Abstract
Guillain-Barre syndrome is an autoimmune disease that provokes neural illness causing acute paralysis neuropathy. This syndrome appears after some bacterial infections produced by Campylobacter jejuni, Streptococcus pyogenes, S. pneumoniae, Haemophilus influenciae, E. coli and current studies showed the appears of this syndrome after SARS-CoV-2 infection. In this study, a in silico analysis was carry out in which to determinate bacterial epitopes than produce the molecule mimicry phenomena and that can produce the immune system activation against this epitope. A conserved amino acid sequence has been encountered with the highest probability to activate the immune system against this bacterial epitope, human gliomedin and ryanodine 3 type receptor. More studies needed to demonstrate in vivo the molecular mimicry in Guillain-Barre syndrome patients.
1. Introduction
Guillain-Barré Syndrome (GBS) is an autoimmune disorder characterized by an acute or subacute presentation of progressive polyradiculoneuropathy [1]. It manifests primarily as acute flaccid paralysis and constitutes an inflammatory polyneuropathy marked by rapid onset, systemic muscular weakness, unstable gait, and hypo- or areflexia [1]. Since the global eradication of poliomyelitis, GBS has emerged as the leading cause of acute or subacute flaccid paralysis worldwide [1,2].
Although its precise etiology remains uncertain, GBS frequently follows gastrointestinal or respiratory infections. Common microbial triggers include Campylobacter jejuni [3], Streptococcus pyogenes [4], Streptococcus pneumoniae [5], Haemophilus influenzae [6], and Escherichia coli [7], along with several viruses implicated in post-infectious immune responses [1,2]. More recently, SARS-CoV-2 has been identified as a potential precipitating factor [8,9,10]. Involvement of the central nervous system (CNS) by SARS-CoV-2 has been associated with symptoms such as headache, altered consciousness, dizziness, acute encephalopathy, seizures, and ataxia [11]. Meanwhile, the peripheral nervous system (PNS) involvement is often manifested through anosmia, ageusia, visual disturbances, neuropathic pain, and skeletal muscle weakness [12].
Several studies have proposed that autoimmune diseases, including GBS, may arise via a mechanism of molecular mimicry, whereby conserved peptide sequences shared between pathogens and host proteins elicit a cross-reactive immune response [13,14,15]. In such cases, the immune system fails to distinguish between foreign and self-antigens, resulting in a humoral and cellular response that inadvertently targets host neural structures [16].
Importantly, GBS can develop in individuals with no prior history of autoimmune or systemic disease, often representing a T cell–mediated immune response directed against neuronal surface proteins. This autoimmune process is thought to be triggered by pathogen-derived peptides that resemble host proteins, activating autoreactive lymphocytes through molecular mimicry [17,18]. As Ang defines it, molecular mimicry is “the theoretical possibility that sequence similarities between foreign and self-peptides are sufficient to lead to cross-activation of autoreactive B cells or T cells by pathogen-derived peptides” [19]. In this context, GBS may be understood as an autoimmune consequence of prior microbial infection in genetically predisposed individuals [19].
The present study aims to investigate the molecular mimicry mechanisms between neuronal surface proteins and microbial epitopes associated with GBS etiology using computational modeling approaches.
2. Materials and Methods
2.1. Review of the Main Microorganisms Related to GBS
A comprehensive review of the scientific literature was conducted to identify microorganisms associated with the onset of Guillain-Barré Syndrome (GBS). Based on this analysis, the microbial agents selected for inclusion in the present study were enteroviruses, influenza viruses, Campylobacter jejuni, Streptococcus pyogenes, Streptococcus pneumoniae, Haemophilus influenzae, Escherichia coli, SARS-CoV-2, and bacterial pathogens implicated in post–SARS-CoV-2 infections.
2.2. Sequence Alignment—Blast
A sequence homology analysis was conducted to identify conserved domains potentially implicated in the pathogenesis of Guillain-Barré Syndrome (GBS). The BLAST tool v. 2.15 (Basic Local Alignment Search Tool) available at http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 20 September 2022) was used to perform a preliminary alignment between GBS-associated autoantigens and proteins from microorganisms implicated in its etiology. A secondary, more refined alignment was subsequently carried out using the DNASTAR Lasergene software suite (Madison, WI, USA) to investigate proteins exhibiting notable similarity [14]. Multiple sequence alignments were performed via the Clustal Omega server v.1.2 using default parameters [20]. Regions of interest were defined as those exhibiting significant homology between GBS-related autoantigens and microbial proteins. These regions were prioritized based on the degree of sequence identity, both in discrete peptide segments and in broader full-length alignments.
2.3. Antigenic Protein Analysis
The antigenic analysis was performed by the Jameson-Wolf algorithm of human and microbial proteins to determine if the epitope found is on the outer surface of the protein where antigen-antibody formation occurs [21]. The DNASTAR Protean program v.5.01 (Madison, WI, USA) was used for this method.
2.4. Protein Modeling
To evaluate the immunogenic potential of the aligned sequences, the Jameson-Wolf antigenicity algorithm was applied to both human and microbial proteins. This analysis aimed to determine whether the predicted epitopes were located on the protein surface, where antigen-antibody interactions typically occur [21]. The analysis was executed using the DNASTAR Protean software v.5.01 (Madison, WI, USA). The three-dimensional model of human gliomedin and ryanodine 3 type receptor was carried out using the I-TASSER online server v.5.1, the Z-score was use to verified the quality of 3D protein modeling to select the best model [22].
2.5. Prediction of B Cell, Citotoxic T Lymphocytes (CTL) and Helper T Lymphocytes (HTL) Epitopes
CTL epitopes were predicted using the IEDB MHC Class I binding algorithm (http://tools.iedb.org/mhci) (accessed on 20 September 2022), while HTL epitopes were identified using the IEDB MHC Class II binding tool (http://tools.iedb.org/mhcii) (accessed on 20 September 2022). The antigenicity of predicted epitopes was further evaluated using the VaxiJen v2.0 server with a threshold set at 0.4 [23].
2.6. Prediction of the 3D Structures of the Predicted Epitope and HLA-A 0201 Allele Molecular Docking
The three-dimensional structures of the predicted epitopes were generated using the PEP-FOLD3 server [24]. Selected epitope models were subsequently used for molecular docking with HLA-A*0201 (PDB ID: 4U6Y) for MHC Class I and HLA-DR52c (PDB ID: 3C5J) for MHC Class II molecules. Docking simulations were performed using the CABS-dock server [25], and binding affinity was assessed through the HawKRank scoring system [26].
3. Results
3.1. Sequence Alignment—Blast
A conserved six–amino acid consensus sequence (“KGEKGD”) was identified across five microbial species—Campylobacter jejuni, Streptococcus pyogenes, Streptococcus pneumoniae, Haemophilus influenzae, and Escherichia coli—as well as in human neural proteins, specifically gliomedin and the ryanodine receptor type 3. This finding is presented in Table 1.

Table 1.
Alignment of the conserved “KGEKGD” epitope across human neural proteins (gliomedin and ryanodine receptor type 3) and microbial species implicated in Guillain-Barré Syndrome (GBS). The presence of this conserved motif supports a mechanism of molecular mimicry that may underlie the aberrant immune cross-reactivity observed in GBS pathogenesis.
Clustal Omega alignments revealed a high degree of sequence similarity among the human gliomedin isoform 1 precursor (NP_861454.2) and several microbial proteins, including the Streptococcus pneumoniae YSIRK-type signal peptide-containing protein (WP_050096714.1), the Haemophilus influenzae hypothetical protein (WP_041175161.1), the predicted protein from Enterococcus faecalis (EET98925.1), and the Escherichia coli phage tail protein (WP_096855986.1) (Figure 1). Beyond the previously identified six–amino acid epitope “KGEKGD,” additional homologous regions were observed across these alignments. In all sequences, the “KGEKGD” motif appears in its entirety. These findings further support the hypothesis of shared immunogenic elements that may contribute to molecular mimicry in GBS.
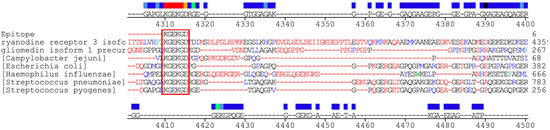
Figure 1.
Multiple sequence alignment of protein sequences, with conserved amino acid regions shaded and the conserved “KGEKG” epitope highlighted in red. Numerous alignment matches were observed, reinforcing the hypothesis of molecular mimicry between the human gliomedin isoform 1 precursor and microbial proteins from Streptococcus pyogenes, Streptococcus pneumoniae, Escherichia coli, Campylobacter jejuni, and Haemophilus influenzae (A multiprotein alignment of the studied species is observed. The consensus region is represented by the color patterns in the upper region, where the green, light blue, blue and black colors indicate a low consensus among the studied proteins in decreasing order. On the other hand, the orange and red colors represent a very high and total consensus, respectively. The pentapeptide KGEKGD is conserved in all proteins, except for a residue in the YadA-like family protein of Haemophilus Influenzae).
3.2. Antigenicity Prediction
Antigenicity analysis was conducted on human gliomedin and ryanodine receptor type 3 proteins, as well as on homologous microbial proteins from S. pyogenes, S. pneumoniae, E. coli, C. jejuni, and H. influenzae, with a focus on the conserved epitope “KGEKDG.” The Jameson-Wolf algorithm yielded a positive antigenicity index for all analyzed proteins, suggesting that the conserved sequence resides within a surface-exposed region likely to participate in antigen–antibody interactions. These results are summarized in Table 2.

Table 2.
Antigenicity predictions based on the Jameson–Wolf index. The conserved residues within the consensus region are surface-exposed, indicating high potential for immunological recognition as B cell epitopes.
The predicted “KGEKGD” epitope demonstrated enhanced potential for B cell activation, attributed to its allergenic properties. Structural modeling confirmed the presence of this epitope within the 3D conformations of the ryanodine receptor type 3 (residues 4112–4379) and gliomedin (residues 203–293), as illustrated in Figure 2. Analysis using BepiPred revealed a strong antigenicity gradient across the “KGEKGD” region, further indicating that the epitope is surface-exposed and therefore accessible for antibody recognition.
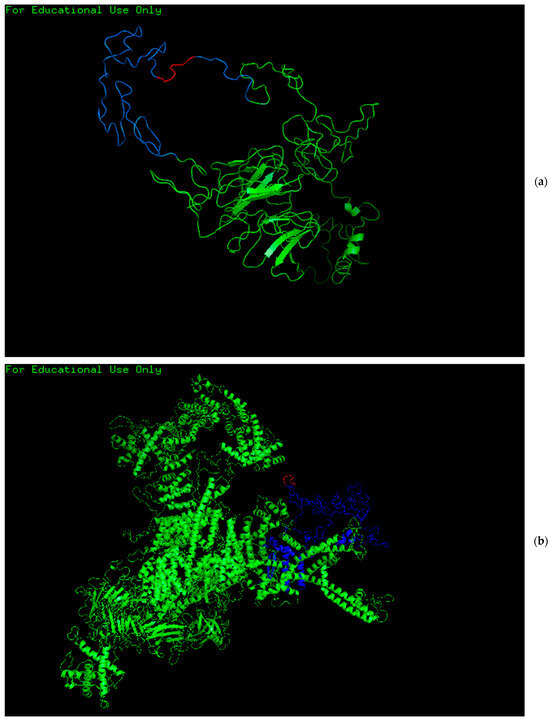
Figure 2.
Three-dimensional representation of gliomedin (a) and ryanodine receptor type 3 (b), highlighting the structural location of the “KGEKGD” epitope (in red). In both protein models, the epitope is surface-exposed and positioned within regions of predicted high antigenicity (in blue), supporting its potential role in B cell-mediated immune recognition. The other remaining parts of the proteins are in green.
3.3. The CTL and HTL Epitopes
The predicted “KGEKGD” epitope exhibits strong antigenic and allergenic properties, enabling it to elicit a robust autoimmune response through recognition by numerous MHC class I and class II alleles (see Table 3).

Table 3.
Immunopeptidomic profiling of the “KGEKGD” epitope, detailing the MHC class I and class II alleles predicted to bind this peptide.
3.4. Prediction of the 3D Structures of the Predicted Epitope and HLA-A 0201 and HLA DR52c Allele Molecular Docking
The three-dimensional structure of the predicted epitope is presented in Figure 3A. The sOPEP energy score for the epitope prediction was calculated as −4.3258 kcal/mol. Peptide–protein docking analysis with the HLA-A*02:01 allele revealed a binding interface energy of −7.64 kcal/mol (Figure 3B). Key interacting residues within the HLA molecule—PHE372, TRP167, GLU55, THR163, and GLU58—formed hydrogen bonds with the peptide residues LYS1, GLY2, GLU3, LYS4, GLY5, and ASP6, as illustrated in Figure 3C. Similarly, docking with the HLA-DR52c allele demonstrated a receptor–peptide binding energy of −12.58 kcal/mol (Figure 3D). The interacting HLA residues GLU9, TYR253, GLN245, GLN249, and ALA57 engaged in hydrogen bonding with the same peptide residues (LYS1, GLY2, GLU3, LYS4, GLY5, ASP6), as shown in Figure 3E.
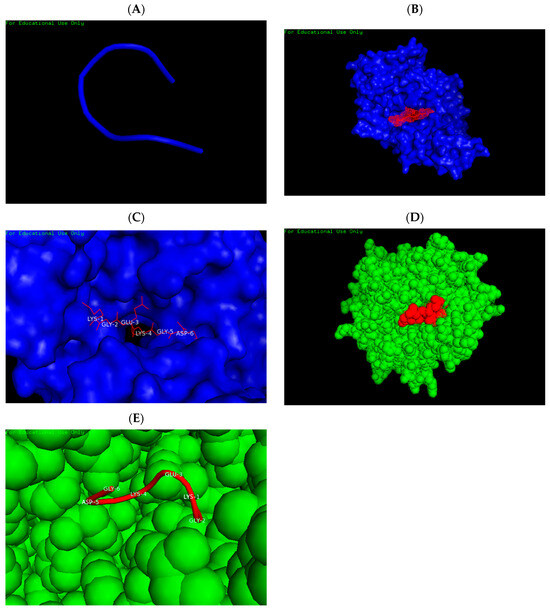
Figure 3.
Structural prediction and molecular docking of the “KGEKGD” epitope with HLA alleles. (A) Representative 3D model of the epitope derived from the top cluster. (B) Docking between the HLA-A02:01 molecule (blue) and the epitope (red) showing a binding free energy of −7.64 kcal/mol. (C) Interaction interface highlighting hydrogen bonds formed between peptide residues LYS1, GLY2, GLU3, LYS4, GLY5, ASP6 (red) and HLA-A02:01 residues (blue). (D) Docking of the HLA-DR52c allele (green) with the epitope (red), exhibiting a binding free energy of −12.58 kcal/mol. (E) Interaction between the same peptide residues (LYS1, GLY2, GLU3, LYS4, GLY5, ASP6; red) and HLA-DR52c residues (green).
4. Discussion
Guillain-Barré Syndrome (GBS) is an acute or subacute neuropathy characterized primarily by muscle weakness or flaccid paralysis [1]. The etiology of this syndrome is typically associated with prior bacterial or viral infections, which induce a lymphocytic immune response against neuronal proteins—an example of molecular mimicry [18]. Several studies have reported the presence of autoantibodies, such as antiglycolipids, in patients with GBS [27]. Additionally, autoantibodies have been identified against neurofascin-155, a glial cell adhesion molecule located in paranodal junctions and axons [28], as well as against gliomedin, a key protein involved in the assembly of adhesion complexes at the nodes of Ranvier [29]. Nevertheless, the precise peptide sequence capable of eliciting an MHC class II-mediated immune response has yet to be conclusively identified [18].
Normal neural function depends on the regulation of membrane potential via the opening of sodium and calcium channels. In this study, we identified a conserved region in channel-regulating proteins, which may plausibly account for the molecular mimicry that initiates GBS. Gliomedin is essential for the structural integrity of the nodes of Ranvier, linking neurofascin and NrCAM—both critical for the clustering of sodium channels in myelinated axons [30]. Similarly, the ryanodine receptor type 3 mediates the intracellular release of calcium in excitable tissues, including muscle and nerve cells [31].
Although the presence of autoantibodies against gliomedin has been previously documented [32,33], the specific epitope responsible for initiating the autoimmune response had not been defined. Our in silico analyses, which included B-cell and T-cell epitope prediction for cytotoxic and helper T lymphocytes, revealed epitopes with high binding affinities to MHC class I and II alleles. These epitopes exhibit strong immunogenic potential, with high antigenicity and allergenic scores, and are conserved among microbial species such as Campylobacter jejuni, Streptococcus pyogenes, Streptococcus pneumoniae, Haemophilus influenzae, and Escherichia coli. Notably, although there is no prior evidence of autoantibodies targeting ryanodine receptor type 3, our findings indicate that the predicted KGEKGD epitope is present in both gliomedin and the ryanodine receptor, suggesting a common immunological trigger.
Additional autoantigens implicated in GBS include neurofascin-186, an isoform of neurofascin-155 localized at the nodes of Ranvier and detected in the sera of GBS patients [33], and contactin-1, which has been observed in approximately 2–4% of patients with the demyelinating subtype of GBS, often correlating with a rapid and severe clinical onset [34] (see the Supplementary Materials).
Interestingly, no epitope homology was observed between human proteins and those from certain viruses, including influenza virus and enteroviruses. This finding diverges from the prevailing scientific consensus, which implicates viral infections as a common precipitating factor in GBS pathogenesis [1,35].
Efforts to identify molecular mimicry in the context of SARS-CoV-2 have yielded similar methodologies. A study by Nuñez-Castilla et al. [36] employed protein alignment, 3D structural modeling, docking simulations, and molecular dynamics to demonstrate mimicry between the SARS-CoV-2 spike protein and erythropoietin. The findings explained antibody-binding region similarities that may account for COVID-19-related thrombocytopenia.
Moreover, Beaudoin et al. [37] conducted a comprehensive structural analysis of spike proteins from three coronaviruses. They generated a representative homo-oligomeric model, screened it using structural alignment algorithms, and performed docking simulations to evaluate molecular mimicry with human antibodies. Similar pipelines have been applied in identifying mimicry across other pathogens by integrating whole proteome analysis, structure prediction using AlphaFold2, B-cell epitope prediction, and surface accessibility profiling [38,39].
This study employed state-of-the-art computational tools in chemical epitope prediction. Epitope validation was performed using the IEDB suite, which incorporates advanced machine learning frameworks such as NetMHCpan-4.1 and NetMHCIIpan-4.0, both trained on mass spectrometry-derived ligand datasets [40]. Additionally, widely established methods for structural modeling, molecular docking, and immunoinformatics were applied to enhance the reliability of the findings.
Nevertheless, it is important to underscore that the current study is grounded in computational immunopeptidomics and, as such, may not fully replicate the complexity of human immunological responses. Experimental validation in biological models remains essential to substantiate these results and confirm the role of molecular mimicry in the pathogenesis of Guillain-Barré Syndrome. Within the limits present in this study, it has been possible to determine that the KGEKGD consensus epitope or sequence is present in human gliomedin proteins and the ryanodine type 3 receptor, which are present on the surface of the neuronal and axonal endings, which regulate the membrane potential, and are also present in microorganisms such as S. pyogenes, S. pneumoniae, E. coli, C. jejuni and H. influenciae, which could generate a molecular mimicry causing lymphocyte autoreactivity, thus, causing an autoimmune neuropathy such as that referred to Guillain-Barre syndrome.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/immuno5030028/s1, Autoantigens found in Guillain Barre syndrome by literature.
Author Contributions
G.A.O.-P.: conceptualization, research design and execution, data analysis, and manuscript preparation. L.A.P.-S.: data analysis and manuscript writing. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to acknowledge the Vice Rectory of Research of the Universidad Católica de Santa Maria for financial and physical support. RESOLUCION No. 27494-R-2020. This research was the result of the postdoctoral studies of Dr. Gustavo Alberto Obando-Pereda.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors gratefully acknowledge Paul Galvez Murillo for his valuable assistance with the grammatical revision and critical review of the manuscript. This work was supported by the Vicerectorado de Investigación of Universidad Católica de Santa María, Arequipa, Perú.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Esposito, S.; Longo, M.R. Guillain-Barre syndrome. Autoimmun. Rev. 2017, 16, 96–101. [Google Scholar] [CrossRef]
- Willison, H.J.; Jacobs, B.C.; van Doorn, P.A. Guillain-Barre syndrome. Lancet 2016, 388, 717–727. [Google Scholar] [CrossRef]
- Sookaromdee, P.; Wiwanitkit, V. Antibodies to dengue, Zika, Campylobacter jejuni and gangliosides in Guillain-Barre syndrome. Neurol. India 2019, 67, 335. [Google Scholar] [CrossRef]
- Takahashi, R.; Yuki, N. Streptococcal IdeS: Therapeutic potential for Guillain-Barre syndrome. Sci. Rep. 2015, 5, 10809. [Google Scholar] [CrossRef]
- Lewczuk, P.; Padilla-Docal, B.; Rodríguez-Rey, A.; Noris-García, E.; Coifiu-Fanego, R.B.; González-Hernández, M.; Sánchez-Martínez, C.; Dorta-Contreras, A.J. sICAM-1 intrathecal synthesis and release during the acute phase in children suffering from Coxsackie A9 and S. pneumoniae meningoencephalitis. Arq. Neuropsiquiatr. 2008, 66, 504–508. [Google Scholar]
- Restrepo-Jimenez, P.; Rodriguez, Y.; Gonzalez, P.; Chang, C.; Gershwin, M.E.; Anaya, J.M. The immunotherapy of Guillain-Barre syndrome. Expert Opin. Biol. Ther. 2018, 18, 619–631. [Google Scholar] [CrossRef]
- Jo, Y.S.; Choi, J.Y.; Chung, H.; Kim, Y.; Na, S.J. Recurrent Guillain-Barre Syndrome Following Urinary Tract Infection by Escherichia coli. J. Korean Med. Sci. 2018, 33, e29. [Google Scholar] [CrossRef]
- Toscano, G.; Palmerini, F.; Ravaglia, S.; Ruiz, L.; Invernizzi, P.; Cuzzoni, M.G.; Franciotta, D.; Baldanti, F.; Daturi, R.; Postorino, P.; et al. Guillain-Barré Syndrome Associated with SARS-CoV-2. N. Engl. J. Med. 2020, 382, 2574–2576. [Google Scholar] [CrossRef]
- Arnaud, S.; Budowski, C.; Ng Wing Tin, S.; Degos, B. Post SARS-CoV-2 Guillain-Barré syndrome. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol. 2020, 131, 1652–1654. [Google Scholar] [CrossRef]
- Finsterer, J.; Scorza, F.A.; Fiorini, A.C. SARS-CoV-2–associated Guillain-Barre syndrome in 62 patients. Eur. J. Neurol. 2021, 2025, 28. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA Neurol. 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef]
- Dreyfus, D.H.; Farina, A.; Farina, G.A. Molecular mimicry, genetic homology, and gene sharing proteomic “molecular fingerprints” using an EBV (Epstein-Barr virus)-derived microarray as a potential diagnostic method in autoimmune disease. Immunol. Res. 2018, 66, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Obando-Pereda, G.A. GAKG-RGEKG an Epitope That Provokes Immune Cross-Reactivity between Prevotella sp. and Human Collagen: Evidence of Molecular Mimicry in Chronic Periodontitis. Autoimmune Dis. 2016, 2016, 5472320. [Google Scholar] [PubMed]
- Rojas, M.; Restrepo-Jiménez, P.; Monsalve, D.M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Leung, P.S.C.; Ansari, A.A.; Gershwin, M.E.; Anaya, J.M. Molecular mimicry and autoimmunity. J. Autoimmun. 2018, 95, 100–123. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, K. Guillain-Barre syndrome during COVID-19 pandemic: An overview of the reports. Neurol. Sci. 2020, 41, 3149–3156. [Google Scholar] [CrossRef]
- Sudo, M.; Miyaji, K.; Spath, P.J.; Morita-Matsumoto, K.; Yamaguchi, Y.; Yuki, N. Polyclonal IgM and IgA block in vitro complement deposition mediated by anti-ganglioside antibodies in autoimmune neuropathies. Int. Immunopharmacol. 2016, 40, 11–15. [Google Scholar] [CrossRef]
- Kuwabara, S. Guillain-Barre syndrome: Epidemiology, pathophysiology and management. Drugs 2004, 64, 597–610. [Google Scholar] [CrossRef]
- Ang, C.W.; Jacobs, B.C.; Laman, J.D. The Guillain-Barre syndrome: A true case of molecular mimicry. Trends Immunol. 2004, 25, 61–66. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, B.; Yu, J.; He, X.; Yang, F.; Liu, X.; Li, Y.; Liu, Y.; Xie, Q.; Yang, Z. Analysis of the Genome Sequence and Prediction of B-Cell Epitopes of the Envelope Protein of Middle East Respiratory Syndrome-Coronavirus. IEEEACM Trans. Comput. Biol. Bioinform. 2018, 15, 1344–1350. [Google Scholar]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; A Greenbaum, J.; Jespersen, M.C.; Jurtz, V.; Kim, H.; Sette, A.; Yan, Z.; Nielsen, M.; Andreatta, M.; Dhanda, S.K.; et al. IEDB-AR: Immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Kurcinski, M.; Badaczewska-Dawid, A.; Kolinski, M.; Kolinski, A.; Kmiecik, S. Flexible docking of peptides to proteins using CABS-dock. Protein Sci. 2020, 29, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Feng, T.; Liu, H.; Li, D.; Chen, F.; Kang, Y.; Sun, H.; Zhu, F. HawkRank: A new scoring function for protein-protein docking based on weighted energy terms. J. Cheminform. 2017, 9, 66. [Google Scholar]
- Uchibori, A.; Chiba, A. Autoantibodies in Guillain-Barre Syndrome. Brain Nerve 2015, 67, 1347–1357. [Google Scholar]
- Ng, J.K.M.; Doppler, K.; Stengel, H.; Sommer, C.; Appeltshauser, L.; Meinl, E.; Grosskreutz, J. Neurofascin-155 IgM autoantibodies in patients with inflammatory neuropathies. J. Neurol. Neurosurg. Psychiatry 2018, 89, 1145–1151. [Google Scholar]
- Ebrahim Soltani, Z.; Rahmani, F.; Rezaei, N. Autoimmunity and cytokines in Guillain-Barré syndrome revisited: Review of pathomechanisms with an eye on therapeutic options. Eur. Cytokine Netw. 2019, 30, 1–14. [Google Scholar] [CrossRef]
- Koch, M.; Maertens, B.; Franzke, C.-W.; Hopkins, D.; Keene, D.R.; Bruckner-Tuderman, L.; Greenspan, D.S. Cleavage and oligomerization of gliomedin, a transmembrane collagen required for node of ranvier formation. J. Biol. Chem. 2007, 282, 10647–10659. [Google Scholar]
- Meissner, G. The structural basis of ryanodine receptor ion channel function. J. Gen. Physiol. 2017, 149, 1065–1089. [Google Scholar] [CrossRef] [PubMed]
- Kira, J.I.; Yamasaki, R.; Ogata, H. Anti-neurofascin autoantibody and demyelination. Neurochem. Int. 2019, 130, 104360. [Google Scholar] [CrossRef] [PubMed]
- Devaux, J.J. Antibodies to gliomedin cause peripheral demyelinating neuropathy and the dismantling of the nodes of Ranvier. Am. J. Pathol. 2012, 181, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Querol, L.; Lleixà, C. Novel Immunological and Therapeutic Insights in Guillain-Barré Syndrome and CIDP. Neurotherapeutics 2021, 18, 2222–2235. [Google Scholar] [CrossRef]
- Nascimento, O.J.M.; da Silva, I.R.F. Guillain-Barre syndrome and Zika virus outbreaks. Curr. Opin. Neurol. 2017, 30, 500–507. [Google Scholar] [CrossRef]
- Cickovski, T.; Mondal, A.M.; Stebliankin, V.; Balbin, C.A.; Baral, P.; Chapagain, P.; Sobhan, M.; Siltberg-Liberles, J.; Nunez-Castilla, J.; Narasimhan, G.; et al. Potential Autoimmunity Resulting from Molecular Mimicry between SARS-CoV-2 Spike and Human Proteins. Viruses 2022, 14, 1415. [Google Scholar]
- Blundell, T.L.; Thomas, S.E.; Bannerman, B.P.; Vedithi, S.C.; Copoiu, L.; Hala, S.; Jamasb, A.R.; van Tonder, A.J.; Alsulami, A.F.; Beaudoin, C.A.; et al. Predicted structural mimicry of spike receptor-binding motifs from highly pathogenic human coronaviruses. Comput. Struct. Biotechnol. J. 2021, 19, 3938–3953. [Google Scholar]
- Rich, K.D.; Srivastava, S.; Muthye, V.R.; Wasmuth, J.D. Identification of potential molecular mimicry in pathogen-host interactions. PeerJ 2023, 11, e16339. [Google Scholar] [CrossRef]
- Bigdeli, A.; Ghaderi-Zefrehei, M.; Lesch, B.J.; Behmanesh, M.; Arab, S.S. Bioinformatics analysis of myelin-microbe interactions suggests multiple types of molecular mimicry in the pathogenesis of multiple sclerosis. PLoS ONE 2024, 19, e0308817. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).