthesis of silver nanoparticles using Bacillus sp. GP-23 and evaluation of their antifungal activity towards Fusarium oxysporum. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 106, 170–174. [Google Scholar] [CrossRef]
- Henry, P.M.; Kirkpatrick, S.C.; Islas, C.M.; Pastrana, A.M.; Yoshisato, J.A.; Koike, S.T.; Daugovish, O.; Gordon, T.R. The Population of Fusarium oxysporum f. sp. fragariae, Cause of Fusarium Wilt of Strawberry, in California. Plant Dis. 2017, 101, 550–556. [Google Scholar] [CrossRef]
- Lievens, B.; Rep, M.; Thomma, B.P. Recent developments in the molecular discrimination of formae speciales of fusarium oxysporum. Pest. Manag. Sci. 2008, 64, 781–788. [Google Scholar] [CrossRef]
- García-Bastidas, F.; Ordóñez, N.; Konkol, J.; Al-Qasim, M.; Naser, Z.; Abdelwali, M.; Salem, N.; Waalwijk, C.; Ploetz, R.C.; Kema, G.H.J. First Report of Fusarium oxysporum f. sp. cubense Tropical Race 4 Associated with Panama Disease of Banana outside Southeast Asia. Plant Dis. 2014, 98, 694. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Su, C.-C.; Chao, C.-P.; Chen, C.-Y.; Chang, C.-J.; Huang, J.-W.; Chang, P.-F.L. A molecular diagnosis method using real-time PCR for quantification and detection of Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 2012, 135, 395–405. [Google Scholar] [CrossRef]
- Tassanakajon, A.; Rimphanitchayakit, V.; Visetnan, S.; Amparyup, P.; Somboonwiwat, K.; Charoensapsri, W.; Tang, S. Shrimp humoral responses against pathogens: Antimicrobial peptides and melanization. Dev. Comp. Immunol. 2018, 80, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.; Müller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef]
- Tincho, M.; Gabere, M.N.; Pretorius, A. In Silico Identification and Molecular Validation of Putative Antimicrobial Peptides for HIV Therapy. J. AIDS Clin. Res. 2016, 7. [Google Scholar] [CrossRef]
- Williams, M.E.; Tincho, M.; Gabere, M.; Uys, A.; Pretorius, M.M.A.A. Molecular Validation of Putative Antimicrobial Peptides for Improved Human Immunodeficiency Virus Diagnostics via HIV Protein p24. J. AIDS Clin. Res. 2016, 7, 2. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32 (Suppl. 1), D590–D592. [Google Scholar] [CrossRef]
- Wang, J.T.L.; Zaki, M.J.; Toivonen, H.; Shasha, D. Data Mining in Bioinformatics; Springer: London, UK, 2005; pp. 3–8. [Google Scholar]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38 (Suppl. 1), D774–D780. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Roddy, J. Reducing False Sequence Annotation Due to Alignment Overextension. In Proceedings of the Conference on Undergraduate Research (UMCUR), Missoula, MT, USA, 27 April 2018. [Google Scholar]
- Fang, Q. Predicting Functional Alterations Caused By Non-Synonymous Variants in CHO Using Models Based on Phylogenetic Tree and Evolutionary Preservation. Ph.D. Thesis, University of Sheffield, Sheffield, UK, 2018. [Google Scholar]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The Ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33 (Suppl. 1), D501–D504. [Google Scholar]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kemmish, H.; Fasnacht, M.; Yan, L. Fully automated antibody structure prediction using BIOVIA tools: Validation study. PLoS ONE 2017, 12, e0177923. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Potterton, L.; Yuan, F.; van Vlijmen, H.; Karplus, M. Evaluation of comparative protein modeling by MODELLER. Proteins Struct. Funct. Bioinf. 1995, 23, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Zouhir, A.; Ben Hamida, J.; Fliss, I. BACTIBASE: A new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E.; Grigolava, M.; Chubinidze, M.; Gogoladze, G.; Vishnepolsky, B.; et al. DBAASP v.2: An enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2008, 37 (Suppl. 1), D933–D937. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Eswar, N.; Sali, A. Protein structure modeling. In From Molecules to Medicines; Springer: Berlin/Heidelberg, Germany, 2009; pp. 139–151. [Google Scholar]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- De Lano, W.L. PyMOL. Available online: http://www.pymol.org (accessed on 31 March 2020).
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33 (Suppl. 2), W363–W367. [Google Scholar] [CrossRef]
- Sayle, R.A.; Milner-White, E.J. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 1995, 20, 374–376. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Perez-Nadales, E.; Di Pietro, A. The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum. Mol. Plant Pathol. 2015, 16, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Gujjula, K.R. Prediction and Comparison of HIV-1 Protease Inhibitor Binding Energies by Various Molecular Docking Methods. Ph.D. Thesis, National Institute of Technology Rourkela, Orissa, India, 2008. [Google Scholar]
- Kim, B.C.; Kim, Y.S.; Chung, J.Y.; Jurng, J.S.; Song, M.Y. Single-Stranded Nucleic Acid Aptamers Specifically Binding to Klebsiella Pneumoniae and Method for Detecting K. pneumonia Using the Same. U.S. Patent No. 9,284,550, 15 March 2016. [Google Scholar]
- Madera, M.; Gough, J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002, 30, 4321–4328. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P.S. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, srep01607. [Google Scholar] [CrossRef]
- Liu, L.; Oza, S.; Hogan, D.; Perin, J.; Rudan, I.; Lawn, E.J.; Cousens, S.; Mathers, C.; Black, E.R. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: An updated systematic analysis. Lancet 2015, 385, 430–440. [Google Scholar] [CrossRef]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W.I. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Silva, C.M.; Carneiro, F.; O’Neill, A.; Cabral, J.S.M.; Guebitz, G.; Cavaco-Paulo, A. Cutinase? A new tool for biomodification of synthetic fibers. J. Polym. Sci. Part A Polym. Chem. 2005, 43, 2448–2450. [Google Scholar] [CrossRef]
- Alexandrakis, G.; Jalali, S.; Gloor, P. Diagnosis of Fusarium keratitis in an animal model using the polymerase chain reaction. Br. J. Ophthalmol. 1998, 82, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Ahn, J.-Y.; Moon, S.-H.; Lee, J. Biodegradation and detoxification of organophosphate insecticide, malathion by Fusarium oxysporum f. sp. pisi cutinase. Chemosphere 2005, 60, 1349–1355. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of Peptide Hydrophobicity in the Mechanism of Action of α-Helical Antimicrobial Peptides. Antimicrob. Agents Chemother. 2006, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Tachi, T.; Epand, R.F.; Epand, R.M.; Matsuzaki, K. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide-lipid interactions and selective toxicity. Biochemistry 2002, 41, 10723–10731. [Google Scholar] [CrossRef] [PubMed]
- Jodoin, J.; Hincke, M.T. Histone H5 is a potent Antimicrobial Agent and a template for novel Antimicrobial Peptides. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Abraham, T.; Prenner, E.J.; Lewis, R.N.; Mant, C.T.; Keller, S.; Hodges, R.S.; McElhaney, R.N. Structure–activity relationships of the antimicrobial peptide gramicidin S and its analogs: Aqueous solubility, self-association, conformation, antimicrobial activity and interaction with model lipid membranes. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 1420–1429. [Google Scholar] [CrossRef]
- Bakare, O.O.; Keyster, M.; Pretorius, A. Identification of biomarkers for the accurate and sensitive diagnosis of three bacterial pneumonia pathogens using in silico approaches. BMC Mol. Cell Biol. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins Struct. Funct. Bioinform. 2004, 57, 702–710. [Google Scholar] [CrossRef]
- Bacalum, M.; Radu, M. Cationic antimicrobial peptides cytotoxicity on mammalian cells: An analysis using therapeutic index integrative concept. Int. J. Peptide Res. Ther. 2015, 21, 47–55. [Google Scholar] [CrossRef]
- Mihajlovic, M.; Lazaridis, T. Charge distribution and imperfect amphipathicity affect pore formation by antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
| Profiles | Training Datasets | Testing Datasets | Total |
|---|---|---|---|
| FO | 24 | 8 | 32 |
| True Positive | False Negative | True Negative | False Positive |
| 6 | 2 | 17,236 | 0 |
| Sensitivity (%) | Specificity (%) | Accuracy (%) | MCC |
| 75 | 100 | 99.99 | 0.87 |
| Organism | Name | AMPs | Number of Amino Acid Residues | Bit Scores | E Values |
|---|---|---|---|---|---|
| Selaginella moellendorffii | BOMK-1 | AlaTrpAlaGlyProGlyCysAsnAsnArgLeu----------ValGlyAlaSerGlnHisGlyGlyTyrSerPheAlaTyrGlnGlyGlnThrAlaAlaAlaTyrAsnThrAlaAsnCysArgGlyValAlaHisThrArgPheSerSerLysGlyGluCysLysSerGlySerValGlnAspCysSerGlyPheGlyTrpArgSerIlePheIleGlnCys | 80 | 35.3 | 5 × 10−8 |
| Selaginella moellendorffii | BOMK-2 | TrpAlaGlyProGlyCysAsnAsnArgLeuGlu----------GlyAlaSerGlnHisGlyGlyTyrSerPheAlaTyrGlnGlyGlnThrAlaAlaAlaTyrAsnThrAlaAsnCysGlnGlyValAlaHisThrArgPheSerArgLysGlyGluCysLysSerGlySerValGlnAspCysSerGlyPheGlyTrpAsnSerPhePheIleGlnCys | 80 | 32.8 | 3.3 × 10−7 |
| Selaginella moellendorffii | BOMK-3 | ThrTrpAlaGlyProGlyCysAsnAsnArgLeu----------ValGlyAlaSerGlnHisGlyGlyTyrSerPheGlyTyrGlnGlyGlnThrAlaAlaAlaTyrAsnThrAlaAsnCysGlnGlyValAlaHisThrArgPheSerArgLysGlyGluCysLysSerGlySerValGlnAspCysSerGlyPheGlyTrpAsnSerPhePheIleGlnCys | 80 | 32.7 | 3.6 × 10−7 |
| Selaginella moellendorffii | BOMK-4 | AlaTrpAlaGlyProGlyCysAsnAsnValLeu----------ValArgAlaSerGlnHisGlyGlyTyrSerPheValTyrGlnGlyGlnThrAlaAlaAlaTyrAsnThrAlaAsnCysArgGlyValAlaHisThrArgPheSerArgLysGlyGluCysLysSerGlySerValGlnAspCysSerGlyPheGlyTrpAsnSerPhePheIleGlnCys | 80 | 31.1 | 1.1 × 10−6 |
| Selaginella moellendorffii | BOMK-5 | ThrTrpAlaGlyProGlyCysAsnAsnGlnArg----------ValGlyAlaSerGlnHisGlyGlyTyrSerPheGlyTyrGlnGlyGlnThrAlaAlaAlaTyrAsnThrAlaAsnCysGlnGlyValAlaGlnThrArgPheSerAlaLysGlyGluCysLysSerGlySerValGlnAspCysSerGlyPheGlyTrpAsnSerPhePheIleGlnCys | 80 | 27.6 | 1.4 × 10−5 |
| Selaginella moellendorffii | BOMK-6 | TrpAlaGlyProGlyCysAsnAsnTrpLeuGlu----------AlaSerGlnHisGlyGlyTyrSerValAlaTyrLeuGlyHisAlaAlaAlaAlaTyrAsnThrAlaAsnCysGlnGlyValAlaGlnArgTrpPheArgArgLysGlyHisCysSerSerGlyCysAlaSerGluCysGluGlyPheArgTrpAsnSerIlePheIleGlnCysSerSer | 80 | 26.4 | 3.6 × 10−5 |
| Selaginella moellendorffii | BOMK-7 | TrpAlaGlyProGlyGlyAsnAsnArgLeuGlu----------AlaSerGlnHisGlyGlyTyrSerValValTyrLeuGlyHisAlaAlaAlaAlaTyrAsnThrAlaAsnCysGlnGlyValAlaGlnArgTrpPheArgArgLysGlyHisCysSerSerGlyCysAlaSerGluCysGluGlyPheArgTrpAsnSerIlePheIleGlnCysSerSer | 80 | 25.0 | 0.00011 |
| Setaria italic | BOMK-8 | ThrSerTrpAlaGlyProGlyCysSerGlyGln----------AsnLeuGlnPheTyrAspGlyGlnGluLysSerTyrGlnGlyGlnThrAlaArgLeuTyrThrGluThrGlyCysAlaGlyThrSerTyrLeuValPheGluAspThrGlnAlaCysGlySerGlyCysAlaSerGluCysGluAspPheGlyTrpArgSerIle | 75 | 21.8 | 0.00073 |
| Oryza sativum | BOMK-9 | LysIleGlnValGluAlaLysSerCysCysProGly----------TyrAsnSerCysArgPheAlaGlyGlySerArgAspThrCysAlaLysLeuSerGlyCysLysIleValCysAspGlyAsnCysLysProProTyr | 54 | 23.5 | 0.00079 |
| Zea mays | BOMK-10 | GlyGlyHisProAspGlyAlaIleProCysGlyGlu----------ValPheGlyCysArgGlyTrpGlyTyrCysGlu | 33 | 19.8 | 0.0037 |
| Solanum lycopersicum | BOMK-11 | AlaGlnGlnCysGlyIleGlnAlaGlyGlyAla----------PheGlyTyrCysGlyThrThrAlaThrAlaTyrCysGlyProGlyCysGlnSerGlnCys | 41 | 16.0 | 0.026 |
| Arabidopsis thaliana | BOMK-12 | ValGlnGluTyrGlyCysProAsnCysLysArg----------GlyGluLeuValMetGluCysAsnLys | 30 | 17.7 | 0.034 |
| AMP | Mol. Mass (Da) | Common Amino Acids | Hydrophobicity (%) | Isoelectric Point | Boman Index (Kcal/mol) | Charge | Half-Life (h) |
|---|---|---|---|---|---|---|---|
| BOMK-1 | 8651.30 | G | 35 | 8.70 | 1.89 | +5 | 4.4 |
| BOMK-2 | 8531.35 | G | 35 | 8.28 | 1.69 | +3 | 2.8 |
| BOMK-3 | 8618.44 | G | 33 | 8.28 | 1.71 | +3 | 7.2 |
| BOMK-4 | 8700.36 | G | 37 | 8.49 | 1.70 | +4 | 4.4 |
| BOMK-5 | 8509.84 | G | 34 | 8.28 | 1.51 | +3 | 7.2 |
| BOMK-6 | 8900.35 | GA | 40 | 8.40 | 1.63 | +4 | 2.8 |
| BOMK-7 | 8852.32 | G | 37 | 8.70 | 1.81 | +5 | 2.8 |
| BOMK-8 | 8121.76 | G | 32 | 3.88 | 1.42 | −5 | 7.2 |
| BOMK-9 | 5796.17 | C | 35 | 8.70 | 1.72 | +5 | 1.3 |
| BOMK-10 | 3376.98 | G | 42 | 4.42 | 0.26 | −2 | 30 |
| BOMK-11 | 4042.34 | GC | 41 | 3.75 | 0.45 | −1 | 4.4 |
| BOMK-12 | 3537.55 | C | 40 | 7.08 | 2.04 | 0 | 100 |
| Receptor | M. wt (Da) | Common Amino Acid | Hydrophobicity (%) | Isoelectric Point | Instability Index | Aliphatic Index | Half-Life (Hours) |
|---|---|---|---|---|---|---|---|
| PR-1-like protein | 95,472.44 | SLP | 40 | 10.00 | 71.04 | 82.72 | 1.1 |
| AMPs | C Score | TM Score | RSMD (Å) |
|---|---|---|---|
| BOMK-1 | 0.84 | 0.83 ± 0.08 | 2.0 ± 1.6 |
| BOMK-2 | 0.49 | 0.78 ± 0.10 | 2.6 ± 1.9 |
| BOMK-3 | 0.83 | 0.83 ± 0.08 | 2.0 ± 1.6 |
| BOMK-4 | 0.88 | 0.83 ± 0.08 | 1.9 ± 1.6 |
| BOMK-5 | 0.74 | 0.81 ± 0.09 | 2.2 ± 1.7 |
| BOMK-6 | 0.77 | 0.82 ± 0.09 | 2.1 ± 1.7 |
| BOMK-7 | 0.73 | 0.81 ± 0.09 | 2.3 ± 1.8 |
| BOMK-8 | 0.67 | 0.80 ± 0.09 | 2.2 ± 1.7 |
| BOMK-9 | 0.42 | 0.66 ± 0.13 | 3.6 ± 2.5 |
| BOMK-10 | 0.86 | 0.61 ± 0.14 | 3.5 ± 2.4 |
| BOMK-11 | 0.58 | 0.79 ± 0.09 | 1.4 ± 1.3 |
| BOMK-12 | −0.75 | 0.62 ± 0.14 | 3.3 ± 2.3 |
| PR-1-like protein | −1.44 | 0.54 ± 0.15 | 9.1 ± 4.6 |
| AMPs | Pathdock Geometry Binding Affinity Scores | HDock Binding Energy Scores (Kcal/mol) |
|---|---|---|
| BOMK−1 | 12,828 | −254.09 |
| BOMK−2 | 12,976 | −238.30 |
| BOMK−3 | 13,776 | −263.43 |
| BOMK−4 | 12,652 | −244.88 |
| BOMK−5 | 13,688 | −263.63 |
| BOMK−6 | 14,334 | −242.95 |
| BOMK−7 | 12,776 | −273.34 |
| BOMK−8 | 14,016 | −213.09 |
| BOMK−9 | 11,468 | −239.40 |
| BOMK−10 | 14,806 | −230.06 |
| BOMK−11 | 9958 | −220.11 |
| BOMK−12 | 14,420 | −236.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakare, O.O.; Gokul, A.; Keyster, M. PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach. BioTech 2021, 10, 8. https://doi.org/10.3390/biotech10020008
Bakare OO, Gokul A, Keyster M. PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach. BioTech. 2021; 10(2):8. https://doi.org/10.3390/biotech10020008
Chicago/Turabian StyleBakare, Olalekan Olanrewaju, Arun Gokul, and Marshall Keyster. 2021. "PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach" BioTech 10, no. 2: 8. https://doi.org/10.3390/biotech10020008
APA StyleBakare, O. O., Gokul, A., & Keyster, M. (2021). PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach. BioTech, 10(2), 8. https://doi.org/10.3390/biotech10020008







