Immunophenotyping and Functional Characterization of NK Cells in SARS-CoV-2 Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients’ Selection
2.2. Blood Samples’ Collection
2.3. Methods
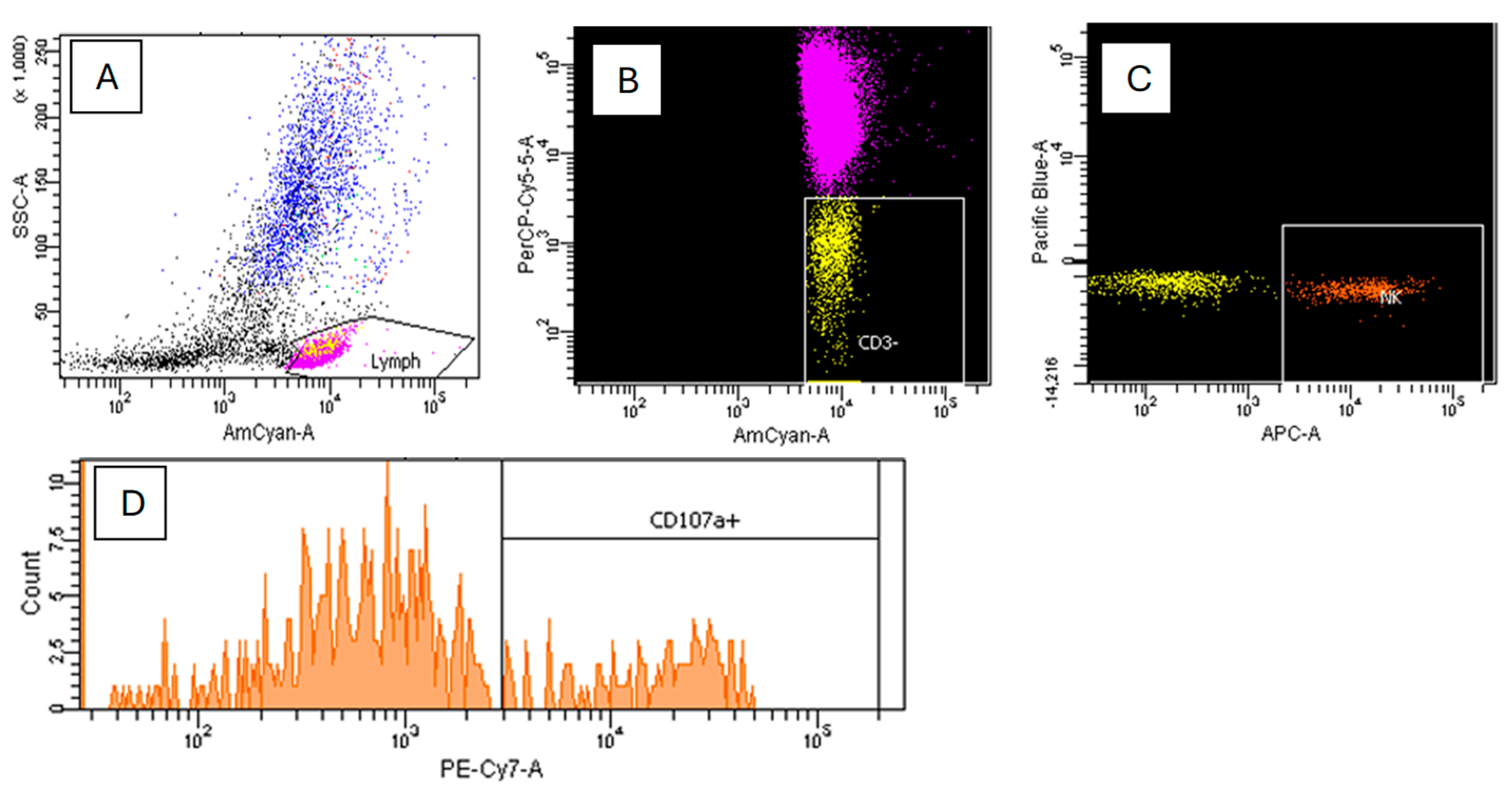
2.3.1. Immunophenotyping
2.3.2. Direct Cytotoxicity Assay of NK Cells
2.4. Statistical Analysis
3. Results
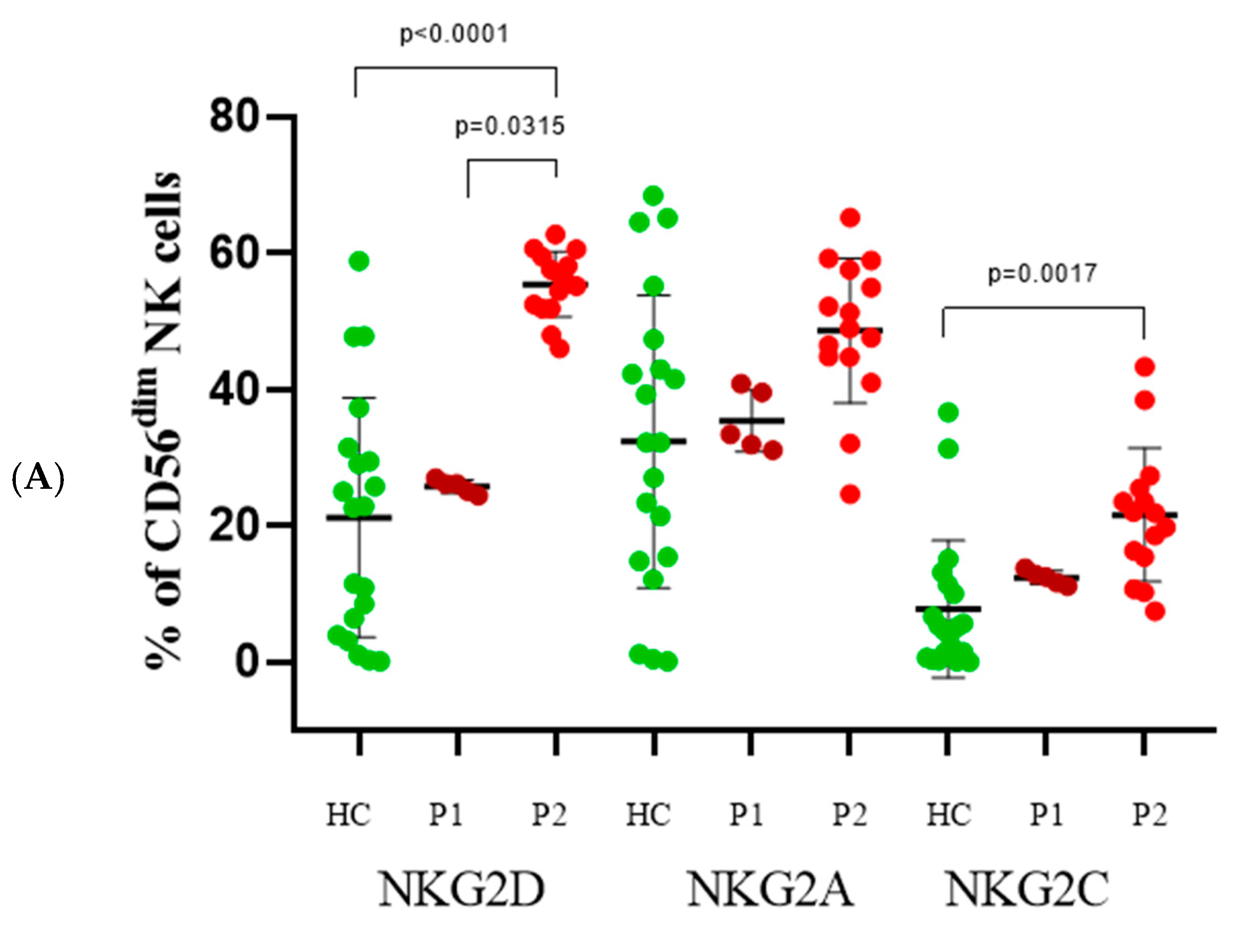
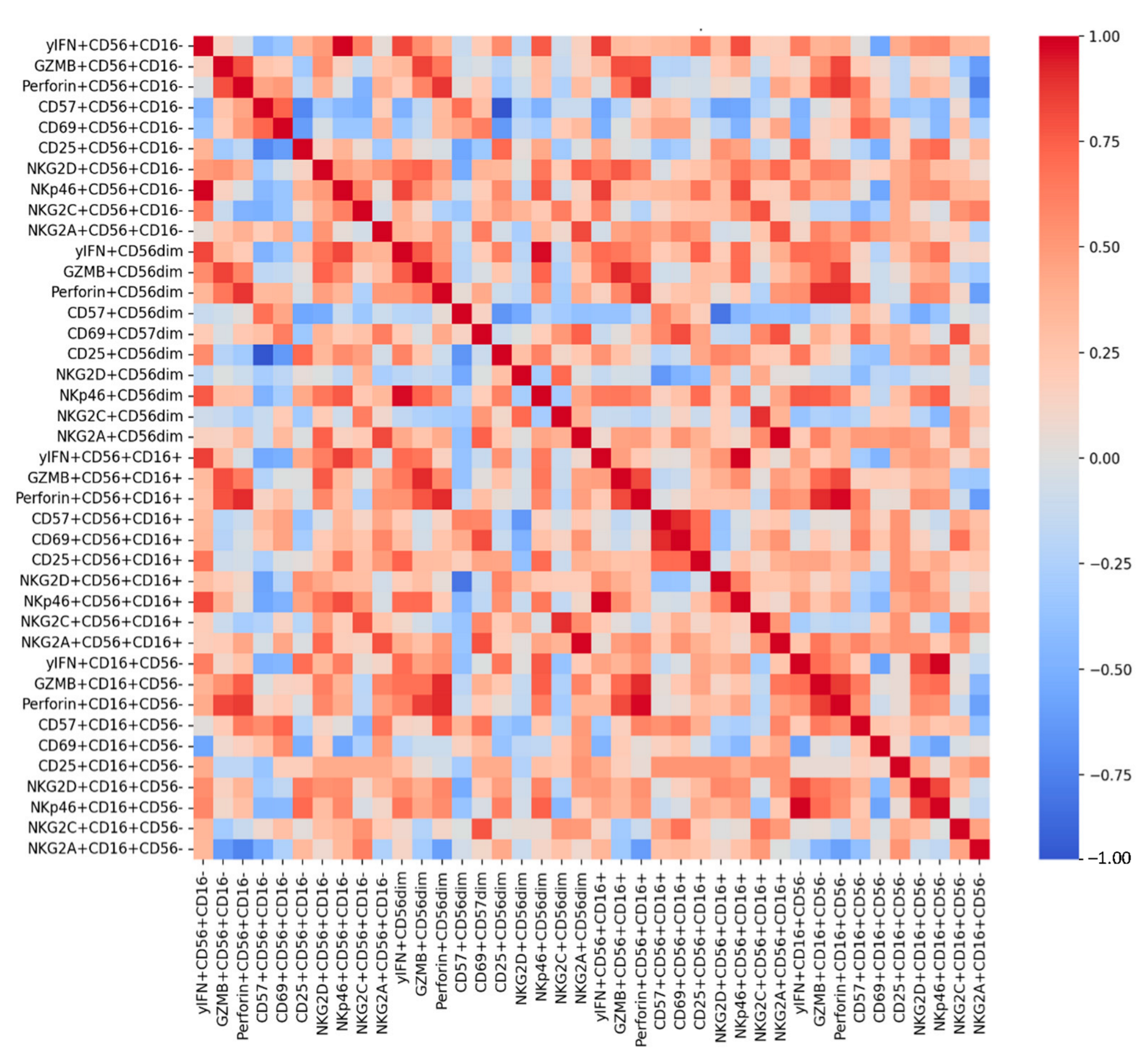
3.1. Distribution of NK Cell Subsets
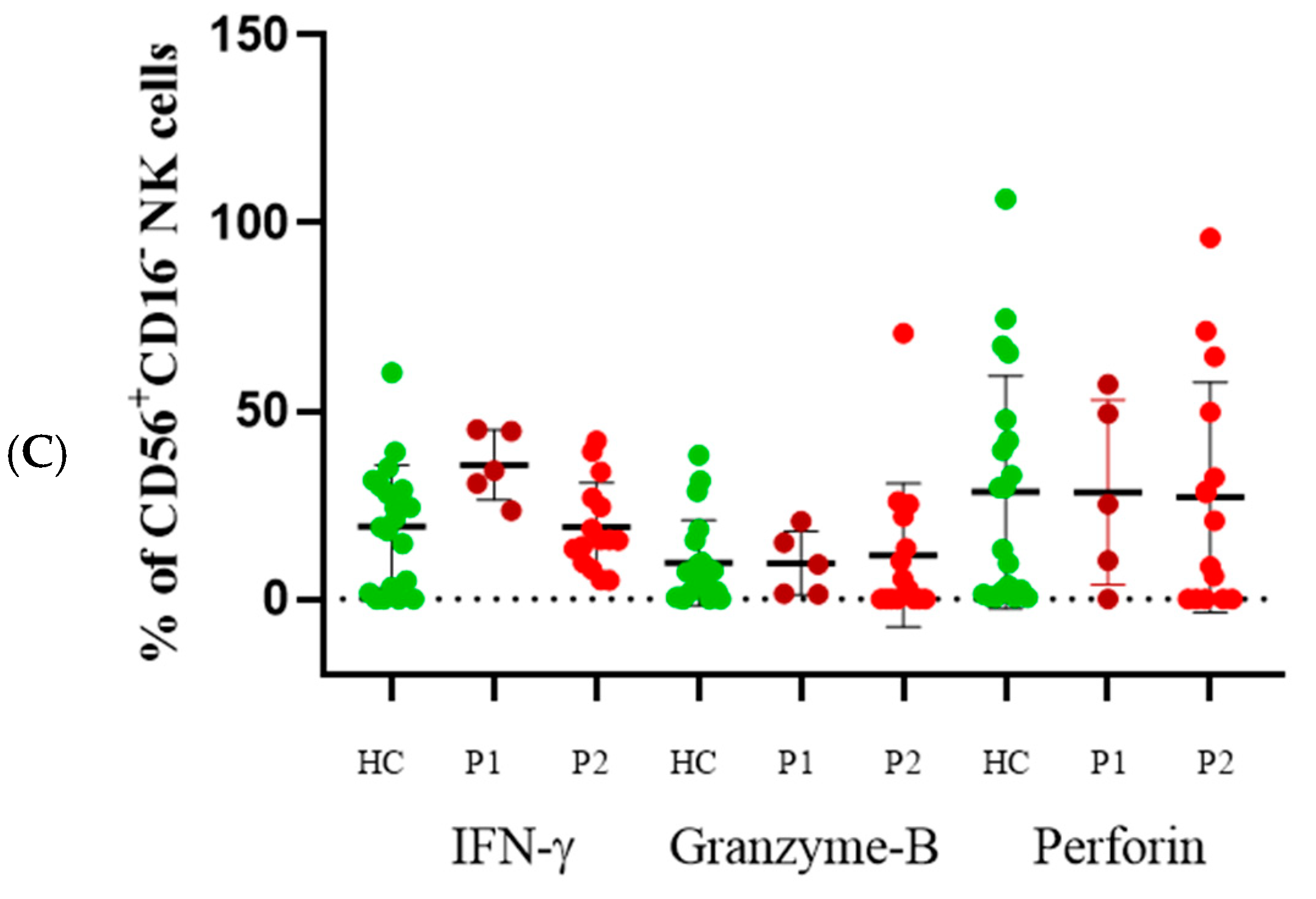
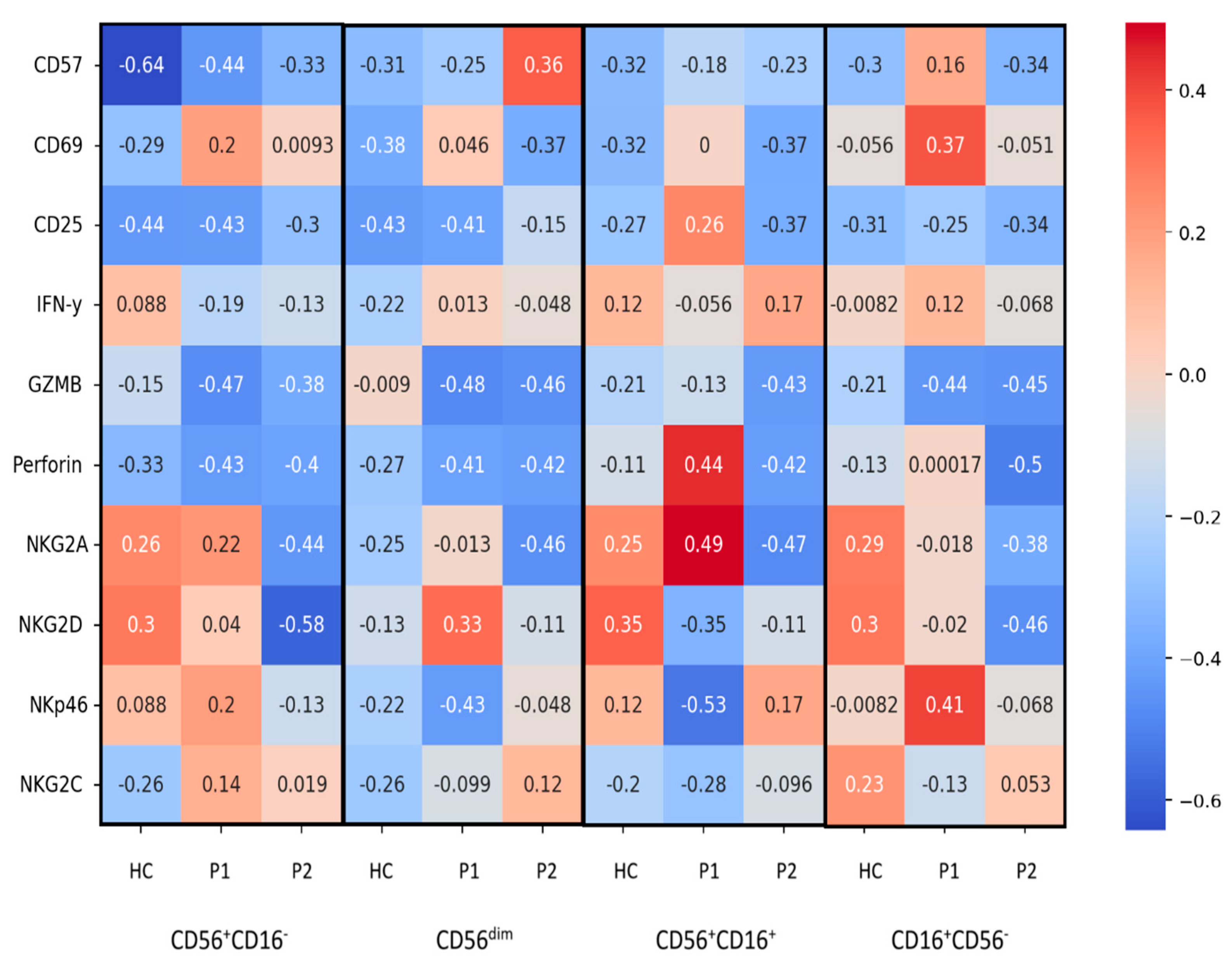
3.2. CD56+CD16− Cells
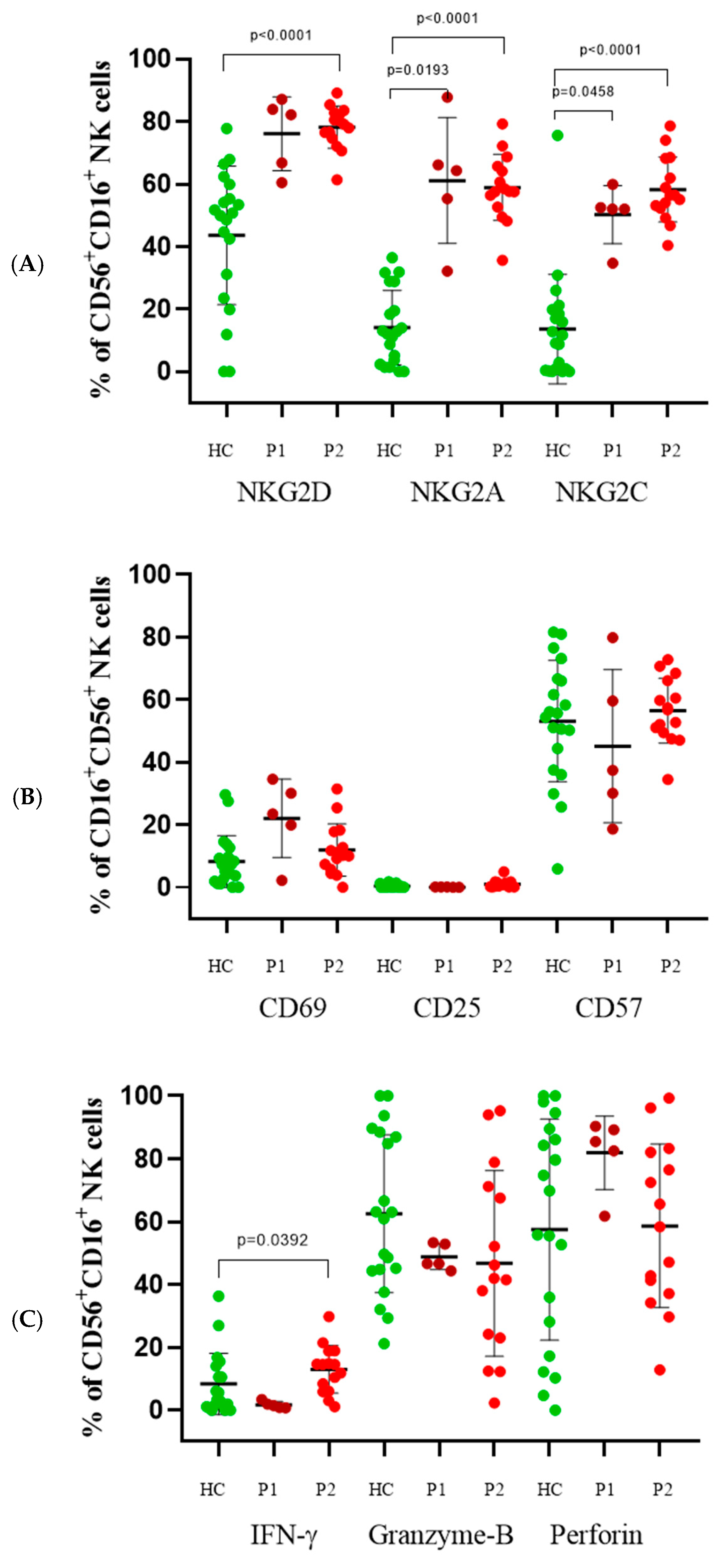
3.3. CD56+CD16+ Cells
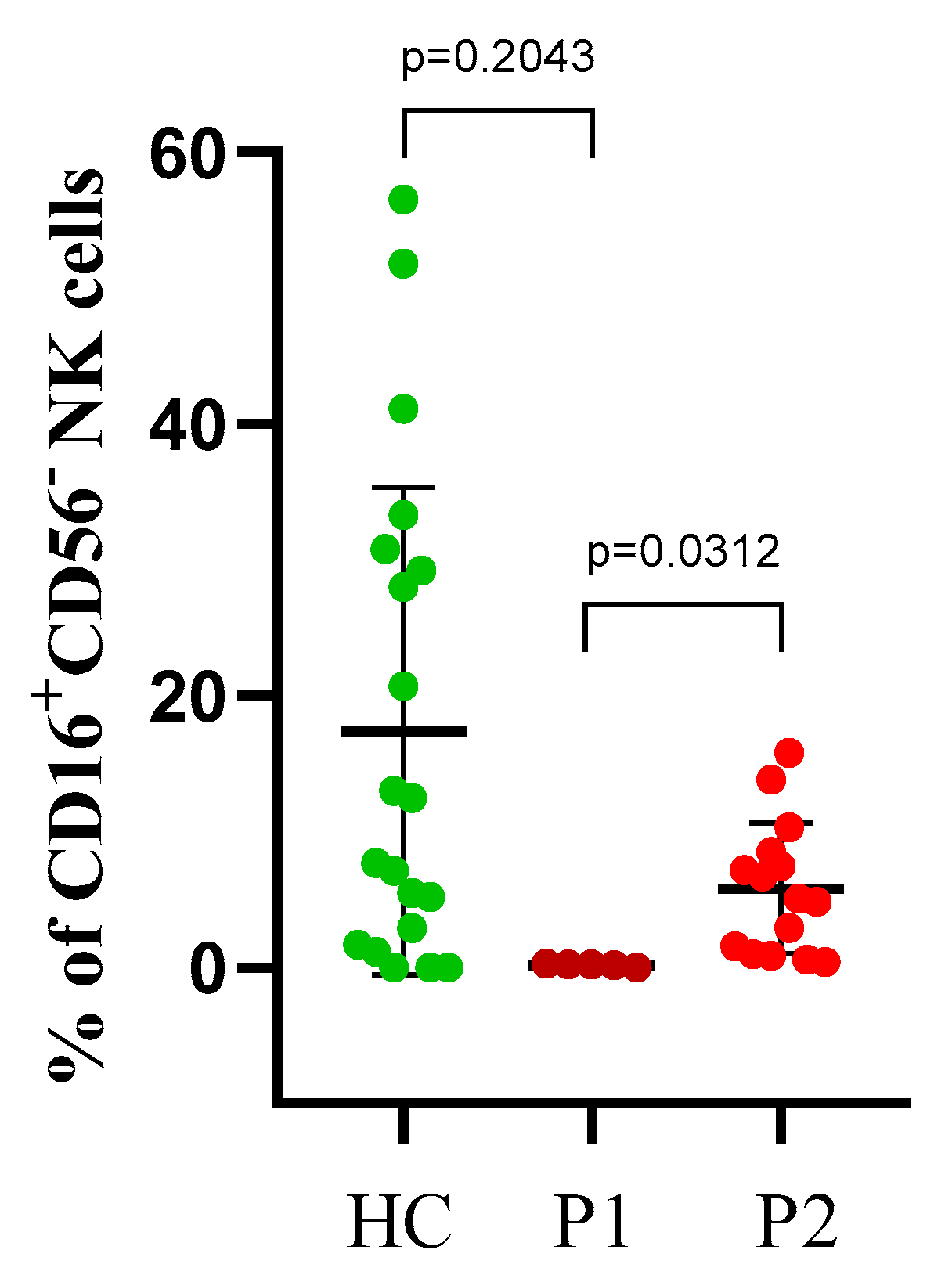
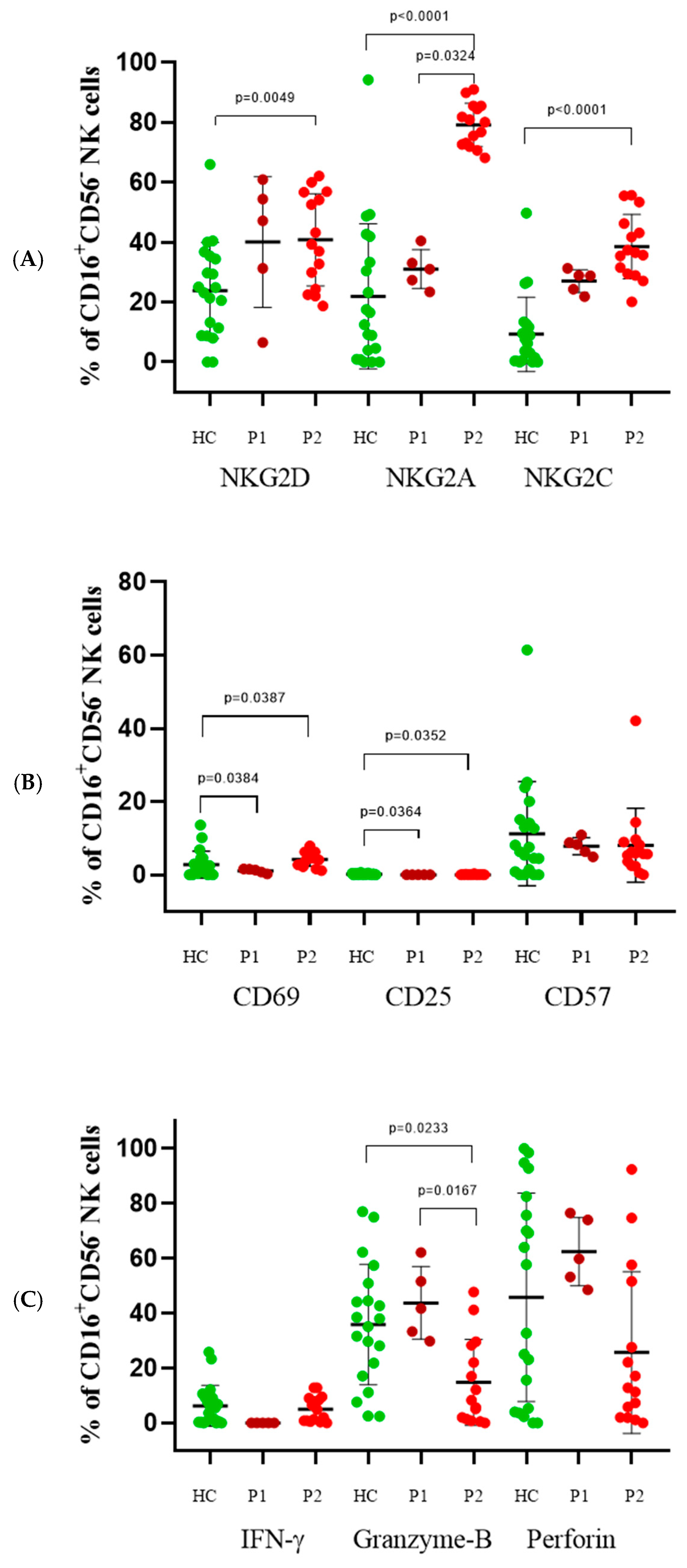
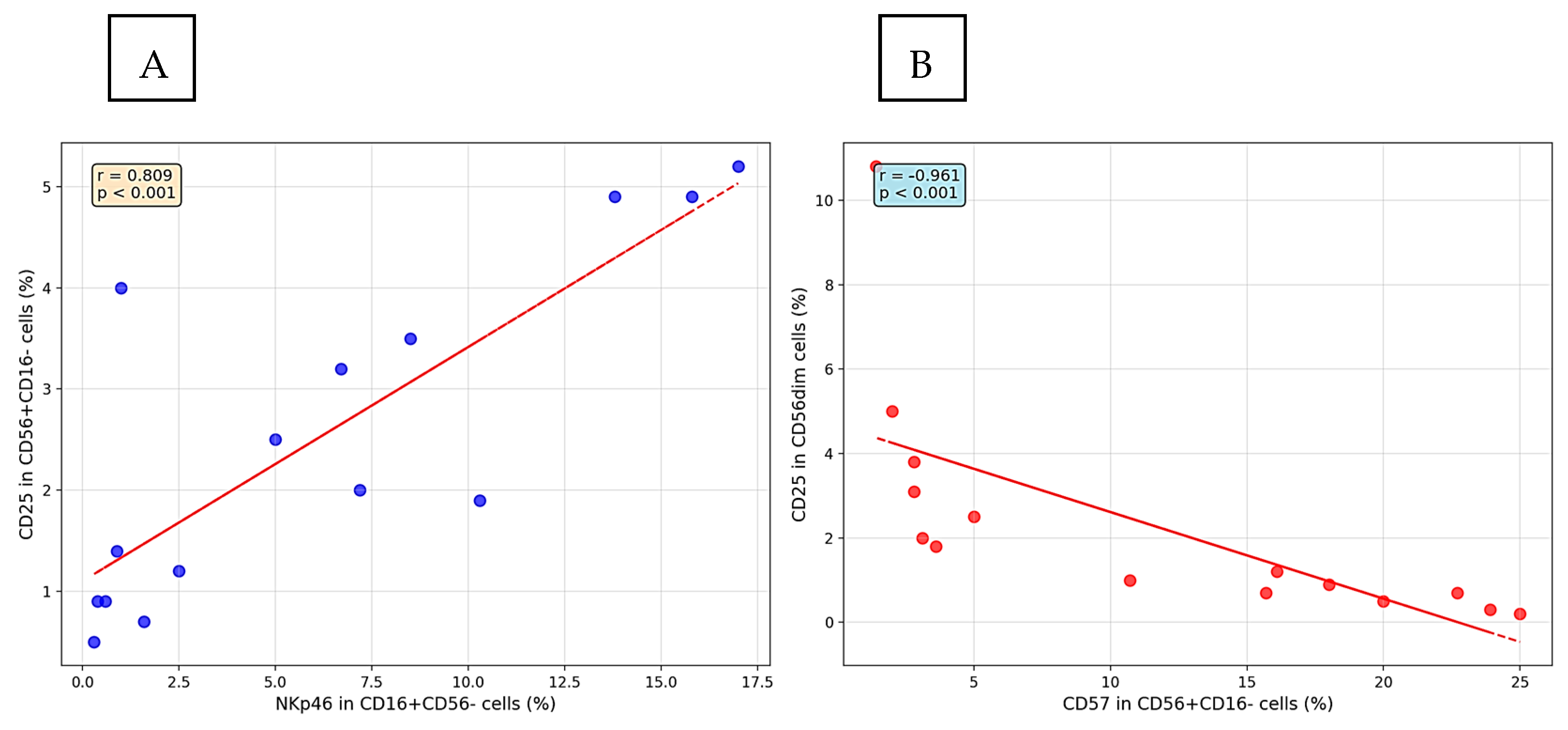
3.4. CD16+CD56− Cells
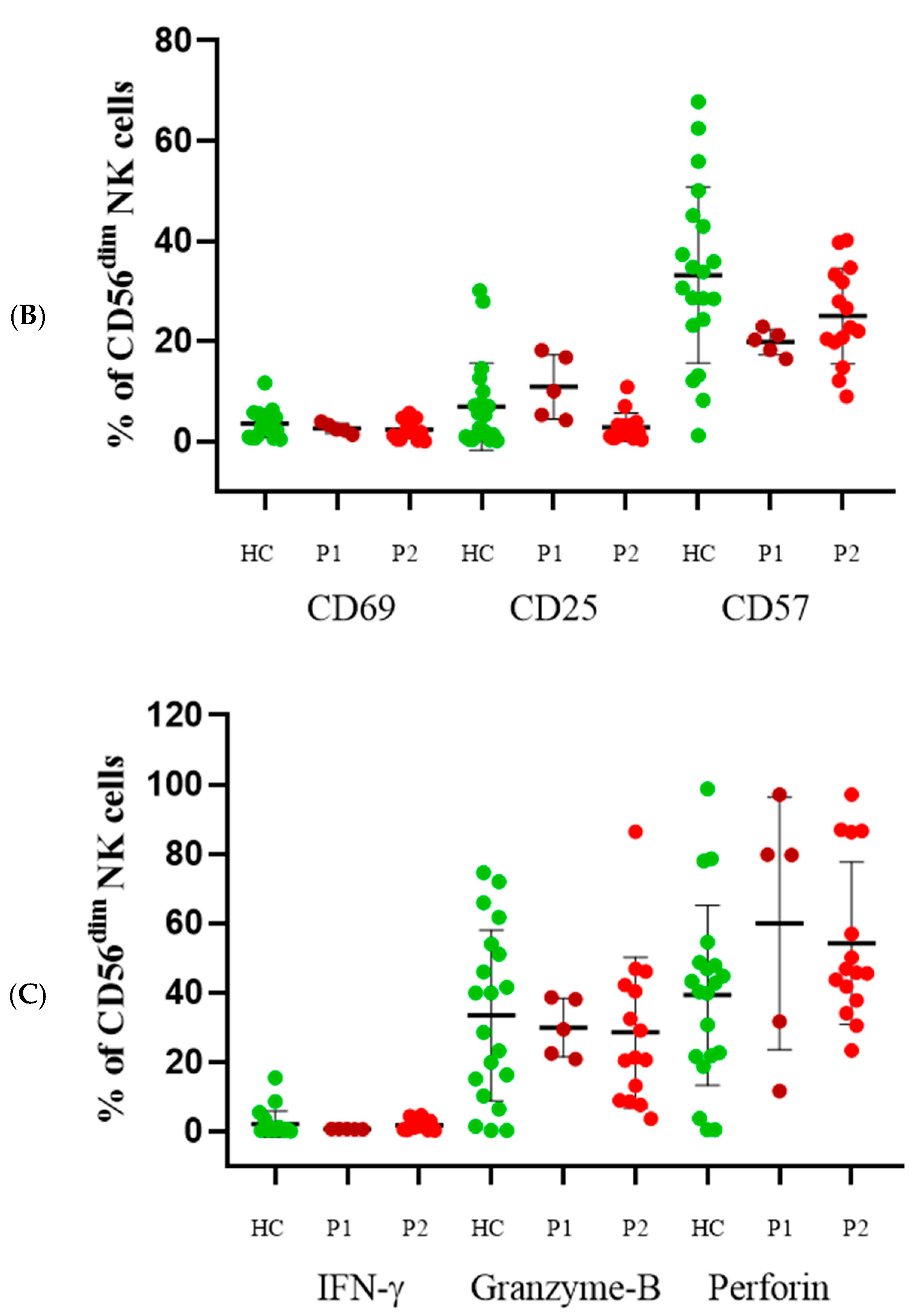
3.5. CD56dim Cells
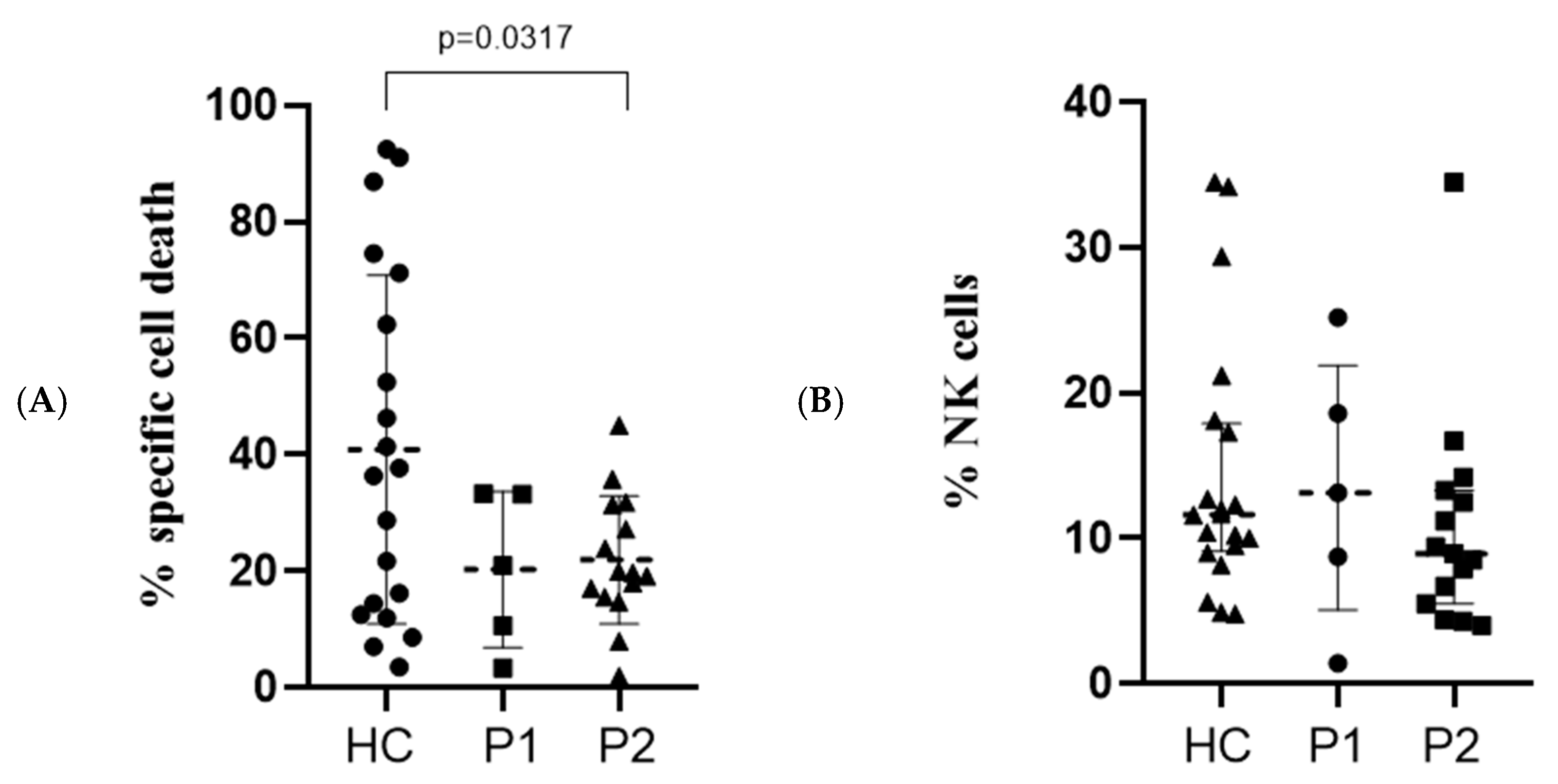
3.6. Direct Cytotoxicity Assay
4. Discussion
- Immune Dysregulation: The reduced cytotoxic potential (GZMB in CD16+CD56− cells) coupled with increased activation markers suggests a state of functional adaptation or partial exhaustion.
- Adaptive-like Features: Elevated NKG2C expression indicates the recruitment of adaptive-like NK cells, possibly in response to prolonged viral or inflammatory stimuli.
- Regulatory Balance: Sustained NKG2A upregulation highlights mechanisms to mitigate hyperinflammation, which is a hallmark of severe COVID-19.
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NK | Natural killer |
| IFN-γ | Interferon-gamma |
| IL-6 | Interleukin-6 |
| KIRs | Killer cell immunoglobulin-like receptors |
| P | Period (of analysis) |
| HC | Healthy subjects |
| PBMCs | Peripheral blood mononuclear cells |
| DMSO | Dimethyl sulfoxide |
| CD | Cluster of differentiation |
| MFI | Median fluorescent intensity |
| PARP1 | poly(ADP-ribose) polymerase 1 |
| DCA | direct cytotoxicity assay |
| E | Effector cells |
| T | Target cells |
| FSC | Forward scatter |
| SSC | Side scatter |
References
- Li, M.; Guo, W.; Dong, Y.; Wang, X.; Dai, D.; Liu, X.; Wu, Y.; Li, M.; Zhang, W.; Zhou, H.; et al. Elevated Exhaustion Levels of NK and CD8+ T Cells as Indicators for Progression and Prognosis of COVID-19 Disease. Front. Immunol. 2019, 11, 580237. [Google Scholar] [CrossRef]
- Maucourant, C.; Filipovic, I.; Ponzetta, A.; Aleman, S.; Cornillet, M.; Hertwig, L.; Strunz, B.; Lentini, A.; Reinius, B.; Brownlie, D.; et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020, 5, eabd6832. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Capone, M.; Vanni, A.; Spinicci, M.; Mencarini, J.; Caporale, R.; Peruzzi, B.; Antonelli, A.; et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Investig. 2020, 130, 4694–4703. [Google Scholar] [CrossRef]
- Varchetta, S.; Mele, D.; Oliviero, B.; Mantovani, S.; Ludovisi, S.; Cerino, A.; Bruno, R.; Castelli, A.; Mosconi, M.; Vecchia, M.; et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2021, 18, 604–612. [Google Scholar] [CrossRef]
- Krämer, B.; Knoll, R.; Bonaguro, L.; ToVinh, M.; Raabe, J.; Astaburuaga-García, R.; Schulte-Schrepping, J.; Kaiser, K.M.; Rieke, G.J.; Bischoff, J.; et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity 2021, 54, 2650–2669. [Google Scholar] [CrossRef]
- Vietzen, H.; Zoufaly, A.; Traugott, M.; Aberle, J.; Aberle, S.W.; Puchhammer-Stöckl, E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA-E variants are risk factors for severe COVID-19. Genet. Med. 2021, 23, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, N.; Litvinova, E.; Samri, A.; Soulié, C.; Morin, V.; Rousseau, A.; Dorgham, K.; Parizot, C.; Bonduelle, O.; Beurton, A.; et al. Identification of natural killer markers associated with fatal outcome in COVID-19 patients. Front. Cell Infect. Microbiol. 2023, 13, 1165756. [Google Scholar] [CrossRef]
- Casado, J.L.; Moraga, E.; Vizcarra, P.; Velasco, H.; Martín-Hondarza, A.; Haemmerle, J.; Gómez, S.; Quereda, C.; Vallejo, A. Expansion of cd56dim cd16neg nk cell subset and increased inhibitory kirs in hospitalized COVID-19 patients. Viruses 2022, 14, 46. [Google Scholar] [CrossRef]
- Lenart, M.; Górecka, M.; Bochenek, M.; Barreto-Duran, E.; Szczepański, A.; Gałuszka-Bulaga, A.; Mazur-Panasiuk, N.; Węglarczyk, K.; Siwiec-Koźlik, A.; Korkosz, M.; et al. SARS-CoV-2 infection impairs NK cell functions via activation of the LLT1-CD161 axis. Front. Immunol. 2023, 14, 1123155. [Google Scholar] [CrossRef]
- Osman, M.; Faridi, R.M.; Sligl, W.; Shabani-Rad, M.T.; Dharmani-Khan, P.; Parker, A.; Kalra, A.; Tripathi, M.B.; Storek, J.; Tervaert, J.W.C.; et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020, 4, 5035–5039. [Google Scholar] [CrossRef]
- Yasin, M.M.; Shehata, I.H.; Elsheikh, N.G.; Elsayed, M.S. Expression of NKG2A inhibitory receptor on cytotoxic lymphocytes as an indicator of severity in Corona Virus Disease 2019 (COVID-19) patients. Egypt. J. Immunol. 2021, 28, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Bergantini, L.; D’alessandro, M.; Cameli, P.; Cavallaro, D.; Gangi, S.; Cekorja, B.; Sestini, P.; Bargagli, E. Nk and T cell immunological signatures in hospitalized patients with COVID-19. Cells 2021, 10, 3182. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Leong, M.W.; Rustagi, A.; Beck, A.; Zeng, L.; Holmes, S.; Qi, L.S.; Blish, C.A. SARS-CoV-2 escapes direct NK cell killing through Nsp1-mediated downregulation of ligands for NKG2D. Cell Rep. 2022, 41, 111892. [Google Scholar] [CrossRef]
- Claus, M.; Pieris, N.; Urlaub, D.; Bröde, P.; Schaaf, B.; Durak, D.; Renken, F.; Watzl, C. Early expansion of activated adaptive but also exhausted NK cells during acute severe SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2023, 13, 1266790. [Google Scholar] [CrossRef]
- Riedhammer, C.; Halbritter, D.; Weissert, R. Peripheral Blood Mononuclear Cells: Isolation, Freezing, Thawing, and Culture. Methods Mol. Biol. 2014, 1304, 53–61. [Google Scholar] [CrossRef]
- Hønge, B.L.; Petersen, M.S.; Olesen, R.; Møller, B.K.; Erikstrup, C. Optimizing recovery of frozen human peripheral blood mononuclear cells for flow cytometry. PLoS ONE 2017, 12, e0187440. [Google Scholar] [CrossRef]
- Kandarian, F.; Sunga, G.M.; Arango-Saenz, D.; Rossetti, M. A Flow Cytometry-Based Cytotoxicity Assay for the Assessment of Human NK Cell Activity. J. Vis. Exp. 2017, 2017, e56191. [Google Scholar] [CrossRef]
- Mata, M.M.; Mahmood, F.; Sowell, R.T.; Baum, L.L. Effects of cryopreservation on effector cells for antibody dependent cell-mediated cytotoxicity (ADCC) and natural killer (NK) cell activity in 51Cr-release and CD107a assays. J. Immunol. Methods 2014, 406, 1–9. [Google Scholar] [CrossRef]
- Schiuma, G.; Beltrami, S.; Bortolotti, D.; Rizzo, S.; Rizzo, R. Innate Immune Response in SARS-CoV-2 Infection. Microorganisms 2022, 10, 501. [Google Scholar] [CrossRef] [PubMed]
- Beer, J.; Crotta, S.; Breithaupt, A.; Ohnemus, A.; Becker, J.; Sachs, B.; Kern, L.; Llorian, M.; Ebert, N.; Labroussaa, F.; et al. Impaired immune response drives age-dependent severity of COVID-19. J. Exp. Med. 2022, 219, e20220621. [Google Scholar] [CrossRef]
- Vavilova, J.D.; Ustiuzhanina, M.O.; Boyko, A.A.; Streltsova, M.A.; Kust, S.A.; Kanevskiy, L.M.; Iskhakov, R.N.; Sapozhnikov, A.M.; Gubernatorova, E.O.; Drutskaya, M.S.; et al. Alterations in the CD56− and CD56+ T Cell Subsets during COVID-19. Int. J. Mol. Sci. 2023, 24, 9047. [Google Scholar] [CrossRef]
- Fernández-Soto, D.; García-Jiménez, Á.F.; Casasnovas, J.M.; Valés-Gómez, M.; Reyburn, H.T. Elevated levels of cell-free NKG2D-ligands modulate NKG2D surface expression and compromise NK cell function in severe COVID-19 disease. Front. Immunol. 2024, 15, 1273942. [Google Scholar] [CrossRef]
- Yaqinuddin, A.; Kashir, J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med. Hypotheses 2020, 140, 109777. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.R.; Arunachalam, J.; Bhardwaj, A.; Saifullah, A.; Lakhchaura, R.; Soni, M.; Bhagawati, G.; Chakrabarti, S. Impact of adaptive natural killer cells, KLRC2 genotype and cytomegalovirus reactivation on late mortality in patients with severe COVID-19 lung disease. Clin. Transl. Immunol. 2022, 11, e1359. [Google Scholar] [CrossRef]
- Bozzano, F.; Dentone, C.; Perrone, C.; Di Biagio, A.; Fenoglio, D.; Parodi, A.; Mikulska, M.; Bruzzone, B.; Giacobbe, D.R.; Vena, A.; et al. Extensive activation, tissue trafficking, turnover and functional impairment of NK cells in COVID-19 patients at disease onset associates with subsequent disease severity. PLoS Pathog. 2021, 17, e1009448. [Google Scholar] [CrossRef] [PubMed]
- Malengier-Devlies, B.; Filtjens, J.; Ahmadzadeh, K.; Boeckx, B.; Vandenhaute, J.; De Visscher, A.; Bernaerts, E.; Mitera, T.; Jacobs, C.; Vanderbeke, L.; et al. Severe COVID-19 patients display hyper-activated NK cells and NK cell-platelet aggregates. Front. Immunol. 2022, 13, 861251. [Google Scholar] [CrossRef]
- Lee, M.; de Los Rios Kobara, I.; Barnard, T.; Torres, X.V.; Tobin, N.; Ferbas, K.; Rimoin, A.W.; O Yang, O.; Aldrovandi, G.M.; Wilk, A.J.; et al. NK cell-monocyte cross-talk underlies NK cell activation in severe COVID-19. bioRxiv 2023. [Google Scholar] [CrossRef]
- Leem, G.; Cheon, S.; Lee, H.; Choi, S.J.; Jeong, S.; Kim, E.S.; Jeong, H.W.; Jeong, H.; Park, S.-H.; Kim, Y.-S.; et al. Abnormality in the NK-cell population is prolonged in severe COVID-19 patients. J. Allergy Clin. Immunol. 2021, 148, 996–1006.e18. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Lee, M.J.; Wei, B.; Parks, B.; Pi, R.; Martínez-Colón, G.J.; Ranganath, T.; Zhao, N.Q.; Taylor, S.; Becker, W.; et al. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J. Exp. Med. 2021, 218, e20210582. [Google Scholar] [CrossRef]
- Ye, W.; Zhang, W.; Wu, S.; Zhu, M.; Xu, Z. Study of surface activation markers on CD3−CD16+ NK cells and their correlation with clinical manifestations in children with infectious mononucleosis. Microbiol. Immunol. 2021, 65, 400–404. [Google Scholar] [CrossRef]
- Herrera, L.; Martin-Inaraja, M.; Santos, S.; Inglés-Ferrándiz, M.; Azkarate, A.; Perez-Vaquero, M.A.; Vesga, M.A.; Vicario, J.L.; Soria, B.; Solano, C.; et al. Identifying SARS-CoV-2 ‘memory’ NK cells from COVID-19 convalescent donors for adoptive cell therapy. Immunology 2022, 165, 234–249. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.C.; Lai, E.Y.; Liu, Y.T.; Wang, Y.F.; Tzeng, Y.S.; Cui, L.; Lai, Y.-J.; Huang, H.-C.; Huang, J.-H.; Ni, H.-C.; et al. NK cell receptor and ligand composition influences the clearance of SARS-CoV-2. J. Clin. Investig. 2021, 131, e146408. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Dhanushkodi, N.; Prakash, S.; Coulon, P.G.; Vahed, H.; Zayou, L.; Quadiri, A.; BenMohamed, L. High Frequencies of Phenotypically and Functionally Senescent and Exhausted CD56+CD57+PD-1+ Natural Killer Cells, SARS-CoV-2-Specific Memory CD4+ and CD8+ T cells Associated with Severe Disease in Unvaccinated COVID-19 Patients. bioRxiv 2022. [Google Scholar] [CrossRef]
- Liu, B.; Yang, G.X.; Sun, Y.; Tomiyama, T.; Zhang, W.; Leung, P.S.C.; He, X.-S.; Dhaliwal, S.; Invernizzi, P.; Gershwin, M.E.; et al. Decreased CD57 expression of natural killer cells enhanced cytotoxicity in patients with primary sclerosing cholangitis. Front. Immunol. 2022, 13, 912961. [Google Scholar] [CrossRef]
- Draghi, M.; Pashine, A.; Sanjanwala, B.; Gendzekhadze, K.; Cantoni, C.; Cosman, D.; Moretta, A.; Valiante, N.M.; Parham, P. NKp46 and NKG2D Recognition of Infected Dendritic Cells Is Necessary for NK Cell Activation in the Human Response to Influenza Infection. J. Immunol. 2007, 178, 2688–2698. [Google Scholar] [CrossRef]
- Kim, J.M.; Yi, E.; Cho, H.; Choi, W.S.; Ko, D.H.; Yoon, D.H.; Hwang, S.-H.; Kim, H.S. Assessment of NK Cell Activity Based on NK Cell-Specific Receptor Synergy in Peripheral Blood Mononuclear Cells and Whole Blood. Int. J. Mol. Sci. 2020, 21, 8112. [Google Scholar] [CrossRef]
- Deguine, J.; Breart, B.; Lemaître, F.; Bousso, P. Cutting Edge: Tumor-Targeting Antibodies Enhance NKG2D-Mediated NK Cell Cytotoxicity by Stabilizing NK Cell–Tumor Cell Interactions. J. Immunol. 2012, 189, 5493–5497. [Google Scholar] [CrossRef]
- Gaddy, J.; Broxmeyer, H.E. Cord blood CD16+56- cells with low lytic activity are possible precursors of mature natural killer cells. Cell. Immunol. 1997, 180, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, H.; Zheng, X.; Wei, H.; Sun, R.; Tian, Z. High Expression of NKG2A/CD94 and Low Expression of Granzyme B Are Associated with Reduced Cord Blood NK Cell Activity. Cell. Mol. Immunol. 2007, 4, 377–382. [Google Scholar]


| NK Cell Subpopulations | P1 | P2 | HC | |||
|---|---|---|---|---|---|---|
| % From NK Cells | SD | % From NK Cells | SD | % From NK Cells | SD | |
| CD56+CD16+ | 18.35 | ±7.75 | 38.89 | ±13.51 | 31.28 | ±20.92 |
| CD56dim | 4 | ±3.1 | 9.98 | ±5.33 | 12.1 | ±11.46 |
| CD56+CD16− | 3.5 | ±0.7 | 8.66 a | ±4.72 | 6.53 a | ±5.86 |
| CD16+CD56− | 1.45 b | ±0.05 | 3.92 b | ±1.65 | 1.63 | ±1.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrov, S.; Bozhkova, M.; Ivanovska, M.; Kalfova, T.; Baldzhieva, A.; Todev, A.; Kirova, D.; Kicheva, Y.; Stoynov, S.; Murdjeva, M.; et al. Immunophenotyping and Functional Characterization of NK Cells in SARS-CoV-2 Infection. Immuno 2025, 5, 35. https://doi.org/10.3390/immuno5030035
Petrov S, Bozhkova M, Ivanovska M, Kalfova T, Baldzhieva A, Todev A, Kirova D, Kicheva Y, Stoynov S, Murdjeva M, et al. Immunophenotyping and Functional Characterization of NK Cells in SARS-CoV-2 Infection. Immuno. 2025; 5(3):35. https://doi.org/10.3390/immuno5030035
Chicago/Turabian StylePetrov, Steliyan, Martina Bozhkova, Mariya Ivanovska, Teodora Kalfova, Alexandra Baldzhieva, Angel Todev, Dilyana Kirova, Yoana Kicheva, Stoyno Stoynov, Marianna Murdjeva, and et al. 2025. "Immunophenotyping and Functional Characterization of NK Cells in SARS-CoV-2 Infection" Immuno 5, no. 3: 35. https://doi.org/10.3390/immuno5030035
APA StylePetrov, S., Bozhkova, M., Ivanovska, M., Kalfova, T., Baldzhieva, A., Todev, A., Kirova, D., Kicheva, Y., Stoynov, S., Murdjeva, M., & Taskov, H. (2025). Immunophenotyping and Functional Characterization of NK Cells in SARS-CoV-2 Infection. Immuno, 5(3), 35. https://doi.org/10.3390/immuno5030035








