Abstract
Asthma is a respiratory condition often stemming from childhood, characterized by difficulty breathing and/or chest tightness. Current treatment options for both adults and children include beta-2 agonists, inhaled corticosteroids (ICS), and leukotriene modifiers (LTM). Despite recommendations by the Global Initiative for Asthma, a substantial number of patients are unresponsive to treatment and unable to control symptoms. Pharmacogenomics have increasingly become the front line of precision medicine, especially with the recent use of candidate gene and genome- wide association studies (GWAS). Screening patients preemptively could likely decrease adverse events and therapeutic failure. However, research in asthma, specifically in pediatrics, has been low. Although numerous adult trials have evaluated the impact of pharmacogenomics and treatment response, the lack of evidence in children has hindered progress towards clinical application. This review aims to discuss the impact of genetic variability and response to asthmatic medications in the pediatric population.
1. Introduction
Asthma is a multifactorial respiratory condition characterized by the narrowing of one’s airways and affects over 300 million people, including 7% of children in the US [1]. Approximately 90% of all asthma cases stem from childhood [2], with the biggest burden on the quality of life in children ages 10–14 [3]. Symptoms such as difficulty breathing, shortness of breath, wheezing, tightness of the chest, etc. are early markers to suspect and diagnose asthma. Left untreated or uncontrolled, many childhood asthma cases can continue into adulthood, decreasing quality of life and potentially increasing the risk of developing chronic obstructive pulmonary disease (COPD). Managing asthma alone accounts for approximately $80 billion spent annually on healthcare in the US [4]. In 2013, children between the ages of 5–17 in the US missed nearly 13 million days of school, an increase from the 10.4 million days in 2008 [5]. With a projected 100 million additional asthma cases by 2025 [6], the importance of finding appropriate treatment and gaining control of symptoms is even more crucial. Common treatment options for asthma are bronchodilators, inhaled corticosteroids (ICS), and leukotriene modifiers (LTM). Goals of therapy include providing symptomatic relief by optimizing airway function, reducing future exacerbations, and minimizing adverse effects from medications. According to the 2021 Global Initiative for Asthma (GINA) guidelines, initial pediatric treatment recommendations include using short acting beta agonists (SABA) and/or low doses of ICS as the first-line of therapy. In situations where symptoms are not adequately controlled, these guidelines provide recommendations when considering additional medications [7].
Despite this, up to 70% of patients are still unable to find proper relief [8]. Improper diagnoses and lack of adherence have been partially blamed; however, genetics have been implicated as well. Providing appropriate therapeutic recommendations could further limit the cases of adverse events from these medications. Up to 3% of pediatric hospital admissions are attributed to asthmatic adverse reactions, ranging from headaches to adrenal suppression [9]. With widespread intra-variability among medication responses, pharmacogenomics has an emerging place in asthmatic treatment. Groundbreaking discoveries in allele variations and polymorphisms, such as single nucleotide polymorphisms (SNPs), provide opportunities for personalized treatment. Pharmacogenomics highlights the importance of optimizing a patient’s response to drug therapy while minimizing adverse reactions. Specifically, breakthrough research completed in 2014 within Cystic Fibrosis patients has allowed therapeutic recommendations to be made for those with the G551D-CFTR variant [10].
Although asthma affects both children and adults, most studies have looked more closely at children, since environmental (i.e., workplace) and social (i.e., smoking) factors are minimized in this patient population. However, there have been no clinically relevant findings that have warranted specific therapeutic recommendations for any asthmatic patient population. Herein is a review of the current pharmacogenomic pediatric findings for beta-2 agonists, ICS, and LTM in asthmatic treatment.
2. Beta-2 Agonists
Beta-2 agonists are a class of bronchodilators commonly used to treat asthma and COPD. Two classes make up these beta-2 agonists: SABAs and long-acting beta agonists (LABAs). As rescue inhalers, SABAs, such as albuterol and levalbuterol, are commonly prescribed to provide immediate relief from asthma symptoms. Onset is typically less than five min and lasts 3–6 h [11]. Conversely, LABAs, such as salmeterol and formoterol may be added alongside ICS in situations where controller inhalers are necessary. Onset is approximately 5–15 min and lasts an average of 12 h [11]. These agonists bind to the β2 adrenergic receptor found abundantly in the smooth muscles located in the lungs. Activation of these G-protein coupled receptors (GPCR) increases cAMP concentrations which in turn leads to bronchodilation (Figure 1). The β2 adrenergic receptor is encoded by the ARB2 gene and has garnered significant clinical interest.
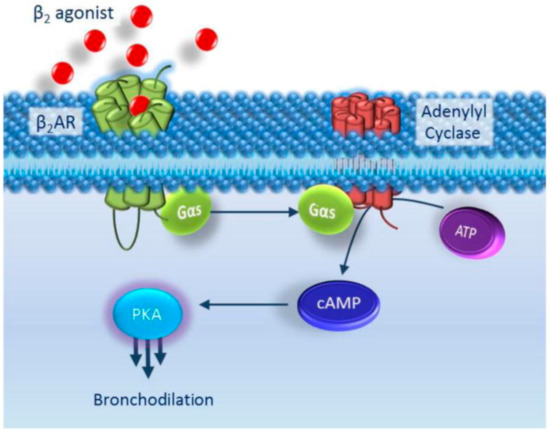
Figure 1.
Beta-2 Agonists Mechanism of Action. Adapted from [11].
2.1. SABA and ADRB2 Variations
The most studied variation of the beta-2 adrenergic receptor is the ADRB2 gene (Table 1). This gene is found on chromosome 5q31-q32, an area highly associated with asthma. The amino acid at position 16 of the beta-2 receptor is thought to modulate downregulation and airway responsiveness [12,13]. Studies on children [14,15] and adults [16] have shown that arginine (Arg) at this position displays better bronchodilation response when treated with a SABA [13]. Choudhry et al. studied 684 Puerto Ricans and Mexican children with an average age of 12 and 13, respectively. The presence of Arg at position 16 produced the greatest bronchodilation (10.46 ± 2.44% Arg/Arg, 6.13 ± 0.74% Arg/Gly change in FEV1), compared to Gly16Gly when administered albuterol in Puerto Ricans (p = 0.002) [14]. In contrast to these children, there was no genetic association found among Mexican children [14]. Martinez et al. reported that Arg16Arg and Arg16Gly were 5.3 and 2.3 times more likely, respectively, to produce greater bronchodilation compared to Gly16Gly when given albuterol. Included in this study were 496 children with an average of 10.8 years, with at least one non-Hispanic parent [17]. Positive bronchodilation was defined as >15.3% predicted FEV1.

Table 1.
SABA and ADRB2 rs1042713 Arg16Gly (+46 A > G).
Another pediatric study found favorable results with Arg16Arg alone as opposed to other genotypes when administered fenoterol. In 100 children with an average age of 9.6, Arg16Arg showed better bronchodilation response (108.68 ± 15.62% post BD FEV1) compared to either Arg16Gly and Gly16Gly (101.86 ± 14.03% post BD FEV1) [18], when administered this β2 agonist. A positive response was defined as ≥12% increase in FEV1. Studies done by Salah [19] and Finkelstein [15] also found that those with Gly16Gly genotypes produced a reduced response, while Arg16Arg displayed a favorable response to albuterol.
An adult study found that the patients with Arg16Arg displayed a better response, measured by peak expiratory flow rate (PEFR), when albuterol was replaced by ipratropium bromide in comparison to the patients with Gly16Gly [20]. Likewise, a pediatric study conducted by Carroll et al. found similar results [21]. Children with the Gly16Gly variation showed a better response, demonstrated by a shorter duration of continuous albuterol treatment (3 ± 0.9 days) and length of ICU stay (43 ± 25 h), compared to those with the Arg variants (4.8 ± 1.9 days and 74 ± 34 h, respectively) [21]. In a prospective case series study, Giubergia [22] observed changes in desensitization over four weeks among 117 Argentinian children treated with albuterol. Although the results were not statistically significant, those with the Arg16Arg genotype displayed a decline in responsiveness to long term treatment.
Focusing on alleles, Jovicic et al. reported that children with the minor G allele demonstrated a better response when given albuterol (GG: 14.4 ± 6.1% and GA: 10.4 ± 5.8% change in FEV1 p = 0.044) [23]. Additionally, they found that children with severe asthma were likely to be homozygote for the G allele (p = 0.01) compared to those with the major A allele, and thus would benefit most when treated with albuterol [23].
The amino acid position 27 on the β2 receptor is also believed to have a role in bronchodilation, however with conflicting reports regarding the SABA response (Table 2) [24,25]. Alghobashy reported greater FEV1/ forced vital capacity (FVC) and FEV1 in children receiving albuterol or terbutaline with the Gln27Glu variants compared to those with Gln27Gln (p = 0.008, p < 0.001, respectively) [26]. However, Giubergia [22] found that Argentinian children with the Gln27Gln and Glu27Glu variants displayed a decline in responsiveness over time compared to those with Gln27Glu (p = 0.001). This decline in pre- and post-albuterol treatment was hypothesized to be from receptor desensitization. In contrast, a smaller study analyzed 31 African American children admitted to the hospital with status asthmaticus. All patients were given the initial treatment of IV steroids and inhaled albuterol with the addition of IV terbutaline, phosphodiesterase inhibitor, and/or aminophylline dependent on response. Approximately 38% of the children with the Gln27Gln variant needed additional terbutaline treatment compared to the ~50% of children with the Gln27Glu variant (p = 533). Only one of the 21 patients with the Gln27Gln variant receiving IV terbutaline needed additional aminophylline treatment, whereas five of the 10 with Gln27Glu did (p = 0.002) [27].

Table 2.
SABA and ADRB2 rs1042714 Gln27Glu (+79 C > G).
Due to conflicting reports on polymorphisms that cause bronchodilation, no clear association at position 27 and a response to SABA therapy has been ascertained [14,15,17,18,21,23]. Some studies [25] have reported no association between SABA (i.e., albuterol) response and ADRB2 polymorphisms. It has been hypothesized that there is a more complex role at position 16 and 27. Jovicic suggested haplotype differences impact therapeutic outcomes more than SNPs, as seen in declining dFEV1 among patients with +46 A/+79 C (p = 0.026) [23].
2.2. SABA and Other Gene Variations
Contradictory findings regarding the ADRB2 gene have promoted interest into other areas (Table 3). The arginase 1 (ARG1) gene is found on chromosome 6q23 and encodes for arginase. Nitric oxide synthase (NOS) uses L-arginine, metabolized by arginase to regulate endogenous nitric oxide (NO), a known bronchodilator. Litonjua found that rs2781659, rs2781663, rs2781665, and rs2749935 SNPs were the most prevalent in bronchodilator response among the Childhood Asthma Management Program (CAMP) [28], Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol (LOCCS) [29], Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma (LODO) [30], and the Asthma Trial studies [31]. CAMP was used to initially screen for SNPs and focused on children and their parents while the other three focused only on adults. The SNP, rs2781659, was ultimately found to be most significant for bronchodilation (Bonferroni-corrected p value = 0.047) [31]. In replication studies, the AA genotype produced the most significant bronchodilation from albuterol in adults compared to either the AG or GG genotypes. Bronchodilation was defined as the percent difference from baseline in FEV1: Asthma trial = 41.39, LOCCS = 7.35, LODO = 11.53. However, Scaparrotta could not find an association with ARG1s, rs2781659 and bronchodilation among asthmatic children given fenoterol (p = 0.02) [18].

Table 3.
SABA and genetic variations.
The thyroid hormone receptor β (THRB) gene is found on chromosome 3p24.2 and encodes for a portion of the thyroid hormone receptor. Thyroid hormones are necessary for lung development in addition to other organs. THRB variants have been associated with altered airway flow and impact smooth muscle in the lungs, though data on adults are scarce. Initial findings by Duan et al. demonstrated that the SNP, rs892940, was associated with bronchodilation in the CAMP study (population-based p value 0.09) and was further replicated in two out of three adult trials (Sepracor asthma trial; LODO; and LOCCS): namely the Sepracor asthma trial and LODO (combined p value 0.0012). Specifically, children with the minor A allele showed better bronchodilation (summary OR 1.34, 95% CI 1.12–1.59) to inhaled albuterol than those with the major allele G [32]. Scaparrotta aimed to confirm that rs892940 (G > A) impacted bronchodilation in children treated with inhaled fenoterol, but could not find any clear association [18]. Duan also identified SNPs in the vitamin D receptor (VDR) and Wilms’ tumor 1 (WT1) in both the CAMP and the LODO study. However, bronchodilation response was insignificant or moderate at best when taking both studies together [32]. In a separate GWAS using participants from the CAMP study, Duan found variants in the COL22A1 and CLOCK region in primary and secondary replication cohorts that were associated with bronchodilator response when using as-needed albuterol [33].
SPATA13-AS1 is an anti-sense RNA that has an unknown mechanism and is thought to modulate the expression of ASEF2. This protein is known to have similar function as the guanine nucleotide exchange factor, activating Rho-family GTPase and inactivating RhoA. GTPase activation involves smooth muscle contraction while RhoA inactivation promotes smooth muscle dilation. Padhukasahasram identified SPATA13-AS1 in African American and European populations diagnosed with asthma. The initial discovery group comprised of African Americans between the ages of 12–56 living in south Michigan with no history of pulmonary disease or heart failure. Further replication groups included both healthy and asthmatic African Americans and European Americans with and without genome-wide data [34]. Initially, SPATA13-AS1 and sulfotransferase family 4A member 1 (SULT4A1) were both thought to affect SABA response. However, only SPATA13-AS1 was found to be statistically significant for bronchodilation when considering all the groups combined (p = 7.38 × 10−7). SPATA13-AS1 variations (rs9507294, rs912142, rs2248119, rs9551086, and rs9553225) and their effect size were also explored. In African Americans with genome-wide data, the SABA response to albuterol produced the greatest bronchodilation at 10.53 ± 12.93 change in FEV1 from baseline [34].
A GWAS done on CAMP, LOCCS, LODO, and three others found that variations in serine-rich 2-like (SPATS2L) protein coding gene were involved in bronchodilation [35]. Although little is known about exact function of this gene, Himes determined through replication studies that SNP, rs296137 produced the lowest combined p value of 9.7 × 10−7. Specifically, those with the TT variant had a measured bronchodilation of 16% while CC and TC measured at 10.3% and 11.2%, respectively. This gave rise to the notion that the improvements in bronchodilation seen with the TT variants were due to decreased SPATS2L transcription, causing an increased beta-2 receptor concentration [35].
Israel et al. conducted a GWAS with 724 participants of the SNP Health Association Resource Asthma Resource Project (SHARP) and identified five SNPs that were positively associated with bronchodilation. However, only four of these (rs350729, rs1840321, rs1384918, and rs1319797) were statistically significant for bronchodilator response after receiving albuterol [36]. These SNPs were located on chromosome 2p16.2 which is near the ankyrin repeat (ASB3) gene. Involvement in muscle cell proliferation led the authors to hypothesize that the ASB3 gene expression modulates pulmonary function. It was reported that those homozygous for the minor allele would be expected to produce diminished responses compared to those homozygous for the major allele [36].
The corticotrophin-releasing hormone receptor 2 (CRHR2) has been reported to cause bronchodilation through smooth muscle relaxation [37]. With the goal of finding bronchodilation changes in CRHR2 variants, Poon reviewed 607 individuals in the CAMP study. Initial findings revealed a statistical significance in rs255100, rs7793837, and rs2267715. Homozygotes for the minor alleles had a reduced bronchodilation response to albuterol (p ≤ 0.05) [37]. This was not the case in the adult studies where different variants were found to produce bronchodilation. Minor alleles for SNPs rs2284220, rs7793837, and rs2267716 showed the lowest albuterol response compared to their respective major alleles [37].
Multiple cohorts were involved in a GWAS and meta-analysis study of 949 African American children done by Spear to determine variations in albuterol responsiveness (Study of African Americans, Asthma, Genes and Environments, SAGE I and II; Genes-Environments and Admixture in Latino Americans, GALA I and II; African Americans from the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity, SAPPHIRE; and African Americans from the Severe Asthma Research Program, SARP). Bronchodilation was measured by changes in FEV1 before and after receiving albuterol. Identified in the study was the SNP, rs73650726, on chromosome 9q21 with the major A allele producing decreased bronchodilation in both SAGE I and II compared to the minor G allele (β = −3.8, p = 7.69 × 10−9) [38]. In the meta-analysis of SAGE I, II, and GALA II, SNPs rs7903366, rs7070958, and rs7081864 were found in the Protein Kinase CGMP-Dependent Type 1 (PRKG1) gene. This gene encodes for cGMP-dependent protein kinase which mediates bronchodilation through the nitric oxide (NO) pathway. However, these SNPs were not found in subsequent replication studies (GALA I, SAPPHIRE, and SHARP). Supporting initial findings by Padhukasahasram, Spear et al.’s additional analysis reported statistical significance of SNP, rs9551086 (p = 0.02), in the SPATA13 gene, whereas statistical significance was not reached for genes ADRB2, ADCY9, CRHR2, ARG1, THRB, CRHR2, and other SNPs of SPATA13 [38].
2.3. LABA with or without ICS and ADRB2 Variations
In cases where controller medications are needed, the 2021 GINA guidelines recommend using ICS/formoterol as opposed to SABA monotherapy [3]. As of 2010, LABA monotherapy has been contraindicated in asthma due to an increased number of adverse events and deaths [7]. Bleecker conducted a study in 2008 using salmeterol with or without fluticasone propionate to analyze the pharmacogenetic effects in individuals ages 12 and older [39]. In this randomized, double-blind, parallel study, it was reported that changes in peak expiratory flow (PEF) were similar, noninferior, and no increase in exacerbations were detected between both treatment groups of all ADRB2 genotypes at position 16 (Table 4). Further analysis among African American patients also revealed no differences in asthma control measures. Similar findings were observed by Bleecker and colleagues, where participants were assigned a budesonide and formoterol combination or fluticasone with salmeterol [40]. Improvements in baseline morning PEF and FEV1 were seen consistently across all genotypes regardless of treatment group (p < 0.001) [40]. Additional studies by Bleecker, Giubergia, and Wang found no pharmacogenetic implications in LABA response [41,42,43], unlike studies done by Palmer [44] and Zuurhout [45]. These latter two studies found that those with the Arg16Arg genotype had an increased risk of exacerbations in children taking a LABA and ICS (OR 3.40, p = 0.022 and OR 12.13, p = 0.004).

Table 4.
ICS with or without LABA and ADRB2 rs1042713 Arg16Gly (+46 A > G).
In contrast to the findings of Bleecker [39,40], Wechsler [46] examined two different adult studies: the Salmeterol or Corticosteroids (SOCS) as well as Salmeterol and ICS (SLIC). Those in the SCOS study were randomized to receive either placebo, triamcinolone, or salmeterol whereas those in SLIC were randomized to receive a combination of salmeterol and steady triamcinolone, tapered triamcinolone, or placebo. Wechsler concluded that those with Arg16 did not respond as well when given salmeterol. Although the increase in morning PEF was greater among Arg16Arg individuals in the SLIC study, levels were not sustained through the end of the trial, as seen with the Gly16Gly individuals (difference in morning PEF −36.8, p = 0.05). Those with Gly16 showed greater changes in morning PEF compared to Arg16Arg (p = 0.005) [46]. These results were comparable to the findings of a study done by Lipworth [47]. Children with persistent asthma with Arg16Arg had increased exacerbations and more school absences among those receiving salmeterol versus montelukast (difference in score −0.40, p = 0.005) [47]. Similarly, Basu [48] compared the albuterol and salmeterol response in participants ages 3 to 22. They found that the Arg16 variants were at higher risk of exacerbations (OR 1.30, p = 0.003) regardless of medication choice. Specifically, the Arg16Arg and Arg16Gly individuals had a higher risk of exacerbations compared to the Gly16Gly individuals (OR 1.74, p = 0.02) [48].
One pediatric study identified associations between Arg16 variants and exacerbations from ICS and LABA therapy. Turner evaluated 4226 children from five trials (BREATHE, PACMAN, GALA II, PASS, and PAGES) and found those with Arg16 had increased risk of exacerbations given ICS and LABA, but not in combination with leukotriene receptor antagonists (LTRAs) (OR 1.52, p = 0.0021) [49]. In addition, the OR was the highest in those administered ICS + LABA compared to SABA and ICS monotherapy, ICS + LTRA, and ICS + LABA + LTRA (OR 1.01, p = 0.95; OR 1.11, p = 0.18; OR 1.11, p = 0.52; OR 0.94, p = 0.65, respectively) [49].
A prospective case-control study done with Egyptian children found significance at ADRB2 codon 16 and 27 (p < 0.05). Pulmonary function tests were assessed using FEV1 and FEV1/FVC. At position 16, those with Gly16Gly showed a decreased response compared to those with Arg16Arg or Arg16Gly. The same decrease in response in those with Glu27Glu compared to those with Gln27Gln or Gln27Glu [26]. However, other pediatric studies could not find pharmacogenetic associations at position 27 [39,42,44].
3. Inhaled Corticosteroids (ICS)
Depending on age and severity, the GINA guidelines recommend using ICS monotherapy, or in combination as the preferred controller in addition to a rescue inhaler. ICS are commonly prescribed to treat asthma and, to an extent, COPD. Highly effective in reducing inflammation, ICS can significantly improve asthma symptoms and reduce morbidity in both children and adults. The mechanism of action involves decreasing inflammatory mediators such as mast cells, cytokines, and enzymes such as COX-2. Inhalation routes allow for minimal systemic absorption; thus, efficacy and safety are robust. However, side effects are not negligible, especially when used in children for an extended period. Osteoporosis, glaucoma, and metabolic disturbances are possible adverse events of systemic exposure (Figure 2).
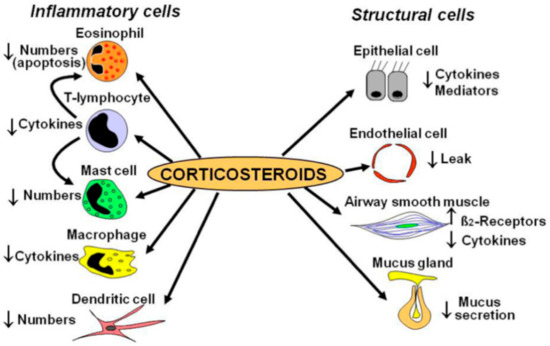
Figure 2.
ICS Mechanism of Action. Adapted from [50].
Tantisira conducted a GWAS in 2011 and identified the glucocorticoid-induced transcript 1 (GLCCI1) gene and the SNPs, rs37972 and rs37973 (Table 5) [51]. Non-Hispanic white children selected from the CAMP study were given budesonide, and the response was measured by the change in FEV1 from baseline. Although little is known about GLCCI1, those homozygote for the mutant T allele at SNP, rs37972, yielded a poorer response compared to those who were heterozygote (CT) or homozygote (CC) (OR, 2.36; 95% CI, 1.27–4.41) [51]. Additionally, linkage disequilibrium was found with the SNP, rs37973 (r2 = 0.99). Individuals that were homozygote for the wild-type A allele had a better response to budesonide compared to those homozygotes for the mutant G allele (3.2 ± 1.6% vs. 9.4 ± 1.1% increase in FEV1) [51]. Similar findings were seen in a trial conducted by Thompson et al. [52]. Included in that study were 402 European children on long-term ICS treatment (>6 months). Higher steroid doses and increased hospitalization were seen in SNP, rs37973, homozygous for the mutant allele compared to those heterozygous or homozygous wild-type [52]. Unlike Tantisira [51], Vijverberg saw no association with individuals having SNP, rs37972 and ICS response in Northern European children [53]. Additionally, Vijverberg utilized hospital visits and oral steroid use as markers for exacerbation and concluded that the mutant T allele was not associated with an increased risk when treated with budesonide [53]. In contrast, Huang et al. found associations in the GLCCI1 gene in 263 Chinese children. SNPs, rs37969 GG, rs37972 CC, and rs37973 AA produced favorable changes measured by maximal mid-expiratory flow (p ≤ 0.05) [54]. Stress-induced phosphoprotein 1 (STIP1) was also explored in this study and was shown to suppress inflammatory mediators. Although a significant association between SNP rs2236647 CC and the risk of developing asthma was found, there was no association with any SNP and ICS response [54].

Table 5.
ICS and genetic variations.
It was hypothesized that histone deacetylase I and 2 (HDAC1, HDAC2) play a positive role in airway responsiveness and inflammation. Kim studied 35 adults and 70 children with the HDAC1 SNP, rs1741981 [55]. Bronchodilation was measured as a % change in FEV1 from baseline [55]. Adults with the CC genotype produced diminished responses compared to the CT or TT genotypes when administered systemic corticosteroids (p = 0.018). The same was true for the children given inhaled corticosteroids. Those with the CC genotype had a FEV1 of 14.1% compared to either the CT or TT (19.4%, p = 0.035).
The corticotropin-release hormone (CRH) is a known mediator to stress and its corresponding receptor, corticotropin-releasing hormone receptor 1 (CRHR1), is hypothesized to have a role in ICS response [56]. In comparison to the unfavorable responses seen in the GLCCI1 variants, Tantisira observed an increase in response in those with the CRHR1 variants among three cohorts of both children and adults treated with an ICS. Response was measured by dFEV1 over a span of eight weeks in the CAMP replication study. Those with the rs242941 mutant TT genotypes displayed a change of 17.80 + 6.77 in FEV1 compared to a change of 7.57 + 1.50 in those with the wild-type CC. This was not seen in the initial adult study treated with flunisolide. SNP, rs1876828 variant, was also associated with triamcinolone responses in the adult replication study but was statistically insignificant in children [54].
Variants in the T-box transcription factor (TBX21), which encodes for transcription factor T-bet, also displayed improvements in ICS response. Children with glutamine at amino acid position 33 in place of histidine (rs2240017) displayed better PC20 [57]. PC20 is the provocative concentration resulting in a 20% drop of FEV1 post methacholine challenge. After four years, PC20 for the 33Q individuals given budesonide was measured to be 27.7 mg/mL, placing them in the non-asthmatic range [57]. Tantisira also conducted a study with a focus on the Fc fragment of IgE low affinity II receptor (FCER2) gene [58]. FCER2 encodes for CD23, causing IgE mediated responses which have been associated with severe asthmatic exacerbations. In this study, 311 Caucasian children were assigned to inhaled budesonide and observed over four years. The SNPs identified to be associated with increased IgE levels and exacerbations were: rs4996974, rs7249320, and rs28364072. Children with the SNP, rs28364072, homozygous for the mutant C allele were at a higher risk of severe exacerbations in both African American and Caucasian populations (HR 3.08 and 3.95, respectively) [58]. Koster replicated these findings in two pediatric cohorts treated with ICS. Children’s homozygotes for the variant C allele had a higher risk of severe exacerbations (OR 2.38, 95% CI 1.47–3.85, p = 0.0004), increase in hospital visits (OR 1.91, 95% CI 1.08–3.40, p = 0.03), and were more likely to use higher ICS doses (OR 2.46, 95% CI 1.38–4.39, p = 0.002 [59]. These findings support the need for earlier detection of FCER2 polymorphisms to identify those resistant to steroid treatment and provide timely anti-IgE treatment.
Located on chromosome 5q31-q32, nuclear receptor subfamily 3, group C, member 1 (NR3C1) is a protein-coding gene for glucocorticoid receptors. Keskin studied 82 children with a mean age of 9.6, given inhaled fluticasone propionate specifically evaluating the role of NR3C1 [60]. Of the 82 children, 26 had the rs41423247, GG genotype and displayed a better response compared to those with the CG or CC variations, measured by FEV1 (24.2% vs. 7.9%, p = 0.006) [60]. Stockman also conducted a study involving 734 Caucasian children receiving inhaled fluticasone propionate. Variations among cytochrome p450 (CYP) 3A4, 3A5, and 3A7, which are involved in fluticasone metabolism, were assessed for symptom control using the asthma control score. CYP3A5 and CYP3A7 were not found to have an association, however, CYP3A4 *22 children displayed better asthma control due to the reduced activity of the metabolic enzyme, leading to increased therapeutic outcomes [61]. These findings were not seen in a follow-up study of 64 children treated with beclomethasone [62]. They observed that children with CYP3A5 *3/*3, commonly found among Caucasian populations, displayed better asthma control, compared to either *1/*1 or *1/*3 variations [62]. Pharmacogenomics testing on these CYP enzymes has the potential to provide guidance on future regimens. Children with the CYP3A4 *1/*1 or *1/*3 variation may be advised to use a non-beclomethasone medication, such as budesonide, as their initial ICS. Similarly, those with the CYP3A4 *22 variation may be advised to start therapy with fluticasone.
4. Leukotriene Modifiers (LTM)
Leukotrienes are involved in inflammatory processes and bronchoconstriction, both of which are implicated in asthmatic symptoms. Leukotriene modifiers are considered second line after ICS and LABA for chronic asthma according to the GINA guidelines [3]. Administered orally, these LTMs include zileuton, montelukast, and zafirlukast which are subsequently classified into leukotriene synthesis inhibitors or leukotriene receptor antagonists. Zileuton inhibits the 5-lipoxygenase (5-LO) enzyme, halting the conversion of arachidonic acid into LTA4. Montelukast, zafirlukast, and pranlukast are cysteinyl-leukotriene receptor (CysLTR) inhibitors, antagonizing the effects of LTC4, LTD4, and LTE4 [63,64].
Similar to bronchodilators and ICS, limitations of leukotriene modifiers are highlighted by genetic variations, emphasizing inter-patient variability leading to exacerbated symptoms [65,66]. In addition, nearly half of the side effects in children taking a LTM are attributed to psychiatric disturbances such as hallucinations and agitation [64]. Genes such as arachidonate 5-lipoxygenase (ALOX5) and cysteinyl-LTs (CysLTs) previously identified and replicated in adult studies have not yet been fully confirmed in children. Identifying variants associated with positive leukotriene modifier response can further benefit children who cannot gain symptom control with ICS or beta agonists.
4.1. Genes That Affect Montelukast Response
Montelukast, favored in the pediatric community for its once-a-day dosing, is an oral medication for chronic asthma and exercise-induced bronchoconstriction (EIB). It binds to the CysT1 receptor (CysLTR1), antagonizing the effects of LTD4, and decreasing bronchoconstriction [66].
Both pharmacodynamic and pharmacokinetic studies have shown to potentially impact montelukast responsiveness in children (Table 6).

Table 6.
Montelukast and genetic variations.
Thromboxane A2 (TBXA2), an arachidonic acid derivative, binds to the thromboxane A2 receptor (TBXA2R) and causes pulmonary smooth muscle constriction and platelet aggregation. To determine an association between the TBXA2R gene and LTM response, Kim et al. conducted a study on 695 Korean children with exercise-induced bronchospasms given montelukast. They found that those with the TBXA2R +795 CC or CT variations were 2.5 times more likely to be unresponsive to treatment (p = 0.063) [67]. Responders were defined as >10% in FEV1 after montelukast treatment. In addition, those with both the TBXA2R +795 CT or CC and +924 TT variation were less likely to respond to montelukast treatment (OR 3.67, p = 0.041) [67].
In comparison, Klotsman found a statistically significant association of responsiveness in the ALOX5 variants among children 15 years of age and older. ALOX5 encodes for 5-lipoxygenase (5-LO) and is found on chromosome 10q11.21 [68]. 5-LO is involved in the conversion of arachidonic acid into LTA4 and is the rate limiting step in leukotriene synthesis (Figure 3). Two out of five variants were significantly associated with a positive response to montelukast. Those with the SNP rs4987105 TT genotype produced a mean PEF of 94.8 while those with CC genotypes were 33.7 (p = 0.01). Those with the SNP rs4986832 AA genotype produced a mean PEF of 102.4 while those with the GG genotype were 34.9 (p = 0.01) [69]. Conversely, Telleria et al. studied the promoter region of the ALOX5 gene. Five copies of the transcription factor binding sequence GGGCGG (rs59439148) were recognized as the major allele, with variants hypothesized to produce reduced activity [70]. They found greater improvements with homozygotes wild type (5/5) and heterozygotes (5/4), compared to homozygote variants (4/4) after 6 months of montelukast treatment. Those with the 4/4 repeats displayed higher rates of exacerbations, worsening FEV1, and greater need for rescue inhaler compared to those with the 5/5 or 5/4 repeats (p ≤ 0.05) [70].
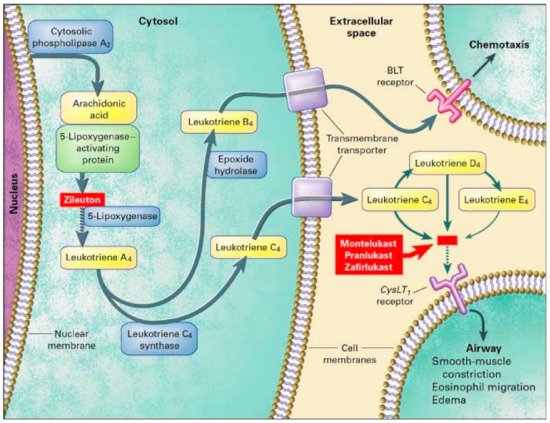
Figure 3.
Leukotriene Modifiers Mechanism of Action. Adapted from [68].
Similar to CystLTR1, cysteinyl- leukotriene (CystLT) receptor 2 (CysLTR2) have recently been found on smooth muscle cells. Although the variant function of this gene has yet to be identified, it has been associated with asthmatic treatment response. SNPs, rs912277 TC and rs912278 CC, were found to produce greater PEF (p = 0.021 and p = 0.02, respectively) in a study done by Klotsman [69]. Participants were at least 15 years of age and observed over 12 weeks. TT genotypes in both SNPs produced a mean PEF of 38.8 and 29.1, respectively. In the same study, they assessed leukotriene C4 synthase (LTC4S). LTC4S converts leukotriene A4 to leukotriene C4, increasing CysLT levels contributing to asthmatic inflammation (Figure 3). However, no association was found in montelukast treatment in either FEV1 or PEF in SNP, rs730012 [69]. Furthermore, Kang could not establish statistical significance in those with the LTC4S polymorphisms between responders and non-responders (p = 0.702) in 100 Korean asthmatic children [71]. Lee claimed that montelukast responsiveness is associated with total IgE and PC20 levels, opposed to FEV1 [72]. This study also observed CysLTR1 variations with exercise-induced bronchoconstriction (EIB). However, no correlation to montelukast responsiveness, measured by >10% increase in FEV1 post exercise challenge, was found [72]. Whelan investigated 13 children, ages 10–16 and found a positive correlation in montelukast responsiveness and LTC4S. Statistical significance in those with the LTC4S AC genotype produced a greater decrease in fraction of exhaled nitric oxide (FENO) (slope −3.13%/day), which was correlated with a decrease in airway inflammation. Those with the AA genotype did not produce any noticeable change in FENO, suggesting montelukast use in these patients may not be beneficial [73].
A study by Kang on 10 Korean asthmatic children investigated prostaglandin D2 receptor (PTGDR), a derivative of arachidonic acid implicated in eosinophilia migration [71]. Over the span of eight weeks children receiving montelukast with the rs803010 TT genotype had a greater FEV1 fall post-exercise challenge compared to those with the TC or CC genotype (63.8% vs. 36.2% p = 0.038). No difference in response was found when analyzing both PTGDR and LTC4S variants [71].
Lipworth observed 62 children with ADRB2 Arg16Arg variants and found montelukast reduced school absences and symptoms compared to those treated with salmeterol. All children also received inhaled fluticasone throughout the one-year study. It was concluded that the use of montelukast and an ICS may be more beneficial than the recommended ICS and LABA combination [47] (Table 7).

Table 7.
Montelukast Combination and ADRB2.
Organic anion transporter polypeptide 2B1 (OATP2B1) are drug transporters found in the liver, intestine, and kidney and are responsible for reuptake of various substrates. This transporter is encoded by the gene solute carrier organic anion transporter family member 2B1 (SLCO2B1). OATP2B1 SNPs rs12422149 and rs2306168 have been associated with montelukast absorption [74]. Mougey utilized the Asthma Symptoms Utility Index (ASUI) to measure drug responsiveness and found that those with the rs12422149 GG genotype demonstrated greater improvement at three months and six months compared to the those with the AG genotype [74]. Similarly, a study by Li with 50 Chinese children with an average age of 4.4 years old found lower montelukast clearance in the SLCO2B1 rs12422149 GG genotype compared to the GA and AA (0.77 ± 0.21 vs. 0.94 ± 0.26, p = 0.020) [75]. Those with the GG genotype displayed better responsiveness to montelukast due to lower drug clearance leading to increased plasma concentrations. Li also examined CYP2C8 variations, however no association was found with montelukast clearance in Chinese children [75]. These results may favor genetic testing to identify SLCO2B1, rs12422149 GG variants, which presumably should see the greatest effect from monteluakast treatment among LTM medications [76]. Those without the SLCO2B1, rs12422149 GG variants may see better efficacy on a different LTM [77].
4.2. Genes Affecting Other Leukotriene Modifier Responses
Zileuton is a 5-LO inhibitor that leads to decreased leukotriene synthesis, inflammation, and bronchoconstriction (Figure 3). Those on zileuton should monitor their ALT regularly [78]. Data on zileuton responsiveness is scarce as most studies utilized montelukast as the drug of choice. Tcheurekdjlan looked at Puerto Rican and Mexican youths with an average age of 12.3 taking a leukotriene modifier and albuterol. Bronchodilator responsiveness, measured by FEV1, was found to be greater in those taking LTM compared to those did not (p = 0.001) [79]. In addition, leukotriene A4 hydrolase (LTA4H) was investigated. LTA4H converts LTA4 to LTB4, which contributes to neutrophilic asthma characterized by severe airway obstruction. The LTA4H SNP rs2540491 minor A allele produced a change of 7–10% in FEV1 compared to the major G allele which produced a change of −0.31% in FEV1 (Table 8). These results were seen in the Puerto Rican population, but not in the Mexican population. The LTA4H SNP rs2540487 GA heterozygotes also produced a change of more than 10% in FEV1 (p < 0.001) compared to homozygotes major or minor (2.5 % change in FEV1, p = 0.180 and 0.679, respectively) [79]. Arachidonate 5-lipoxygenase activating protein (ALOX5AP) modulates downstream leukotriene synthesis along with ALOX5, converting arachidonic acid into leukotriene A4 (Figure 3). No association was found between ALOX5AP, rs10507391 and rs955196 variants with leukotriene modifier bronchodilation. Interactions between LTA4H, ALOX5AP, and their variants were also analyzed for bronchodilation. The presence of ALOX5AP, rs10507391 major A allele and LTA4H minor allele variants contributed to bronchodilation, whereas the presence of ALOX5AP, rs9551963 minor C allele and LT4AH minor allele variants contributed to bronchodilation. These findings were significant in Puerto Rican populations but not Mexican populations [79].

Table 8.
Leukotriene Modifier and Genetic Variants.
5. Conclusions
A tailored approach to asthmatic treatment may be warranted based on genetic variability. Beta-2 agonists have been the most widely studied in both adult and pediatric populations. However, results surrounding ADRB2 gene variations at both position 16 and 27 are conflicting, promoting interest into the impact of diverse patient populations. Among the ICS studies, budesonide and fluticasone have shown to be associated with conflicting treatment response. Similarly, montelukast and zileuton have shown to be associated with inconsistent treatment response among the leukotriene modifiers. GINA guidelines also recommend adding on biologics such as omalizumab (Xolair) or dupilumab (Dupixent) [80]. These biologics are indicated in children aged six and older with severe symptoms, despite optimizing therapy with high dose ICS-LABA-oral corticosteroids.
Pharmacogenetic testing in asthmatic adults has shown cost effectiveness and increased quality of life when adding a LTM to patients identified as exhibiting reduced ICS response [80]. Current guidelines recommend increasing ICS-LABA dosing to reduce exacerbations, or alternatively using a LTM as a controller therapy. Identifying children who may be genetically predisposed to ICS unresponsiveness may incline providers to initiate alternative LTM therapy earlier in the patient’s course of treatment. There are no studies to date on the effectiveness of pharmacogenomic testing for asthmatic biologics. However, future trials on the stepwise therapy model may be questioned as some children may be better suited for biologics rather than LTMs. Potential testing on certain genetic markers, such as drug transporter or CYP450 enzymes, can additionally guide clinicians when choosing initial medications among drug classes. Future studies should be conducted to evaluate the effectiveness of pharmacogenomic testing in children. Although preemptive pharmacogenomic testing for asthma is not currently recommended, due to the lack of evidence and accuracy, the hope for universal testing at an early age has the potential to eliminate prolonged medication trial and error. Unfortunately, limitations in many of these studies include the small sample size and lack of diversity. With recent discussions on race and ethnicity, there is a need for further stratification based on genetic ancestry beyond the non-Hispanic white population. Some studies utilized measurable outcomes such as PEF and FEV1, whereas others used exacerbation rates and asthma scores, which further complicates drug responsiveness. Additionally, many studies have included both adults and children. These results cannot be extrapolated specifically to children. Larger pediatric asthmatic studies [81,82] could improve confidence for the translation of genotype to phenotype, with the common goal of developing the best course of treatment for each individual child.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank the School of Pharmacy at the Massachusetts College of Pharmacy and Health Sciences for financial support of this project. C.L. would also like to thank Roseann Gammal and Hyun Kim for their invaluable insights and endless support throughout the research process.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Centers for Disease Control and Prevention. 2019 National Health Interview Survey (NHIS) Data. Available online: https://www.cdc.gov/asthma/nhis/2019/data.htm/ (accessed on 25 October 2021).
- Weiss, S.T.; Litonjua, A.A.; Lange, C.; Lazarus, R.; Liggett, S.B.; Bleecker, E.R.; Tantisira, K.G. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics 2006, 6, 311–326. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The Global Asthma Report 2018; Global Asthma Network: Auckland, New Zealand. 2018. Available online: globalasthmareport.org (accessed on 31 January 2022).
- Nurmagambetov, T.; Kuwahara, R.; Garbe, P. The Economic Burden of Asthma in the United States 2008-2013. Ann. Am. Thorac Soc. 2018, 15, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Asthma-Related Missed School Days among Children Aged 5–17 Years. Available online: https://www.cdc.gov/asthma/asthma_stats/missing_days.htm/ (accessed on 20 August 2021).
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of Asthma in Children and Adults. Front Pediatr. 2019, 18, 246. [Google Scholar] [CrossRef] [PubMed]
- Global Initiative for Asthma. Global Initiative for Asthma—GINA. Available online: https://ginasthma.org/ (accessed on 2 September 2021).
- Gustafsson, P.M.; Watson, L.; Davis, K.J.; Rabe, K.F. Poor asthma control in children: Evidence from epidemiological surveys and implications for clinical practice. Int. J. Clin. Pract. 2006, 60, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.M.; Mason, J.R.; Bird, K.A.; Kirkham, J.J.; Peak, M.; Williamson, P.R.; Nunn, A.J.; Turner, M.A.; Pirmohamed, M.; Smyth, R.L. Adverse drug reactions causing admission to a paediatric hospital. PLoS ONE 2012, 7, e50127. [Google Scholar] [CrossRef]
- Clancy, J.P.; Johnson, S.G.; Yee, S.W.; McDonagh, E.M.; Caudle, K.E.; Klein, T.E.; Cannavo, M.; Giacomini, K.M. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clin. Pharmacol. Ther. 2014, 95, 592–597. [Google Scholar] [CrossRef]
- Billington, C.K.; Penn, R.B.; Hall, I.P. β2 Agonists. Handb. Exp. Pharmacol. 2017, 237, 23–40. [Google Scholar]
- Carroll, C.L.; Stoltz, P.; Schramm, C.M.; Zucker, A.M. B2-adrenergic receptor polymorphisms affect response to treatment in children with severe asthma exacerbations. Chest 2009, 135, 1186–1192. [Google Scholar] [CrossRef]
- Ortega, V.E.; Meyers, D.A.; Bleecker, E.R. Asthma pharmacogenetics and the development of genetic profiles for personalized medicine. Pharmgenom. Pers. Med. 2015, 8, 9–22. [Google Scholar]
- Choudhry, S.; Ung, N.; Avila, P.C.; Ziv, E.; Nazario, S.; Casal, J.; Torres, A.; Gorman, J.D.; Salari, K.; Rodriguez-Santana, J.R.; et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am. J. Respir. Crit. Care. Med. 2005, 171, 563–570. [Google Scholar] [CrossRef]
- Finkelstein, Y.; Bournissen, F.G.; Hutson, J.R.; Shannon, M. Polymorphism of the ADRB2 gene and response to inhaled beta- agonists in children with asthma: A meta-analysis. J. Asthma 2009, 46, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.J.; Thomason, D.B.; Mohamed, M.H.; Eberle, L.V.; Self, T.H.; Johnson, J.A. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin. Pharmacol. Ther. 1999, 65, 519–525. [Google Scholar] [CrossRef]
- Martinez, F.D.; Graves, P.E.; Baldini, M.; Solomon, S.; Erickson, R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J. Clin. Invest 1997, 100, 3184–3188. [Google Scholar] [CrossRef] [PubMed]
- Scaparrotta, A.; Franzago, M.; Marcovecchio, M.L.; Di Pillo, S.; Chiarelli, F.; Mohn, A.; Stuppia, L. Role of THRB, ARG1, and ADRB2 Genetic Variants on Bronchodilators Response in Asthmatic Children. J. Aerosol. Med. Pulm. Drug Deliv. 2019, 32, 164–173. [Google Scholar] [CrossRef]
- Salah, K.; Morsy, S.; Atta, A. Effects of β2-adrenergic receptor polymorphisms on asthma severity and response to salbutamol in Egyptian children. Egypt J. Pediatr. Allergy Immunol. 2012, 10, 81–86. [Google Scholar]
- Israel, E.; Chinchilli, V.M.; Ford, J.G.; Boushey, H.A.; Cherniack, R.; Craig, T.J.; Deykin, A.; Fagan, J.K.; Fahy, J.V.; Fish, J.; et al. Use of regularly scheduled albuterol treatment in asthma: Genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet 2004, 364, 1505–1512. [Google Scholar] [CrossRef]
- Turner, S.W. Genetic predictors of response to therapy in childhood asthma. Mol. Diagn. Ther. 2009, 13, 127–135. [Google Scholar] [CrossRef]
- Giubergia, V.; Gravina, L.P.; Castanos, C.; Chertkoff, L.; Grenoville, M. Influence of beta2-adrenoceptor polymorphisms on the response to chronic use of albuterol in asthmatic children. Pediatr. Pulmonol. 2008, 43, 421–425. [Google Scholar] [CrossRef]
- Jovicic, N.; Babic, T.; Dragicevic, S.; Nestorovic, B.; Nikolic, A. ADRB2 gene polymorphisms and salbutamol responsiveness in Serbian children with asthma. Balk. J. Med. Genet. 2018, 21, 33–38. [Google Scholar] [CrossRef]
- Perez-Garcia, J.; Espuela-Ortiz, A.; Lorenzo-Diaz, F.; Pino-Yanes, M. Pharmacogenetics of Pediatric Asthma: Current Perspectives. Pharmgenom. Pers. Med. 2020, 13, 89–103. [Google Scholar] [CrossRef]
- Hikino, K.; Kobayashi, S.; Ota, E.; Mushiroda, T.; Urayama, K.Y.; Kobayashi, T. A meta-analysis of the influence of ADRB2 genetic polymorphisms on albuterol (salbutamol) therapy in patients with asthma. Br. J. Clin. Pharmacol. 2021, 87, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Alghobashy, A.A.; Elsharawy, S.A.; Alkholy, U.M.; Abdalmonem, N.; Abdou, M.A.; Basset, M.A.A.; Pasha, H.F. B2 adrenergic receptor gene polymorphism effect on childhood asthma severity and response to treatment. Pediatr. Res. 2018, 83, 597–605. [Google Scholar] [CrossRef]
- Elbahlawan, L.; Binaei, S.; Christensen, M.L.; Zhang, Q.; Quasney, M.W.; Dahmer, M.K. β2-Adrenergic receptor polymorphisms in African American children with status asthmaticus. Pediatr. Crit. Care Med. 2006, 7, 15–18. [Google Scholar] [CrossRef]
- The Childhood Asthma Management Program (CAMP): Design, rationale, and methods. Control Clin. Trials. 1999, 20, 91–120. [CrossRef]
- Peters, S.P.; Anthonisen, N.; Castro, M.; Holbrook, J.T.; Irvin, C.G.; Smith, L.J.; Wise, R.A. Randomized comparison of strategies for reducing treatment in mild persistent asthma. N. Engl. J. Med. 2007, 356, 2027–2039. [Google Scholar] [PubMed]
- American Lung Association Asthma Clinical Research Centers. Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am. J. Respir. Crit. Care Med. 2007, 175, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Litonjua, A.A.; Lasky-Su, J.; Schneiter, K.; Tantisira, K.G.; Lazarus, R.; Klanderman, B.; Lima, J.J.; Irvin, C.G.; Peters, S.P.; Hanrahan, J.P.; et al. ARG1 is a novel bronchodilator response gene: Screening and replication in four asthma cohorts. Am. J. Respir. Crit. Care Med. 2008, 178, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.L.; Du, R.; Lasky-Su, J.; Klanderman, B.J.; Partch, A.B.; Peters, S.P.; Irvin, C.G.; Hanrahan, J.P.; Lima, J.J.; Blake, K.V.; et al. A polymorphism in the thyroid hormone receptor gene is associated with bronchodilator response in asthmatics. Pharm. J. 2013, 13, 130–136. [Google Scholar] [CrossRef]
- Duan, Q.L.; Lasky-Su, J.; Himes, B.E.; Qiu, W.; Litonjua, A.A.; Damask, A.; Lazarus, R.; Klanderman, B.; Irvin, C.G.; Peters, S.P.; et al. A genome-wide association study of bronchodilator response in asthmatics. Pharm. J. 2014, 14, 41–47. [Google Scholar] [CrossRef][Green Version]
- Padhukasahasram, B.; Yang, J.J.; Levin, A.M.; Yang, M.; Burchard, E.G.; Kumar, R.; Kwok, P.Y.; Seibold, M.A.; Lanfear, D.E.; Williams, L.K. Gene-based association identifies SPATA13-AS1 as a pharmacogenomic predictor of inhaled short-acting beta-agonist response in multiple population groups. Pharm. J. 2014, 14, 365–371. [Google Scholar] [CrossRef][Green Version]
- Himes, B.E.; Jiang, X.; Hu, R.; Wu, A.C.; Lasky-Su, J.A.; Klanderman, B.J.; Ziniti, J.; Senter-Sylvia, J.; Lima, J.J.; Irvin, C.G.; et al. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 2012, 8, e1002824. [Google Scholar] [CrossRef] [PubMed]
- Israel, E.; Lasky-Su, J.; Markezich, A.; Damask, A.; Szefler, S.J.; Schuemann, B.; Klanderman, B.; Sylvia, J.; Kazani, S.; Wu, R.; et al. Genome-wide association study of short-acting β2-agonists. A novel genome-wide significant locus on chromosome 2 near ASB3. Am. J. Respir. Crit. Care Med. 2015, 191, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.H.; Tantisira, K.G.; Litonjua, A.A.; Lazarus, R.; Xu, J.; Lasky-Su, J.; Lima, J.J.; Irvin, C.G.; Hanrahan, J.P.; Lange, C.; et al. Association of corticotropin-releasing hormone receptor-2 genetic variants with acute bronchodilator response in asthma. Pharmacogenet. Genom. 2008, 18, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Spear, M.L.; Hu, D.; Pino-Yanes, M.; Huntsman, S.; Eng, C.; Levin, A.M.; Ortega, V.E.; White, M.J.; McGarry, M.E.; Thakur, N.; et al. A genome-wide association and admixture mapping study of bronchodilator drug response in African Americans with asthma. Pharm. J. 2019, 19, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, E.R.; Nelson, H.S.; Kraft, M.; Corren, J.; Meyers, D.A.; Yancey, S.W.; Anderson, W.H.; Emmett, A.H.; Ortega, H.G. Beta2-receptor polymorphisms in patients receiving salmeterol with or without fluticasone propionate. Am. J. Respir. Crit. Care Med. 2010, 181, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, E.R.; Postma, D.S.; Lawrance, R.M.; Meyers, D.A.; Ambrose, H.J.; Goldman, M. Effect of ADRB2 polymorphisms on response to long-acting b2-agonist therapy: A pharmacogenetic analysis of two randomised studies. Lancet 2007, 370, 2118–2125. [Google Scholar] [CrossRef]
- Bleecker, E.R.; Yancey, S.W.; Baitinger, L.A.; Edwards, L.D.; Klotsman, M.; Anderson, W.H.; Dorinsky, P.M. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J. Allergy Clin. Immunol. 2006, 118, 809–816. [Google Scholar] [CrossRef]
- Giubergia, V.; Gravina, L.; Castanos, C.; Chertkoff, L. Influence of beta(2)-adrenergic receptor polymorphisms on asthma exacerbation in children with severe asthma regularly receiving salmeterol. Ann. Allergy Asthma Immunol. 2013, 110, 156–160. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Liu, R.; He, J.; Wu, D.; Wang, Y.; Zhang, J. ADRB2 Arg16Gly polymorphism and pulmonary function response of inhaled corticosteroids plus long-acting beta agonists for asthma treatment: A systematic review and meta-analysis. Can. Respir. J. 2018, 5712805. [Google Scholar] [CrossRef]
- Palmer, C.N.; Lipworth, B.J.; Lee, S.; Ismail, T.; Macgregor, D.F.; Mukhopadhyay, S. Arginine-16 beta2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. Thorax 2006, 61, 940–944. [Google Scholar] [CrossRef]
- Zuurhout, M.J.; Vijverberg, S.J.; Raaijmakers, J.A.; Koenderman, L.; Postma, D.S.; Koppelman, G.H.; Maitland-van der Zee, A.H. Arg16 ADRB2 genotype increases the risk of asthma exacerbation in children with a reported use of long-acting β2-agonists: Results of the PACMAN cohort. Pharmacogenomics 2013, 14, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Wechsler, M.E.; Lehman, E.; Lazarus, S.C.; Lemanske, R.F.; Boushey, H.A., Jr.; Deykin, A.; Fahy, J.V.; Sorkness, C.A.; Chinchilli, V.M.; Craig, T.J.; et al. Beta-Adrenergic receptor polymorphisms and response to salmeterol. Am. J. Respir. Crit. Care Med. 2006, 173, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Lipworth, B.J.; Basu, K.; Donald, H.P.; Tavendale, R.; Macgregor, D.F.; Ogston, S.A.; Palmer, C.N.; Mukhopadhyay, S. Tailored second-line therapy in asthmatic children with the Arg(16) genotype. Clin. Sci. (Lond.). 2013, 124, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Basu, K.; Palmer, C.N.; Tavendale, R.; Lipworth, B.J.; Mukhopadhyay, S. Adrenergic beta2- receptor genotype predisposes to exacerbations in steroid-treated asthmatic patients taking frequent albuterol or salmeterol. J. Allergy Clin. Immunol. 2009, 124, 1188–1194.e3. [Google Scholar] [CrossRef]
- Turner, S.; Francis, B.; Vijverberg, S.; Pino-Yanes, M.; Maitland-van der Zee, A.H.; Basu, K.; Bignell, L.; Mukhopadhyay, S.; Tavendale, R.; Palmer, C.; et al. Pharmacogenomics in Childhood Asthma Consortium. Childhood asthma exacerbations and the Arg16 β2-receptor polymorphism: A meta-analysis stratified by treatment. J Allergy Clin. Immunol. 2016, 138, 107–113.e5. [Google Scholar] [CrossRef]
- Barnes, P.J. Inhaled Corticosteroids. Pharmaceuticals 2010, 3, 514–540. [Google Scholar] [CrossRef]
- Tantisira, K.G.; Lasky-Su, J.; Harada, M.; Murphy, A.; Litonjua, A.A.; Himes, B.E.; Lange, C.; Lazarus, R.; Sylvia, J.; Klanderman, B.; et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N. Engl. J. Med. 2011, 365, 1173–1183. [Google Scholar] [CrossRef]
- Thompson, B.; Hawcutt, D.; Carr, D.; Jorgensen, A.; Smyth, R.; Pirmohamed, M. S31 variation at GLC1CI1: Association with increased steroid dose but not adrenal suppression in asthmatic children. Thorax 2012, 67 (Suppl. 2), A17. [Google Scholar] [CrossRef][Green Version]
- Vijverberg, S.J.; Tavendale, R.; Leusink, M.; Koenderman, L.; Raaijmakers, J.A.; Postma, D.S.; Koppelman, G.H.; Turner, S.W.; Mukhopadhyay, S.; Palmer, C.N.; et al. Pharmacogenetic analysis of GLCCI1 in three north European pediatric asthma populations with a reported use of inhaled corticosteroids. Pharmacogenomics 2014, 15, 799–806. [Google Scholar] [CrossRef]
- Huang, J.; Hu, X.; Zheng, X.; Kuang, J.; Liu, C.; Wang, X.; Tang, Y. Effects of STIP1 and GLCCI1 polymorphisms on the risk of childhood asthma and inhaled corticosteroid response in Chinese asthmatic children. BMC Pulm. Med. 2020, 20, 303. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, S.H.; Kim, Y.K.; Hong, S.J.; Min, K.U.; Cho, S.H.; Park, H.W. A polymorphism in the histone deacetylase 1 gene is associated with the response to corticosteroids in asthmatics. Korean J. Intern. Med. 2013, 28, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Tantisira, K.G.; Lake, S.; Silverman, E.S.; Palmer, L.J.; Lazarus, R.; Silverman, E.K.; Liggett, S.B.; Gelfand, E.W.; Rosenwasser, L.J.; Richter, B.; et al. Corticosteroid pharmacogenetics: Association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 2004, 13, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Tantisira, K.G.; Hwang, E.S.; Raby, B.A.; Silverman, E.S.; Lake, S.L.; Richter, B.G.; Peng, S.L.; Drazen, J.M.; Glimcher, L.H.; Weiss, S.T. TBX21: A functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc. Natl. Acad. Sci. USA 2004, 101, 18099–18104. [Google Scholar] [CrossRef] [PubMed]
- Tantisira, K.G.; Silverman, E.S.; Mariani, T.J.; Xu, J.; Richter, B.G.; Klanderman, B.J.; Litonjua, A.A.; Lazarus, R.; Rosenwasser, L.J.; Fuhlbrigge, A.L.; et al. FCER2: A pharmacogenetic basis for severe exacerbations in children with asthma. J. Allergy. Clin. Immunol. 2007, 120, 1285–1291. [Google Scholar] [CrossRef]
- Koster, E.S.; Maitland-van der Zee, A.H.; Tavendale, R.; Mukhopadhyay, S.; Vijverberg, S.J.; Raaijmakers, J.A.; Palmer, C.N. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy 2011, 66, 1546–1552. [Google Scholar] [CrossRef]
- Keskin, O.; Uluca, Ü.; Birben, E.; Coşkun, Y.; Ozkars, M.Y.; Keskin, M.; Kucukosmanoglu, E.; Kalayci, O. Genetic associations of the response to inhaled corticosteroids in children during an asthma exacerbation. Pediatr. Allergy Immunol. 2016, 27, 507–513. [Google Scholar] [CrossRef]
- Stockmann, C.; Fassl, B.; Gaedigk, R.; Nkoy, F.; Uchida, D.A.; Monson, S.; Reilly, C.A.; Leeder, J.S.; Yost, G.S.; Ward, R.M. Fluticasone propionate pharmacogenetics: CYP3A4*22 polymorphism and pediatric asthma control. J. Pediatr. 2013, 162, 1227–1227, 1227.e1–2. [Google Scholar] [CrossRef]
- Stockmann, C.; Reilly, C.A.; Fassl, B.; Gaedigk, R.; Nkoy, F.; Stone, B.; Roberts, J.K.; Uchida, D.A.; Leeder, J.S.; Sherwin, C.M.; et al. Effect of CYP3A5*3 on asthma control among children treated with inhaled beclomethasone. J. Allergy Clin. Immunol. 2015, 136, 505–507. [Google Scholar] [CrossRef]
- Lima, J.J. Treatment heterogeneity in asthma: Genetics of response to leukotriene modifiers. Mol. Diagn. Ther. 2007, 11, 97–104. [Google Scholar] [CrossRef]
- Choi, J. Leukotriene Receptor Antagonists. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554445/ (accessed on 5 November 2021).
- Drazen, J.M.; Israel, E.; O’Byrne, P.M. Treatment of asthma with drugs modifying the leukotriene pathway. N. Engl. J. Med. 1999, 340, 197–206. [Google Scholar] [CrossRef]
- Bäck, M. Functional characteristics of cysteinyl-leukotriene receptor subtypes. Life Sci. 2002, 71, 611–622. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.Y.; Kim, H.B.; Jin, H.S.; Yu, J.H.; Kim, B.J.; Kim, B.S.; Kang, M.J.; Jang, S.O.; Hong, S.J. TBXA2R gene polymorphism and responsiveness to leukotriene receptor antagonist in children with asthma. Clin. Exp. Allergy 2008, 38, 51–59. [Google Scholar] [CrossRef]
- Mougey, E.; Lang, J.E.; Allayee, H.; Teague, W.G.; Dozor, A.J.; Wise, R.A.; Lima, J.J. ALOX5 polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin. Exp. Allergy 2013, 43, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Klotsman, M.; York, T.P.; Pillai, S.G.; Vargas-Irwin, C.; Sharma, S.S.; van den Oord, E.J.; Anderson, W.H. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet. Genom. 2007, 17, 189–196. [Google Scholar] [CrossRef]
- Telleria, J.J.; Blanco-Quiros, A.; Varillas, D.; Armentia, A.; Fernandez-Carvajal, I.; Jesus Alonso, M.; Diez, I. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir. Med. 2008, 102, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Kwon, J.W.; Kim, B.J.; Yu, J.; Choi, W.A.; Shin, Y.J.; Hong, S.J. Polymorphisms of the PTGDR and LTC4S influence responsiveness to leukotriene receptor antagonists in Korean children with asthma. J. Hum. Genet. 2011, 56, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, H.B.; Kim, J.H.; Kim, B.S.; Kang, M.J.; Jang, S.O.; Seo, H.J.; Hong, S.J. Responsiveness to montelukast is associated with bronchial hyperresponsiveness and total immunoglobulin E but not polymorphisms in the leukotriene C4 synthase and cysteinyl leukotriene receptor 1 genes in Korean children with exercise-induced asthma (EIA). Clin. Exp. Allergy 2007, 37, 1487–1493. [Google Scholar] [CrossRef]
- Whelan, G.J.; Blake, K.; Kissoon, N.; Duckworth, L.J.; Wang, J.; Sylvester, J.E.; Lima, J.J. Effect of montelukast on time-course of exhaled nitric oxide in asthma: Influence of LTC4 synthase A(-444)C polymorphism. Pediatr Pulmonol. 2003, 36, 413–420. [Google Scholar] [CrossRef]
- Mougey, E.B.; Feng, H.; Castro, M.; Irvin, C.G.; Lima, J.J. Absorption of montelukast is transporter mediated: A common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet. Genom. 2009, 19, 129–138. [Google Scholar] [CrossRef]
- Li, Q.; Wang, K.; Shi, H.Y.; Wu, Y.E.; Zhou, Y.; Kan, M.; Zheng, Y.; Hao, G.X.; Yang, X.M.; Yang, Y.L. Developmental Pharmacogenetics of SLCO2B1 on Montelukast Pharmacokinetics in Chinese Children. Drug Des. Devel. Ther. 2019, 13, 4405–4411. [Google Scholar] [CrossRef]
- Karonen, T.; Neuvonen, P.J.; Backman, J.T. CYP2C8 but not CYP3A4 is important in the pharmacokinetics of montelukast. Br. J. Clin. Pharmacol. 2012, 73, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Aquilante, C.L.; Niemi, M.; Gong, L.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 8. Pharmacogenet. Genom. 2013, 23, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Bouchette, D.; Preuss, C.V. Zileuton. Available online: https://www.ncbi.nlm.nih.gov/books/NBK448202/ (accessed on 21 August 2021).
- Tcheurekdjian, H.; Via, M.; De Giacomo, A.; Corvol, H.; Eng, C.; Thyne, S.; Chapela, R.; Rodriguez-Cintron, W.; Rodriguez-Santana, J.R.; Avila, P.C.; et al. Genetics of Asthma in Latino Americans Study. Genetics of asthma in Latino Americans study. ALOX5AP and LTA4H polymorphisms modify augmentation of bronchodilator responsiveness by leukotriene modifiers in Latinos. J. Allergy Clin. Immunol. 2010, 126, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.C.; Gay, C.; Rhett, M.D.; Stout, N.; Weiss, S.T.; Fuhlbrigge, A.L. Pharmacogenomic test that predicts response to inhaled corticosteroids in adults with asthma likely to be cost-saving. Pharmacogenomics 2015, 16, 591–600. [Google Scholar] [CrossRef]
- Devonshire, A.L.; Rajesh, K. Pediatric asthma: Principles and treatment. Allergy Asthma Proc. 2019, 40, 389–392. [Google Scholar] [CrossRef]
- Vitale, C.; Maglio, A.; Pelaia, C.; Vatrella, A. Long-term treatment in pediatric asthma: An update on chemical pharmacotherapy. Expert Opin. Pharmacother. 2017, 18, 667–676. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).