Insights for Deriving Induced Pluripotent Stem Cells in Marsh Deer (Blastocerus dichotomus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Biopsies
2.2. Isolation of Somatic Cells
2.3. Pluripotency Induction
2.3.1. Electroporation Using the PiggyBac System
2.3.2. Lentiviral Transduction Using the STEMCCA System
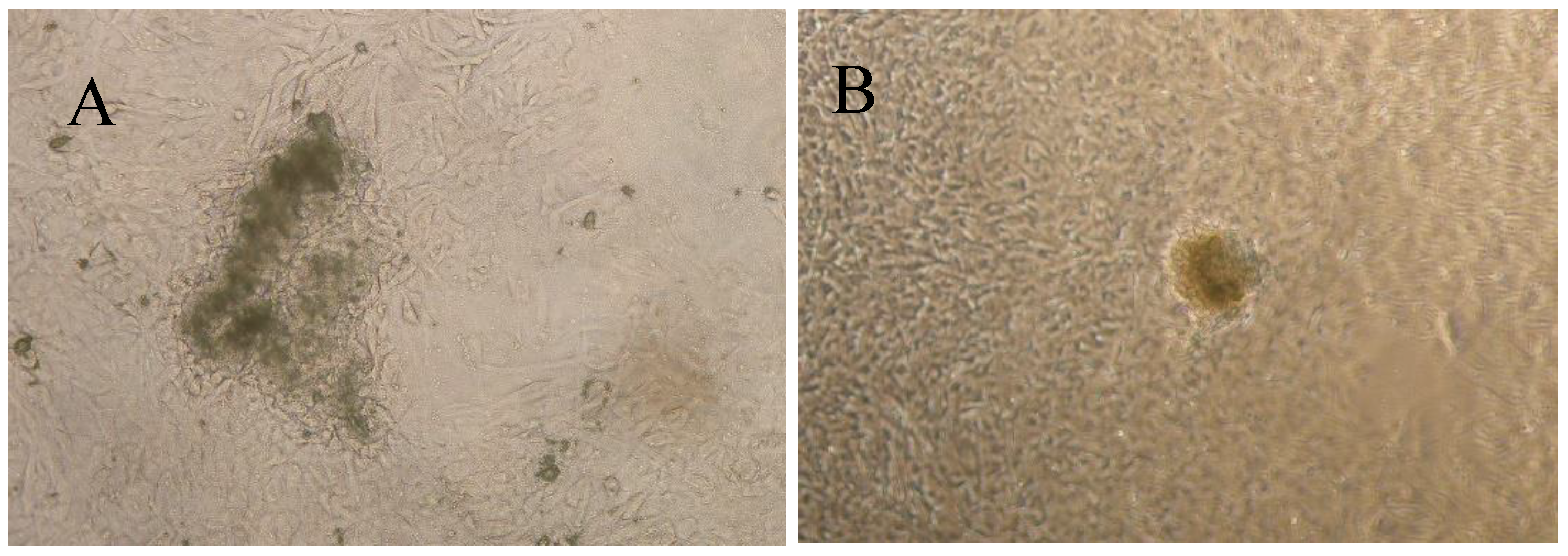
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schipper, J.; Chanson, J.S.; Chiozza, F.; Cox, N.A.; Hoffmann, M.; Katariya, V.; Lamoreux, J.; Rodrigues, A.S.L.; Stuart, S.N.; Temple, H.J.; et al. The status of the world’s land and marine mammals: Diversity, threat, and knowledge. Science 2008, 322, 225–230. [Google Scholar] [CrossRef]
- The IUCN Red List of Threatened Species. Available online: https://www.iucnredlist.org/ (accessed on 31 March 2025).
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. A Primer of Conservation Genetics; Cambridge University Press: Cambridge, UK, 2004; p. 220. [Google Scholar]
- Keller, L.F.; Biebach, I.; Ewing, S.R.; Hoeck, P.E.A. The genetics of reintroductions: Inbreeding and genetic drift. In Reintroduction Biology: Integrating Science and Management; Ewen, J.G., Armstrong, D.P., Parker, K.A., Seddon, P.J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 360–394. [Google Scholar]
- Weber, M.; Gonzalez, S. Latin American deer diversity and conservation: A review of status and distribution. Écoscience 2003, 10, 443–454. [Google Scholar] [CrossRef]
- Piovezan, U.; Tiepolo, L.M.; Tomas, W.M.; Duarte, J.B.; Varela, D.; Marinho-Filho, J.S. Marsh deer Blastocerus dichotomus (Illiger, 1815). In Neotropical Cervidology: Biology and Medicine in Latin American Deer; Duarte, J.M.B., González, S., Eds.; Funep/IUCN: Jaboticabal, Brazil, 2010; pp. 66–76. [Google Scholar]
- González, S.; Lessa, E.P. Historia de la mastozoología en Uruguay. In Historia de la Mastozoología en Latinoamérica, las Guayanas y el Caribe; Ortega, J., Martínez, J.L., Tirira, D.G., Eds.; Editorial Murciélago Blanco y Asociación Ecuatoriana de Mastozoología: Quito, Ecuador; México D.F., Mexico, 2014; pp. 381–404. [Google Scholar]
- González, S.; Aristimuño, M.P.; Moreno, F. New record in Uruguay of the marsh deer (Blastocerus dichotomus Illiger, 1815) redefines its southern geographic distribution area. Front. Ecol. Evol. 2024, 12, 1419234. [Google Scholar] [CrossRef]
- González, S.; Duarte, J.M.B. Speciation, evolutionary history and conservation trends of Neotropical deer. Mastozool. Neotrop. 2020, 27, 37–47. [Google Scholar] [CrossRef]
- Holt, W.V.; Pickard, A.R. Role of reproductive technologies and genetic resource banks in animal conservation. Rev. Reprod. 1999, 4, 143–150. [Google Scholar] [CrossRef]
- Wildt, D.E. Genetic resource banks for conserving wildlife species: Justification, examples and becoming organized on a global basis. Anim. Reprod. Sci. 1992, 28, 247–257. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef]
- Hayashi, K.; Ogushi, S.; Kurimoto, K.; Shimamoto, S.; Ohta, H.; Saitou, M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 2012, 338, 971–975. [Google Scholar] [CrossRef]
- Smith, Z.D.; Sindhu, C.; Meissner, A. Molecular features of cellular reprogramming and development. Nat. Rev. Mol. Cell Biol. 2016, 17, 139–154. [Google Scholar] [CrossRef]
- González, F.; Boué, S.; Belmonte, J.C.I. Methods for making induced pluripotent stem cells: Reprogramming a la carte. Nat. Rev. Genet. 2011, 12, 231–242. [Google Scholar] [CrossRef]
- Wang, D.; Berg, D.; Ba, H.; Sun, H.; Wang, Z.; Li, C. Deer antler stem cells are a novel type of cells that sustain full regeneration of a mammalian organ—Deer antler. Cell Death Dis. 2019, 10, 443. [Google Scholar] [CrossRef]
- Colitti, M.; Allen, P.; Price, J.S. Programmed cell death in the regenerating deer antler. J. Anat. 2005, 207, 339–351. [Google Scholar] [CrossRef]
- Kierdorf, U.; Kierdorf, H. Deer antlers—A model of mammalian appendage regeneration: An extensive review. Gerontology 2011, 57, 53–65. [Google Scholar] [CrossRef]
- Li, C.; Yang, F.; Sheppard, A. Adult stem cells and mammalian epimorphic regeneration—Insights from studying annual renewal of deer antlers. Curr. Stem Cell Res. Ther. 2009, 4, 237–251. [Google Scholar] [CrossRef]
- Rolf, H.J.; Kierdorf, U.; Kierdorf, H.; Schulz, J.; Seymour, N.; Schliephake, H.; Napp, J.; Niebert, S.; Wolfel, H.; Wiese, K.G. Localization and characterization of STRO-1 cells in the deer pedicle and regenerating antler. PLoS ONE 2008, 3, e2064. [Google Scholar] [CrossRef]
- Tat, P.A.; Sumer, H.; Jones, K.L.; Upton, K.; Verma, P.J. The efficient generation of induced pluripotent stem (iPS) cells from adult mouse adipose tissue-derived and neural stem cells. Cell Transplant. 2010, 19, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.R. Long-term survival of fat transplants: Controlled demonstrations. Aesthetic Plast. Surg. 1995, 19, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Yarak, S.; Okamoto, O.K. Human adipose-derived stem cells: Current challenges and clinical perspectives. An. Bras. Dermatol. 2010, 85, 647–656. [Google Scholar] [CrossRef]
- Liao, J.; Cui, C.; Chen, S.; Ren, J.; Chen, J.; Gao, Y.; Li, H.; Jia, N.; Cheng, L.; Xiao, L. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 2009, 4, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Nakada, A.; Hashimoto, Y.; Shigeno, K.; Shionoya, Y.; Nakamura, T. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol. Reprod. Dev. 2009, 77, 2. [Google Scholar] [CrossRef] [PubMed]
- Ezashi, T.; Telugu, B.P.; Alexenko, A.P.; Sachdev, S.; Sinha, S.; Roberts, R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 10993–10998. [Google Scholar] [CrossRef]
- Táncos, Z.; Nemes, C.; Varga, E.; Bock, I.; Rungarunlert, S.; Tharasanit, T.; Techakumphu, M.; Kobolák, J.; Dinnyés, A. Establishment of a rabbit induced pluripotent stem cell (RbiPSC) line using lentiviral delivery of human pluripotency factors. Stem Cell Res. 2017, 21, 16–18. [Google Scholar] [CrossRef]
- Bao, L.; He, L.; Chen, J.; Wu, Z.; Liao, J.; Rao, L.; Ren, J.; Li, H.; Zhu, H.; Qian, L.; et al. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011, 21, 600–608. [Google Scholar] [CrossRef]
- Nagy, K.; Sung, H.K.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C.; et al. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 2011, 7, 693–702. [Google Scholar] [CrossRef]
- Sumer, H.; Liu, J.; Malaver-Ortega, L.F.; Lim, M.L.; Khodadadi, K.; Verma, P.J. NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J Anim. Sci. 2011, 89, 2708–2716. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, Q.; Luo, C.; Chen, S.; Li, X.; Wang, C.; Zhang, X.; Zhang, L.; Zhao, Y.; Shi, D. Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 2012, 21, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; West, F.D.; Jordan, B.J.; Mumaw, J.L.; Jordan, E.T.; Gallegos-Cardenas, A.; Willard, S.T.; Freeman, K.W.; Stice, S.L. Avian-induced pluripotent stem cells derived using human reprogramming factors. Stem Cells Dev. 2012, 21, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Lu, Y.; Jordan, B.J.; Liu, Y.; Yang, J.Y.; Hutcheson, J.M.; Nelson, T.R.; Smith, S.M.; West, F.D. Nonviral mini-circle generation of induced pluripotent stem cells compatible with production of chimeric chickens. Cell. Reprogram. 2014, 16, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.; Liu, P.; Gao, J.; Jin, M.; Xu, T.; Zuo, Y.; Zhang, X.; Chen, H.; Wu, J.; Liu, D. Generation of Arbas Cashmere goat induced pluripotent stem cells through fibroblast reprogramming. Cell. Reprogram. 2015, 17, 297–305. [Google Scholar] [CrossRef]
- Yada, R.C.; Hong, S.G.; Lin, Y.; Winkler, T.; Dunbar, C.E. Rhesus macaque iPSC generation and maintenance. Curr. Protoc. Stem Cell Biol. 2017, 41, 4A.11.1–4A.11.13. [Google Scholar] [CrossRef] [PubMed]
- Ben-Nun, I.F.; Montague, S.C.; Houck, M.L.; Tran, H.T.; Garitaonandia, I.; Leonardo, T.R.; Wang, Y.C.; Charter, S.J.; Laurent, L.C.; Ryder, O.A. Induced pluripotent stem cells from highly endangered species. Nat. Methods 2011, 8, 829–831. [Google Scholar] [CrossRef]
- Verma, R.; Holland, M.K.; Temple-Smith, P.; Verma, P.J. Inducing pluripotency in somatic cells from the snow leopard (Panthera uncia), an endangered felid. Theriogenology 2012, 77, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Liu, J.; Holland, M.K.; Temple-Smith, P.; Williamson, M.; Verma, P.J. Derivation of induced pluripotent stem cells from the tiger (Panthera tigris), an endangered felid. Stem Cells Dev. 2013, 22, 169–176. [Google Scholar]
- Mo, X.; Li, N.; Wu, S. Generation and characterization of bat-induced pluripotent stem cells. Theriogenology 2014, 82, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, C.; Fan, X.; Ma, Y.; Liu, Z.; Ye, X.; Zhang, R.; Lin, H.; Wu, C. The impact of induced pluripotent stem cells in animal conservation. Vet. Res. Commun. 2024, 48, 649–663. [Google Scholar] [CrossRef]
- Clerc, J.; Weller, T.; Schineller, J.B.; Szewczak, J.M. Minimally invasive collection of adipose tissue facilitates the study of eco-physiology in small-bodied mammals. Methods Ecol. Evol. 2017, 8, 109–115. [Google Scholar] [CrossRef]
- Huerta-Leidenz, N.O.; Cross, H.R.; Savell, J.W.; Lunt, D.K.; Baker, J.F.; Pelton, L.S.; Smith, S.B. Comparison of the fatty acid composition of subcutaneous adipose tissue from mature Brahman and Hereford cows. J. Anim. Sci. 1993, 71, 625–630. [Google Scholar] [CrossRef]
- Galicia, M.P.; Thiemann, G.W.; Dyck, M.G.; Ferguson, S.H. Are tissue samples obtained via remote biopsy useful for fatty acid-based diet analyses in a free-ranging carnivore? J. Mammal. 2021, 102, 1067–1078. [Google Scholar] [CrossRef]
- Bressan, F.F.; Bassanezze, V.; de Figueiredo Pessôa, L.V.; Sacramento, C.B.; Malta, T.M.; Kashima, S.; Meirelles, F.V. Generation of induced pluripotent stem cells from large domestic animals. Stem Cell Res. Ther. 2020, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Pelekanos, R.A.; Ellis, R.L.; Horne, R.; Wolvetang, E.J.; Fisk, N.M. Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl. Med. 2012, 1, 83–95. [Google Scholar] [CrossRef]
- Zhao, X. Establishment of highly efficient somatic cell reprogramming system to generate iPSC lines. In Studies of Pluripotency in Embryonic Stem Cells and Induced Pluripotent Stem Cells; Springer: Berlin/Heidelberg, Germany, 2014; pp. 41–52. [Google Scholar]
- Chen, J.; Liu, H.; Liu, J.; Qi, J.; Wei, B.; Yang, J.; Liang, H.; Chen, Y.; Chen, J.; Wu, Y.; et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013, 45, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef]
- Stadtfeld, M.; Apostolou, E.; Ferrari, F.; Choi, J.; Walsh, R.M.; Chen, T.; Ooi, S.S.; Kim, S.Y.; Bestor, T.H.; Shioda, T.; et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012, 44, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.L.; Brena, R.M.; Kolle, G.; Grimmond, S.M.; Berman, B.P.; Laird, P.W.; Pera, M.F.; Wolvetang, E.J. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 2010, 28, 1848–1855. [Google Scholar] [CrossRef]
- Wang, T.; Chen, K.; Zeng, X.; Yang, J.; Wu, Y.; Shi, X.; Qin, B.; Zeng, L.; Esteban, M.A.; Pan, G. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 2011, 9, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Polo, J.M.; Anderssen, E.; Walsh, R.M.; Schwarz, B.A.; Nefzger, C.M.; Lim, S.M.; Borkent, M.; Apostolou, E.; Alaei, S.; Cloutier, J. A molecular roadmap of cellular reprogramming into iPS cells. Cell 2012, 151, 1617–1632. [Google Scholar] [CrossRef]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef]
- Maherali, N.; Ahfeldt, T.; Rigamonti, A.; Utikal, J.; Cowan, C.; Hochedlinger, K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 2008, 3, 340–345. [Google Scholar] [CrossRef]
- Miura, K.; Okada, Y.; Aoi, T.; Okada, A.; Takahashi, K.; Okita, K.; Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Ohnuki, M.; et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 2009, 27, 743–745. [Google Scholar] [CrossRef]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.R.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Pessôa, L.V.F.; Pires, P.R.L.; Del Collado, M.; Pieri, N.C.G.; Recchia, K.; Souza, A.F.; Perecin, F.; da Silveira, J.C.; de Andrade, A.F.C.; Ambrosio, C.E.; et al. Generation and miRNA characterization of equine induced pluripotent stem cells derived from fetal and adult multipotent tissues. Stem Cells Int. 2019, 2019, 1393791. [Google Scholar] [CrossRef]
- Brevini, T.A.L.; Antonini, S.; Cillo, F.; Crestan, M.; Gandolfi, F. Porcine embryonic stem cells: Facts, challenges and hopes. Theriogenology 2007, 68 (Suppl. S1), S206–S213. [Google Scholar] [CrossRef]
- Talbot, N.C.; Blomberg, L.A. The pursuit of ES cell lines of domesticated ungulates. Stem Cell Rev. 2008, 4, 235–254. [Google Scholar] [CrossRef]
- Tapia, N.; Schöler, H.R. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell 2016, 19, 298–309. [Google Scholar] [CrossRef]
- Doss, M.X.; Sachinidis, A. Current challenges of iPSC-based disease modeling and therapeutic implications. Cells 2019, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Madrid, M.; Sumen, C.; Aivio, S.; Saklayen, N. Autologous induced pluripotent stem cell–based cell therapies: Promise, progress, and challenges. Curr. Protoc. 2021, 1, e88. [Google Scholar] [CrossRef]
- Nandal, A.; Telugu, B.P.V.L. Large animal induced pluripotent stem cells as models of human diseases. In Pluripotent Stem Cells; Springer: Cham, Switzerland, 2014; pp. 215–230. [Google Scholar]
- Ma, H.; Morey, R.; O’Neil, R.C.; He, Y.; Daughtry, B.; Schultz, M.D.; Hariharan, M.; Nery, J.R.; Castanon, R.; Sabatini, K.; et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 2014, 511, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, J.; Liu, H.; Lu, D.; Chen, X.; Zenonos, Z.; Liu, P. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. USA 2011, 108, 18283–18288. [Google Scholar] [CrossRef] [PubMed]
- Nie, T.; Deng, W.; Gao, X.; Sun, W.; Hui, X.; Song, H.; Qin, D.; Xu, A.; Li, P.; Liu, P.; et al. Reprogramming mature terminally differentiated adipocytes to induced pluripotent stem cells. Sci. Bull. 2015, 60, 1752–1758. [Google Scholar] [CrossRef]
- Honda, A.; Choijookhuu, N.; Izu, H.; Kawano, Y.; Inokuchi, M.; Honsho, K.; Lee, A.R.; Nabekura, H.; Ohta, H.; Tsukiyama, T.; et al. Flexible adaptation of male germ cells from female iPSCs of endangered Tokudaia osimensis. Sci. Adv. 2017, 3, e1602179. [Google Scholar] [CrossRef]
- Talluri, T.R.; Kumar, D.; Glage, S.; Garrels, W.; Ivics, Z.; Debowski, K.; Behr, R.; Niemann, H.; Kues, W.A. Derivation and characterization of bovine induced pluripotent stem cells by transposon-mediated reprogramming. Cell. Reprogram. 2015, 17, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Zywitza, V.; Rusha, E.; Shaposhnikov, D.; Ruiz-Orera, J.; Telugu, N.; Rishko, V.; Hayashi, M.; Michel, G.; Wittler, L.; Stejskal, J.; et al. Naïve-like pluripotency to pave the way for saving the northern white rhinoceros from extinction. Sci. Rep. 2022, 12, 3100. [Google Scholar] [CrossRef]
- West, F.D.; Terlouw, S.L.; Kwon, D.J.; Mumaw, J.L.; Dhara, S.K.; Hasneen, K.; Dobrinsky, J.R.; Stice, S.L. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010, 19, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Fukuda, T.; Kaneko, T.; Nakagawa, Y.; Tajima, A.; Naito, M.; Ohmaki, H.; Endo, D.; Asano, M.; Nagamine, T.; et al. Induced pluripotent stem cells of endangered avian species. Commun. Biol. 2022, 5, 1049. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Tsukiyama, T.; Kimura, K.; Matsuyama, S.; Minami, N.; Yamada, M.; Imai, H. Generation of naïve bovine induced pluripotent stem cells using piggyBac transposition of doxycycline-inducible transcription factors. PLoS ONE 2015, 10, e0135403. [Google Scholar] [CrossRef] [PubMed]
- Tsukiyama, T.; Osafune, K. A modified EpiSC culture condition containing a GSK3 inhibitor can support germline-competent pluripotency in mice. PLoS ONE 2014, 9, e95329. [Google Scholar] [CrossRef]
- Lou, J.; Li, W.; Chen, P.; Chen, H.; Shakoor, A.; Chen, Y.; Zhang, S. Application of induced pluripotent stem cells in the conservation of endangered animals. Stem Cell Res. Ther. 2025, 16, 261. [Google Scholar] [CrossRef]

| Cell Line | Electroporation Using PiggyBac | Lentiviral with STEMCCA Vector | ||||
|---|---|---|---|---|---|---|
| FBS | KSR | hOSKM | mOSKM | |||
| Protocol 1 * | Protocol 2 ** | Protocol 1 * | Protocol 2 ** | |||
| Antler | 3 | 2 | 1 | 2 | 1 | 1 |
| Fat | 1 | 1 | 1 | 1 | 1 | |
| Skin | 1 | 2 | 2 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rola, L.D.; Bressan, F.F.; Sandoval, E.D.P.; Therrien, J.; Smith, L.C.; Duarte, J.M.B. Insights for Deriving Induced Pluripotent Stem Cells in Marsh Deer (Blastocerus dichotomus). J. Zool. Bot. Gard. 2025, 6, 43. https://doi.org/10.3390/jzbg6030043
Rola LD, Bressan FF, Sandoval EDP, Therrien J, Smith LC, Duarte JMB. Insights for Deriving Induced Pluripotent Stem Cells in Marsh Deer (Blastocerus dichotomus). Journal of Zoological and Botanical Gardens. 2025; 6(3):43. https://doi.org/10.3390/jzbg6030043
Chicago/Turabian StyleRola, Luciana Diniz, Fabiana Fernandes Bressan, Eluzai Dinai Pinto Sandoval, Jacinthe Therrien, Lawrence Charles Smith, and José Maurício Barbanti Duarte. 2025. "Insights for Deriving Induced Pluripotent Stem Cells in Marsh Deer (Blastocerus dichotomus)" Journal of Zoological and Botanical Gardens 6, no. 3: 43. https://doi.org/10.3390/jzbg6030043
APA StyleRola, L. D., Bressan, F. F., Sandoval, E. D. P., Therrien, J., Smith, L. C., & Duarte, J. M. B. (2025). Insights for Deriving Induced Pluripotent Stem Cells in Marsh Deer (Blastocerus dichotomus). Journal of Zoological and Botanical Gardens, 6(3), 43. https://doi.org/10.3390/jzbg6030043







