Impact of hMLH1 −93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) Polymorphisms on Colorectal Cancer Susceptibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
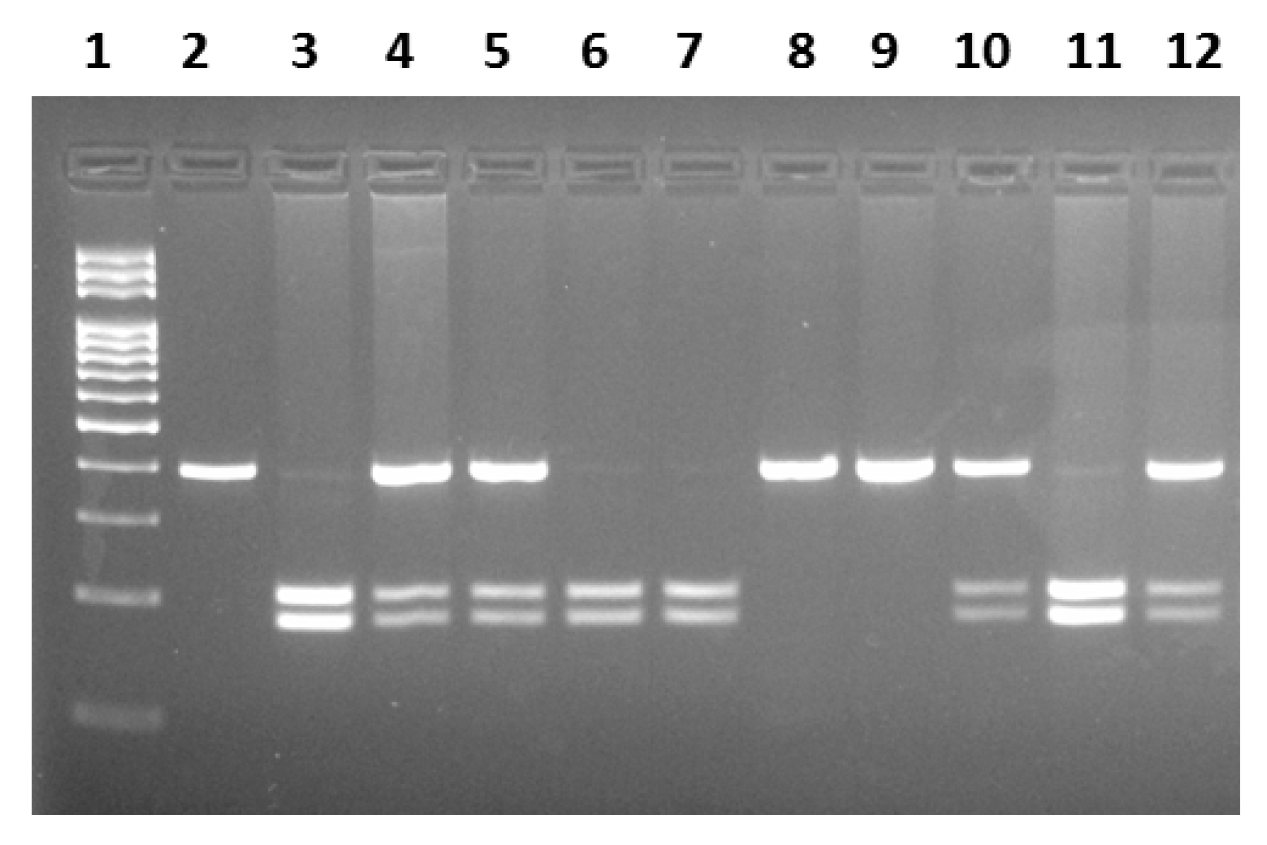
2.2. Genotyping
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | Colorectal cancer |
| SNP | Single-nucleotide polymorphism |
| MMR | Mismatch repair |
| GWAS | Genome-wide association studies |
| PCR | Polymerase chain reaction |
| RFLP | Restriction fragment length polymorphism |
| OR | Odds ratios |
| CI | Confidence interval |
| HNPCC | Hereditary nonpolyposis colorectal cancer |
References
- Matsuda, T.; Fujimoto, A.; Igarashi, Y. Colorectal Cancer: Epidemiology, Risk Factors, and Public Health Strategies. Digestion 2025, 106, 91–99. [Google Scholar] [CrossRef]
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Rustgi, A.K. The Hereditary Nonpolyposis Colorectal Cancer Syndrome: Genetics and Clinical Implications. Ann. Intern. Med. 2003, 138, 560–570. [Google Scholar] [CrossRef]
- De’ Angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’ Angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. Atenei Parmensis 2018, 89, 97–101. [Google Scholar] [CrossRef]
- dos Santos, I.B.; da Costa, A.C.A.; Gellen, L.P.A.; Sales, L.L.S.; Monte, N.; de Moraes, F.C.A.; Santo, M.O.M.; Rodrigues, J.C.G.; de Assumpção, P.P.; Guerreiro, J.F.; et al. Identification of genomic variants associated with colorectal cancer heredity in indigenous populations of the Amazon. Sci. Rep. 2025, 15, 14616. [Google Scholar] [CrossRef] [PubMed]
- Söreide, K.; Janssen, E.A.M.; Söiland, H.; Körner, H.; Baak, J.P.A. Microsatellite instability in colorectal cancer. Br. J. Surg. 2006, 93, 395–406. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite Instability in Colorectal Cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer—The stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.-M.; Yang, W.-Z.; Xu, G.-H.; Bai, P.; Qin, H.-J.; Zhang, L.-S.; Zhai, X.-D.; Tang, M.; Deng, W.; Zhang, L.; et al. The association between MLH1 -93 G>A polymorphism of DNA mismatch repair and cancer susceptibility: A meta-analysis. Mutagenesis 2011, 26, 667–673. [Google Scholar] [CrossRef]
- Zare, M.; Jafari-Nedooshan, J.; Jafari, M.; Neamatzadeh, H.; Abolbaghaei, S.M.; Foroughi, E.; Nasiri, R.; Zare-Shehneh, M. Relevance of hMLH1 -93G>A, 655A>G and 1151T>A polymorphisms with colorectal cancer susceptibility: A meta-analysis based on 38 case-control studies. Rev. Assoc. Méd. Bras. 2018, 64, 942–951. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Sima, L.; Shi, L.; Wang, Z.; Ni, C.; Zhang, Z.; Wang, M. Association between MLH1 -93G>A Polymorphism and Risk of Colorectal Cancer. PLoS ONE 2012, 7, e50449. [Google Scholar] [CrossRef][Green Version]
- He, Y.; Xu, X.; Chen, H.; Wang, J.; Xiong, W.; Xu, Y.; Liu, J. The hMLH1 promoter polymorphisms and cancer susceptibility in Asian populations: A meta-analysis. Gene 2013, 523, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 2154. [Google Scholar] [CrossRef]
- Pardini, B.; Corrado, A.; Paolicchi, E.; Cugliari, G.; Berndt, S.I.; Bezieau, S.; Bien, S.A.; Brenner, H.; Caan, B.J.; Campbell, P.T.; et al. DNA repair and cancer in colon and rectum: Novel players in genetic susceptibility. Int. J. Cancer 2020, 146, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Koshy, L.; Anju, A.L.; Harikrishnan, S.; Kutty, V.R.; Jissa, V.T.; Kurikesu, I.; Jayachandran, P.; Nair, A.J.; Gangaprasad, A.; Nair, G.M.; et al. Evaluating genomic DNA extraction methods from human whole blood using endpoint and real-time PCR assays. Mol. Biol. Rep. 2017, 44, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef]
- Mik, M.; Dziki, L.; Malinowska, K.; Trzcinski, R.; Majsterek, I.; Dziki, A. Polymorphism of MSH2 Gly322Asp and MLH1-93G>A in non-familial colon cancer—A case-controlled study. Arch. Med. Sci. 2017, 6, 1295–1302. [Google Scholar] [CrossRef]
- Wang, Y.; Li, G.; Hu, F.; Bi, H.; Wu, Z.; Zhao, X.; Li, Y.; Li, S.; Li, D.; Cui, B.; et al. The prognostic significance of polymorphisms in hMLH1/hMSH2 for colorectal cancer. Med. Oncol. 2014, 31, 975. [Google Scholar] [CrossRef]
- Li, G.; Hu, F.; Yuan, F.; Fan, J.; Yu, Z.; Wu, Z.; Zhao, X.; Li, Y.; Li, S.; Rong, J.; et al. Intronic and promoter polymorphisms of hMLH1/hMSH2 and colorectal cancer risk in Heilongjiang Province of China. J. Cancer Res. Clin. Oncol. 2015, 141, 1393–1404. [Google Scholar] [CrossRef]
- Zhang, L.-L.; Tang, X.-J.; Wang, X.-Y.; Zhu, Y.-W.; Peng, X.-B.; Gong, L. A promoter polymorphism in the hMLH1 gene (-93G/A) associated with sporadic colorectal cancer. Oncol. Lett. 2016, 12, 4035–4040. [Google Scholar] [CrossRef][Green Version]
- Chen, H.; Shen, Z.; Hu, Y.; Xiao, Q.; Bei, D.; Shen, X.; Ding, K. Association between MutL homolog 1 polymorphisms and the risk of colorectal cancer: A meta-analysis. J. Cancer Res. Clin. Oncol. 2015, 141, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Oztas, E.; Cilingir, U.; Akyuz, A.; Yanar, H.T.; Ozhan, G. MLH1 -93G>A and I219V polymorphisms are susceptible to increased risk of sporadic colorectal cancer in a Turkish population. Istanb. J. Pharm. 2017, 47, 63–67. [Google Scholar] [CrossRef]
- Nizam, Z.M.; Abdul Aziz, A.A.; Kaur, G.; Abu Hassan, M.R.; Mohd Sidek, A.S.; Lee, Y.Y.; Mazuwin, M.; Ankathil, R. Contribution of the MLH1 -93G>A Promoter Polymorphism in Modulating Susceptibility Risk in Malaysian Colorectal Cancer Patients. Asian Pac. J. Cancer Prev. 2013, 14, 619–624. [Google Scholar] [CrossRef]
- Allan, J.M.; Shorto, J.; Adlard, J.; Bury, J.; Coggins, R.; George, R.; Katory, M.; Quirke, P.; Richman, S.; Scott, D.; et al. MLH1 -93G>A promoter polymorphism and risk of mismatch repair deficient colorectal cancer. Int. J. Cancer 2008, 123, 2456–2459. [Google Scholar] [CrossRef] [PubMed]
- Pongsavee, M.; Wisuwan, K.; Pongsavee, K. MLH1 rs1800734 Pathogenic Variant among Patients with Colorectal Cancer in the Lower Northeastern Region of Thailand. Asian Pac. J. Cancer Prev. 2023, 24, 2911–2916. [Google Scholar] [CrossRef]
- Liu, T.; Stathopoulos, P.; Lindblom, P.; Rubio, C.; Wasteson Arve, B.; Iselius, L.; Holmberg, E.; Grönberg, H.; Lindblom, A. MSH2 codon 322 Gly to Asp seems not to confer an increased risk for colorectal cancer susceptibility. Eur. J. Cancer 1998, 34, 1981. [Google Scholar] [CrossRef]
- Romanowicz, H.; Bieńkiewicz, J.; Szaflik, T.; Malinowski, J.; Smolarz, B. Association between Gly322Asp polymorphism of hMSH2 (1032G>A, rs4987188) and endometrial cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 2199–2204. [Google Scholar]
- Smolarz, B.; Makowska, M.; Samulak, D.; Michalska, M.M.; Romanowicz, H. Gly322Asp and Asn127Ser single nucleotide polymorphisms (SNPs) of hMSH2 mismatch repair gene and the risk of triple-negative breast cancer in Polish women. Fam. Cancer 2015, 14, 81–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Campbell, P.T.; Curtin, K.; Ulrich, C.M.; Samowitz, W.S.; Bigler, J.; Velicer, C.M.; Caan, B.; Potter, J.D.; Slattery, M.L. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut 2009, 58, 661–667. [Google Scholar] [CrossRef]
- Samowitz, W.S.; Curtin, K.; Wolff, R.K.; Albertsen, H.; Sweeney, C.; Caan, B.J.; Ulrich, C.M.; Potter, J.D.; Slattery, M.L. The MLH1 -93 G>A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes Chromosomes Cancer 2008, 47, 835–844. [Google Scholar] [CrossRef]
- Whiffin, N.; Broderick, P.; Lubbe, S.J.; Pittman, A.M.; Penegar, S.; Chandler, I.; Houlston, R.S. MLH1-93G>A is a risk factor for MSI colorectal cancer. Carcinogenesis 2011, 32, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
| Patients n = 134 (%) | Controls n = 137 (%) | p Value | |
|---|---|---|---|
| Gender Male Female | 76 (56.7) 58 (43.3) | 64 (43.3) 73 (56.7) | 0.256 |
| Age Average Mean | 25–85 60 ± 10.2 | 32–82 60 ± 11.3 | |
| Grade G1 G2 G3 | 11 (8.2) 90 (67.1) 33 (24.7) | ||
| Stage T I T II T III T IV | 3 (2.1) 12 (9.3) 109 (81.5) 10 (7.1) | ||
| Smoking Smokers Non-smokers Unknown | 44 (34.3) 82 (60) 8 (5.7) | 55 (33.5) 98 (59.8) 11 (6.7) | 0.942 |
| Alcohol consumption User Non-user Unknown | 36 (32.1) 90 (62.2) 8 (5.7) | 59 (36) 93 (56.6) 12 (7.4) | 0.410 |
| Genotypes | Patients n = 134 (%) | Controls n = 137 (%) | OR (95%CI) | p Value |
|---|---|---|---|---|
| GG GA AA Dominant GG GA + AA Recessive GG + GA AA | 21 (15.7) 43 (32.1) 70 (52.2) 21 (15.7) 113 (84.3) 64 (47.8) 70 (52.2) | 23 (16.8) 62 (45.2) 52 (38) 23 (16.8) 114 (83.2) 85 (62) 52 (38) | 1 0.760 (0.374–1.542) 1.474 (0.738–2.945) 1 0.921 (0.569–2.070) 1 1.788 (1.102–2.900) | - 0.446 0.270 - 0.803 - 0.018 |
| Allele G A | 85 (53.1) 183 (46.9) | 108 (56.1) 166 (43.9) | 1 1.400 (0.984–1.995) | - 0.062 |
| Males | Patients n = 76 (%) | Controls n = 64 (%) | OR (95%CI) | p Value |
|---|---|---|---|---|
| Genotypes GG GA AA | 14 (18.4) 25 (32.9) 37 (48.7) | 10 (15.6) 28 (43.8) 26 (40.6) | 1 0.638 (0.241–1.690) 1.016 (0.392–2.639) | - 0.364 0.973 |
| Female | n = 58 (%) | n = 73 (%) | ||
| Genotypes GG GA AA | 7 (12.1) 18 (31) 33 (56.9) | 13 (17.8) 34 (46.6) 26 (35.6) | 1 0.983 (0.333–2.901) 2.357 (0.823–6.755) | - 0.976 0.110 |
| Age ≤60 | Patients n = 48 (%) | Controls n = 96 (%) | OR (95%CI) | p Value |
|---|---|---|---|---|
| GG GA AA | 6 (12.5) 16 (33.3) 26 (54.2) | 17 (39.2) 44 (38.2) 35 (22.6) | 1 1.030 (0.346–3.072) 2.105 (0.729–6.075) | - 0.957 0.169 |
| >60 | n = 86 (%) | n = 41 (%) | ||
| GG GA AA | 15 (17.5) 27 (31.4) 44 (51.1) | 6 (26.3) 18 (47.4) 17 (26.3) | 1 0.600 (0.196–1.837) 1.035 (0.345–3.110) | - 0.370 0.951 |
| Genotypes | Smokers n = 44 (%) | Non-Smokers n = 82 (%) | OR (95%CI) | p Value |
|---|---|---|---|---|
| GG GA AA | 7 (16) 13 (29.5) 24 (54.5) | 12 (14.6) 27 (32.9) 43 (52.5) | 1 0.825 (0.263–2.589) 0.957 (0.332–2.755) | - 0.742 0.935 |
| Alcohol consumer n = 36 (%) | Non-drinkers n = 90 (%) | |||
| GG GA AA | 4 (11.1) 10 (27.8) 22 (61.1) | 15 (16.7) 30 (33.3) 45 (50) | 1 1.250 (0.336–4.655) 1.833 (0.544–6.179) | - 0.739 0.328 |
| GG, (%) | GA, (%) | AA, (%) | p Value | |
|---|---|---|---|---|
| Tumor grade G 1 G 2 G 3 | 6 (54.5) 38 (42.2) 11 (33.3) | 4 (36.4) 34 (37.8) 18 (54.5) | 1 (9.1) 18 (20) 4 (12.2) | 0.277 |
| Tumor stage T I T II T III T IV | 1 (33.3) 6 (50) 45 (41.3) 4 (40) | 1 (33.3) 4 (33.3) 51 (46.8) 4 (40) | 1 (33.3) 2 (16.7) 13 (11.9) 2 (20) | 0.033 |
| Genotypes | Patients n = 134 (%) | Controls n = 137 (%) | OR (95%CI) | p Value |
|---|---|---|---|---|
| GG GA AA Dominant GG GA + AA Recessive GG + GA AA | 110 (82) 23 (17.2) 1 (0.8) 110 (82.1) 24 (17.9) 133 (99.2) 1 (0.8) | 122 (89.1) 13 (9.5) 2 (1.4) 122 (89.1) 15 (10,9) 135 (98.6) 2 (1.4) | 1 1.962 (0.948–4.061) 0.555 (0.050–6.201) 1 1.775 (0.886–3.555) 1 0.508 (0.045–5.664) | - 0.066 0.627 - 0.103 - 0.508 |
| Allele G A | 243 (90.7) 25 (9.3) | 257 (93.8) 17 (6.2) | 1 1.555 (0.820–2.951) | - 0.174 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayramov, B.; Karimova, N.; Mehdiyeva, N.; Valiyeva, H.; Karimova, R.; Shirinov, R.; Aslanov, H.; Safarzade, Z.; Isayev, O.; Bayramov, N. Impact of hMLH1 −93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) Polymorphisms on Colorectal Cancer Susceptibility. J. Mol. Pathol. 2025, 6, 15. https://doi.org/10.3390/jmp6030015
Bayramov B, Karimova N, Mehdiyeva N, Valiyeva H, Karimova R, Shirinov R, Aslanov H, Safarzade Z, Isayev O, Bayramov N. Impact of hMLH1 −93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) Polymorphisms on Colorectal Cancer Susceptibility. Journal of Molecular Pathology. 2025; 6(3):15. https://doi.org/10.3390/jmp6030015
Chicago/Turabian StyleBayramov, Bayram, Nigar Karimova, Nigar Mehdiyeva, Hagigat Valiyeva, Rena Karimova, Royal Shirinov, Hazi Aslanov, Zumrud Safarzade, Orkhan Isayev, and Nuru Bayramov. 2025. "Impact of hMLH1 −93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) Polymorphisms on Colorectal Cancer Susceptibility" Journal of Molecular Pathology 6, no. 3: 15. https://doi.org/10.3390/jmp6030015
APA StyleBayramov, B., Karimova, N., Mehdiyeva, N., Valiyeva, H., Karimova, R., Shirinov, R., Aslanov, H., Safarzade, Z., Isayev, O., & Bayramov, N. (2025). Impact of hMLH1 −93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) Polymorphisms on Colorectal Cancer Susceptibility. Journal of Molecular Pathology, 6(3), 15. https://doi.org/10.3390/jmp6030015







