Abstract
Respiratory viruses are the causative agents responsible for seasonal epidemics and occasional pandemic outbreaks and are a leading cause of death worldwide. Type I interferon (IFNα/β) signaling in the lung epithelial cells plays a major role in the innate immunity to respiratory viruses. Gene signatures are a set of differentially expressed genes in a particular disease or condition and are used to diagnose, monitor, and predict disease progression. These signatures can be used to identify regulatory modules and gene regulatory networks (GRNs) in mammalian signal transduction pathways. Considerable progress has been made in the identification of type I interferon-regulated gene signatures in the host response to respiratory viruses, including antiviral, immunomodulatory, apoptosis, and transcription factor signatures. Respiratory virus infections and host defenses require a dramatic change in the metabolic flux of macromolecules involved in nucleotide, lipid, and protein metabolism. The profiling of IFN-stimulated metabolic genes induced in the host response to several respiratory viruses led to the identification of a common gene signature in human lung epithelial cells and in the lungs of mouse models of respiratory virus infection. The regulation of the metabolic gene signature was correlated with the induction of IFN-beta (IFN-β) and IFN-inducible transcription factors at the RNA level in lung epithelial cells. Furthermore, the gene signature was also detected in response to bacterial lipopolysaccharide-induced acute lung injury. A protein interaction network analysis revealed that metabolic enzymes interact with IFN-regulated transcription factors and members of the unfolded protein response (UPR) to form a module and potentially regulate type I interferon signaling, constituting a feedback loop. In addition, components of the metabolic gene expression signature were differentially regulated in the lung tissues of COVID-19 patients compared with healthy controls. These results suggest that the metabolic gene signature is a potential therapeutic target for the treatment of respiratory virus infections and inflammatory diseases.
Keywords:
respiratory virus; innate immunity; gene signature; lung; interferon; metabolism; JAK-STAT; TLR signaling; LPS; poly I:C 1. Introduction
Type I interferons (IFNs) are pluripotent cytokines that play a major role in the host immune response to respiratory viruses, including Respiratory Syncytial Virus (RSV), influenza, and coronaviruses (e.g., SARS-CoV-2), by regulating several hundred genes involved in antiviral defense, immune modulation, and cell growth [1,2]. SARS-CoV-2 infection and the resulting severe coronavirus disease 19 (COVID-19) have caused millions of deaths worldwide [3,4]. A variety of high-throughput studies in the last 3 years have provided novel insights into respiratory virus–host interactions, including gene signatures for antiviral defense, cytokine and chemokine responses, and signal transduction pathways, and the identification of potential drug targets [5,6,7,8,9,10]. Gene expression profiling by microarrays and RNA-seq enabled the global monitoring of cell-, tissue-, and disease-specific gene expression signatures in human lung cells in response to SARS-CoV-2 and in COVID-19 patients through the simultaneous quantitative analysis of differential expression levels of thousands of genes on a single platform [5,6,9,10]. Multiplex antibody assays provided important information on the temporal activation of cytokine levels, including type I and type II interferons (IFN), tumor necrosis factor-alpha (TNF-α), interleukin-1 beta 1(IL-1β), and interleukin 6 (IL-6) in the bronchoalveolar lavage fluid (BALF) of COVID-19 patients [11]. Multilevel proteomics revealed the function of SARS-CoV-2-encoded viral proteins and their interactions with the host genome and global alterations in signaling such as transforming growth factor-beta (TGF-β) and autophagy signaling in A549 human lung adenocarcinoma cells reconstituted with viral entry receptor ACE2 [12]. Metabolic studies revealed high levels of the tryptophan catabolic product kynurenine in the serum of COVID-19 patients [13,14]. Tryptophan is an essential amino acid for cell survival, and the intracellular enzyme that breaks down tryptophan to kynurenine is known as indoleamine-3,5-dioxygenase or IDO1 [15]. Furthermore, elevated levels of cholesterol-25-hydroxylase (CH25H), which is involved in cholesterol metabolism, have been reported in serum samples of COVID-19 patients [13]. Modulating cell metabolism to their advantage is a key tactic of respiratory viruses in their interactions with the host [16,17]. Macromolecules such as nucleotides, carbohydrates, proteins, and fats play major structural and functional roles in cell metabolism and are essential for viral replication and multiplication. Among these macromolecules, proteins composed of amino acids are the most diverse in terms of functions, such as enzyme activities that accelerate metabolic processes and hormones and neurochemicals, which are substances that deliver signals within the body to regulate cell metabolism. Many enzymes involved in gene regulation require vitamins and co-factors such as those generated during metabolism, demonstrating a direct link between metabolism and gene expression [18]. The transcriptional induction of host catabolic enzymes by interferon depletes the essential components of cell survival, such as amino acid, lipid, nucleotide, and polyamine reserves, to limit respiratory virus proliferation [19]. Metabolic reprogramming by type I interferons in response to respiratory virus infection includes increasing the uptake of amino acids and promoting the synthesis of amino acid-derived metabolites, such as polyamines, coenzymes, and nitric oxide, in different cell types [17,19]. Polyamines are essential for cell proliferation and differentiation, whereas nitric oxide is a signaling molecule with various immunoregulatory and antiviral functions [20]. Respiratory virus infection induces glycolysis and inhibits type I interferon production and the antiviral response in plasmacytoid dendritic cells [21]. Another important metabolic effect of IFNs is the regulation of fatty acid metabolism and oxidative phosphorylation [22]. The availability of multiple GEO datasets of human lung epithelial cells infected with several respiratory viruses as well as mouse models of respiratory virus infection has led to the identification of differentially expressed genes (DEGs) or gene signatures [5,6,23,24,25,26,27,28,29]. Bioinformatic analyses of lung gene expression datasets from cell culture and mouse models of respiratory virus infection revealed the induced expression of five key enzymes involved in macromolecule metabolism. Respiratory viruses induce an IFN-regulated metabolic gene signature that includes cholesterol-25-hydroxylase (CH25H), nicotinamide phosphoribosyltransferase (NAMPT), eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2), sterile alpha motif and histidine/aspartic acid domain-containing protein (SAMHD1), and indoleamine-3,5-dioxygenase (IDO1) at the RNA level in human lung epithelial cells. These metabolic enzymes are involved in the feedback regulation of type I interferon signaling. Furthermore, this novel interferon-regulated metabolic gene signature was also activated in response to bacterial lipopolysaccharide (LPS), and it also interacts with proteins involved in the unfolded protein response (UPR) activated specifically by SARS-CoV-2 infection in lung epithelial cells [29]. In this study, the regulation of metabolic gene signatures in human lung epithelial cells and in mouse models in response to respiratory viruses and type I interferon signaling was investigated.
2. Materials and Methods
Gene Expression Datasets and Bioinformatics
Standard bioinformatic methods were used [5,6,26]. Gene expression profiles of human lung cell lines infected with respiratory viruses and COVID-19 patients have been reported previously [5,30,31,32]. Microarray data in Gene Expression Omnibus (GEO) datasets GSE147507, GSE 47960, and GSE 148729 and GEO profiles were retrieved from the PubMed (NIH) resources. ImmGen RNA seq SKYLINE resources were used (http://rstats.immgen.org/Skyline_COVID-19/skyline.html, accessed on 7 August 2023). Double-stranded RNA (poly IC)-induced gene signatures in the mouse lungs were retrieved from GSE 39304 as described previously [33]. Gene signatures of mice infected with reconstructed 1918 influenza were retrieved from GSE70445 as described previously [32]. RSV-induced gene signatures in wild-type (WT) and PKR KO mice were retrieved from GSE 18170 as described previously [34]. LPS-induced gene signatures in wild-type and BPIFA1KO mice were retrieved from GSE 104214 as described previously [35]. The GEO datasets were analyzed with the GeoR2R method (NCBI). Corresponding mock and treatment data and multiple datasets were used. Outliers of expression were excluded in the data analysis. Additional gene expression datasets were also downloaded from the ImmGen browsers and Coronascape. Cluster analysis was performed using gene expression software tools [36]. Protein–protein interactions were visualized in the STRING database [37]. Gene ontology (GO) and signaling pathway analyses were performed in Metascape [38]. Interferon-related data were retrieved from the Interferome database at www.Interferome.org, accessed on 12 September 2023. Gene-specific information such as intracellular location and functions were retrieved from standard bioinformatics websites such as Mouse Genome Informatics (MGI) and Genecards (https://www.genecards.org/, accessed on 7 October 2023).
3. Results
3.1. Profiling a Metabolic Gene Expression Signature in Response to Respiratory Viruses and Type I Interferon Signaling
Bioinformatic studies revealed antiviral, cytokine, chemokine, and apoptosis gene signatures in lung epithelial cells in response to several respiratory viruses, including RSV, influenza, and SARS-CoV-2 [23,24,25,32]. Furthermore, the delineation of gene signatures in the transcriptome facilitated the identification of key hubs, modules, and gene regulatory networks in response to respiratory viruses [26,27]. Previous studies have identified the role of transcription factor subnetworks in the regulation of antiviral and immunomodulatory functions as well as cell growth in response to type I interferon signaling and respiratory viruses [28]. Recent studies have shown a direct link between metabolism and transcription [18,39]. However, the diverse roles of IFN-stimulated metabolic enzymes in the regulation of transcription in response to respiratory viruses remains to be explored. Respiratory syncytial virus (RSV) infections are common in children and not a concern for adults and do not pose a pandemic threat [24]. In contrast, HPIV3 infections are more common in adults with a compromised immune system [40]. The profiling of IFN-stimulated metabolic genes in gene expression datasets of lung epithelial cells infected with RSV and HPIV3 respiratory viruses revealed an induced expression of five key enzymes involved in nucleotide, lipid, and protein metabolism (Figure 1). Some of these metabolic effectors of interferon signaling are well known, including CH25H, which is involved in cholesterol metabolism; IDO1, which is involved in tryptophan catabolism; and EIF2AK2 (PKR), which is involved in the regulation of protein synthesis [15,41,42]. NAMPT and SAMHD1 are metabolic enzymes involved in innate immunity and nucleotide metabolism [43,44]. SAMHD1’s enzyme functions include deoxy-nucleoside triphosphate activity (dNTPase), which reduces cellular dNTP levels to restrict viral replication. These enzymes deplete the metabolic building blocks required for optimal viral replication and multiplication [19]. Recent studies have also revealed the role of these five metabolic enzymes in the control of transcription and interferon-stimulated gene expression (Table 1). CH25H converts cholesterol to 25-hydroxy cholesterol (25-HC), thereby repressing cholesterol synthesis and influencing transcriptional regulation in macrophages [41]. IDO1 is a rate-limiting enzyme in the catabolism of tryptophan as well as a regulator of stress-induced transcription factors and solute transporter gene expression [45]. NAMPT and SAMHD1 regulate NAD+ levels in the cell and are involved in the regulation of transcription [39]. EIF2AK2 is a serine/threonine kinase involved in the regulation of transcription factors, such as NF-κB and IRF-1, and type I interferon production [42]. Consistent with their role in transcription, several of these metabolic enzymes were also identified in the nucleus in addition to their predominant cytoplasmic compartment localization. In contrast, CH25H was exclusively located in the cytoplasm (Table 1).
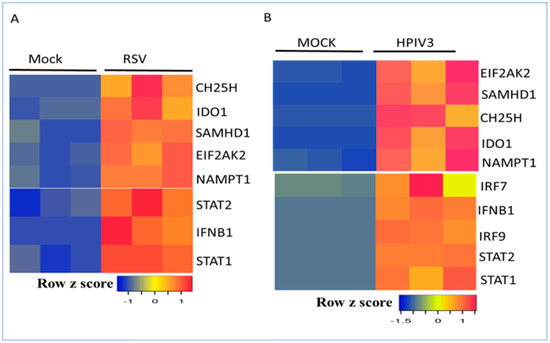
Figure 1.
Regulation of the interferon-regulated metabolic gene signature and transcription factors by RSV and HPIV3 in A549 cells. (A) Human A549 lung cells mock-infected or infected with RSV for 24 h. (B) A549 cells mock-infected or infected with HPIV3 for 24 h. Data from 3 samples for each condition are shown. RNA levels were normalized by the DESeq2 method. Low and high gene expression are represented by blue and red, respectively.

Table 1.
Regulation of an interferon-regulated metabolic gene signature by respiratory viruses.
3.2. Regulation of Metabolic Gene Signature Expression by Coronaviruses
Most gene expression studies on SARS-CoV-2 were performed in well-established human lung epithelial cell lines such as Calu-3, A549, and NHBE. The basal expression of metabolic genes was very low in these cell lines. The infection of Calu-3 cells with SARS-CoV-2 dramatically induced the expression of the metabolic gene signature at the RNA level within 24 h. Mock-infected cells served as controls (Figure 2A). The induction of the metabolic gene signature was correlated with IFN-beta (IFNB1), STAT1, STAT2, and IRF9 at the RNA level (Figure 2B). A time-course analysis revealed that the metabolic gene signature was induced within 12–24 h after virus infection (Figure 2C). These results are consistent with the interpretation that IFN-β production by viral RNA recognition via the Toll-like receptor (TLR) pathway drives autocrine or paracrine IFN-β signaling involving the Jak–Stat pathway and the activation of interferon-stimulated gene expression [33,34]. SARS-CoV-2 was more effective in the induction of IDO1 and CH25H at the RNA level than SARS-CoV-1 in Calu-3 cells. In contrast, SAMHD1 and NAMPT were both induced to the same extent by both coronaviruses (Figure 2D). The productive infection of human lung epithelial cells by SARS-CoV-2 requires the cell membrane proteins ACE2 and TMPRSS2 to facilitate viral entry [46]. SARS-CoV-2 infection of A549 cells failed to induce IFNB1 and interferon-stimulated gene expression. The expression of ACE2 in A549 cells restored SARS-CoV-2-mediated induction of IFNB1, STAT1, STAT2, and IRF9 mRNA expression (Figure 3A,B). These studies revealed the important role of ACE2 in viral entry, activation of type I IFN signaling, and temporal regulation of interferon-stimulated transcription factors and the metabolic gene signature. Although cell culture models of respiratory virus infection provide important details on virus replication and the differential regulation of gene expression, they may not accurately reflect the host–pathogen interactions in vivo, where multiple immune and non-immune cell types in the lung interact in a complex manner. Mouse models of SARS-CoV-2 infection were recently developed to study host–pathogen interactions in vivo [47]. In an aerosol infection mouse model, SARS-CoV-2 induced the type I interferon-regulated transcription factors and metabolic gene signature, confirming the results in human lung epithelial cells (Figure 3C).
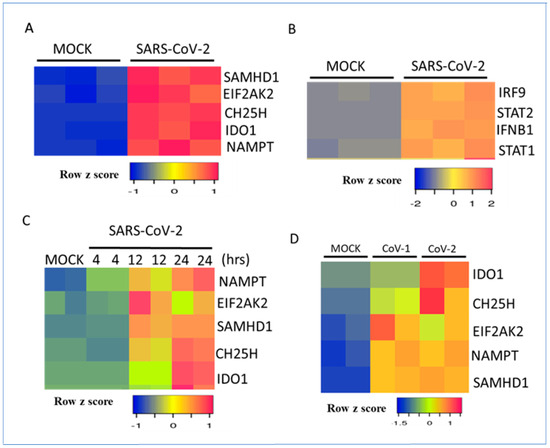
Figure 2.
Regulation of interferon-regulated metabolic genes and transcription factors by SARS-CoV-2 in Calu-3 cells. (A) Heatmap representation of RNA levels of metabolic genes after mock infection or virus infection in Calu-3 cells for 24 h. (B) Heatmap representation of IFN-beta- and IFN-regulated transcription factors at the RNA level after mock infection or virus infection in Calu-3 cells for 24 h. (C) Heatmap representation of RNA levels of metabolic genes after mock infection or virus infection in Calu-3 cells for 4, 12 or 24 h. Data from two or three samples for each condition are shown. (D) Regulation of the metabolic gene signature by SARS-CoV-1 and SARS-CoV-2 after mock infection or virus infection in Calu-3 cells. Data from two samples for each condition are shown. RNA expression levels were normalized by the DEseq2 method.
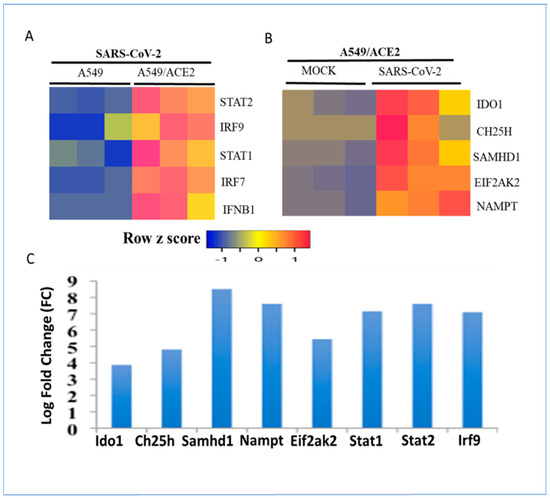
Figure 3.
Regulation of the metabolic gene signature and interferon-stimulated transcription factors in A549 cells expressing ACE2 receptors and in an aerosol model of SARS-CoV-2 infection in mice. (A) Expression levels of IFN-regulated transcription factors in A549 cells with or without expression of ACE2 receptors after infection with SARS-CoV-2 for 24 h. (B) Changes in expression of metabolic regulators in A549 cells expressing ACE2 receptors were mock-infected or infected with the SARS-CoV-2 virus for 24 h. Data from three samples for each condition are shown. RNA expression levels were normalized by the DEseq2 method. (C) Regulation of metabolic gene signature and interferon-regulated transcription factors at the RNA level in the lungs in an aerosol model of SARS-CoV-2 infection in mice. Expression levels of the interferon-regulated gene signature are ranked by log-fold change (logFC).
3.3. Regulation of Metabolic Gene Signature Expression by Influenza Viruses
Influenza virus is a major respiratory pathogen that often causes significant mortality and morbidity, particularly among young children and geriatric patients [48]. Influenza is generally limited to the upper respiratory tract; however, when the lower respiratory tract becomes involved, significant lung damage is observed. This may occur with pandemic strains (such as the 1918 virus that killed more than 50 million worldwide) or pathogenic influenza strains (such as H1N1 and H5N1). Highly pathogenic strains of influenza, such as the reconstructed 1918 influenza virus (H1N1), induce a dramatic increase in inflammatory cytokines and an influx of neutrophils and macrophages, resulting in inflammation, tissue injury, and death in mice [32,49]. Infection of mice with the reconstructed 1918 influenza virus dramatically induced interferon-stimulated antiviral and inflammatory gene expression [49]. These genes include well-known antiviral interferon-stimulated genes (ISGs) such as oligoadenylate synthetase (Oas), guanylate-binding proteins (Gbps), interferon-induced protein with tetratricopeptide (Ifit), and immunoregulatory cytokines and chemokines induced 3 days after virus infection [32]. The metabolic enzyme gene signature was also induced 3 days after virus infection and was temporally correlated with the inducible expression of the transcription factors Stat1, Stat2, and Irf9 (Figure 4A). In contrast to the pandemic influenza virus, the seasonal non-pandemic strains of influenza inhibit type I interferon and inflammatory responses. In these strains, the virus encodes a non-structural protein (NS1) that is a potent inhibitor of interferon regulatory factor 3 (IRF3) and type I interferon production [50]. Furthermore, NS1 inhibits STAT1 activation, resulting in the attenuation of interferon-stimulated gene expression [51]. Induction of the metabolic gene signature in NHBE bronchial epithelial cells was abrogated by the wild-type influenza A virus (IAV). In contrast, IAV infection with a mutant virus containing a deletion of NS1 (IAVNS1) resulted in the induction of the metabolic gene signature in NHBE cells (Figure 4B). This induction was correlated with the induction of IFN-beta (IFNB1) and interferon-inducible transcription factors STAT1, STAT2, and IRF7 (Figure 4B). These studies suggest that multiple factors, such as virus strains, host-encoded factors, virus-encoded factors, and cell type may play an important role in determining viral pathogenesis.
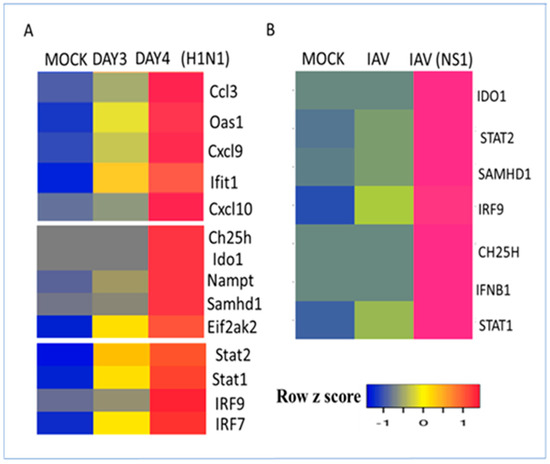
Figure 4.
Regulation of the interferon-regulated metabolic gene signature by influenza viruses. (A) Regulation of interferon-stimulated antiviral genes, metabolic regulators, and transcription factors at the RNA level in mouse lungs by the reconstructed 1918 H1N1 influenza virus. Mice were mock-infected or infected with the virus for 3 or 4 days. Heatmap representation of interferon-regulated gene expression. (B) Human NHBE1 bronchial epithelial cells were mock-infected or infected with the influenza virus (IAV) or influenza virus lacking the NS1 (IAVNS1) for 12 h. Heatmap representation of interferon-regulated gene expression regulated by mock, wild-type IAV, and NS1-deleted IAV. RNA expression values were normalized by the DEseq2 method and are shown as a heatmap. Data from 4 samples for each condition are shown.
3.4. Regulation of Metabolic Gene Signature by Double-Stranded RNA and EIF2AK2 In Vivo
EIF2AK2, also known as protein kinase R (PKR), is a serine/threonine kinase that plays a central role in double-stranded RNA (dsRNA) and type I interferon (IFN) signaling [42,52]. It is activated by the presence of dsRNA, which is a common feature of many respiratory viruses. EIF2AK2 phosphorylates the eukaryotic translation initiation factor 2α (eIF2α), which leads to the inhibition of protein translation. This inhibition of protein translation has a number of downstream effects on IFN signaling, including an increased production of type I interferons, activation of the Jak–STAT pathway, and activation of transcription factors and antiviral genes [34,52,53]. Furthermore, EIF2AK2 levels are induced by type I interferons and poly I:C, which is a double-stranded RNA mimetic and a potent inducer of type I interferons constituting a positive feedback loop [54]. Intratracheal instillation of double-stranded RNA (poly I:C) in mouse lungs induces interferon-stimulated transcription factors and the metabolic gene signature with distinct temporal profiles [33]. For example, EIif2ak2, Ch25h, and Saamhd1 were maximally induced at 18 h, while Ido1 and Naampt were highly induced at 24 h (Figure 5A). These results indicate that double-stranded RNA treatment is necessary and sufficient to induce the metabolic gene signature in vivo in a mouse model of acute lung injury [34]. Consistent with previous results, interferon-inducible transcription factors were also upregulated at 18–24 h (Figure 5A). RSV infection of wild-type and Eif2ak2-knockout (KO) mice revealed that the induction of interferon-stimulated transcription factors and the metabolic gene signature required an intact Eif2ak2 gene (Figure 5B). Furthermore, Eif2ak2-regulated antiviral gene expression was also abrogated and lung injury was enhanced in Eif2ak2-KO mice [34].
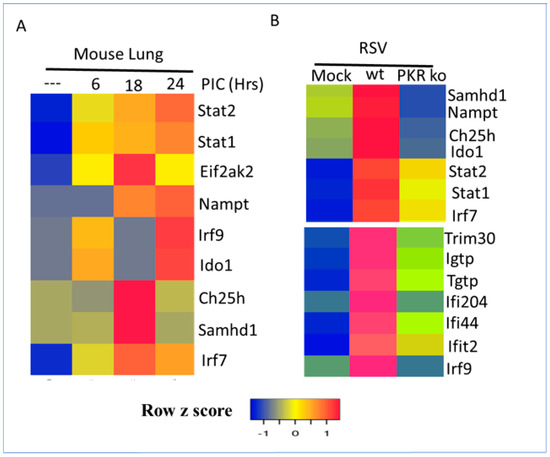
Figure 5.
Regulation of the interferon-regulated metabolic gene signature and transcription factors by double-stranded RNA and protein kinase R (PKR/EIF2AK2) in mice. Mock treatment was indicated by (---) (A) Mice treated with double-stranded RNA mimetic poly I:C for 6, 18, or 24 h. (B) Wild-type or PKR-KO mice were mock-infected or infected with RSV for 24 h. RNA expression values were normalized by the DEseq2 method and are shown as a heatmap. Data from two samples for each condition are shown.
3.5. Regulation of Metabolic Gene Signature by Type I Interferons in Human Lung Epithelial Cells
The host immune response to respiratory viruses is complex and involves a variety of cell types and multiple cytokine signaling pathways. It is important to understand whether type I interferon treatment alone can induce the metabolic gene signature in lung epithelial cells. An interrogation of microarray datasets revealed that IFN-β treatment induced the metabolic gene signature and interferon-induced transcription factors at the RNA level within 6 h in A549 lung cells. Interestingly, IDO1, SAMHD1, and EIF2AK2 levels were further increased after 24 h, whereas CH25H and NAMPT RNA levels were attenuated by 24 h (Figure 6A). Furthermore, IFN-α induced interferon-regulated transcription factors and the metabolic gene signature in both A549 and HTBE human lung epithelial cells (Figure 6B).
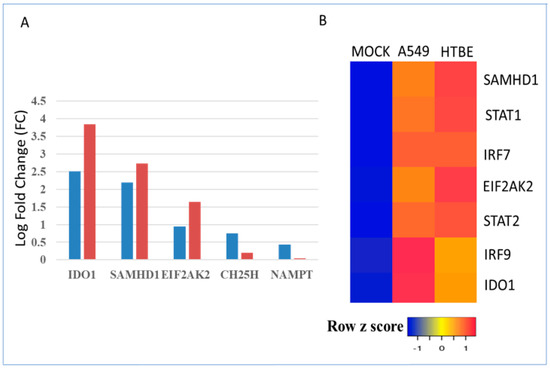
Figure 6.
Regulation of the metabolic gene signature and transcription factors by type I interferon treatment in A549 and HTBE human lung epithelial cells. Indicated in 6A legend as 6 h and 24 h (A) Regulation of the metabolic gene signature by IFN-β treatment for 6 h and 24 h in A549 cells represented by blue- and red-colored bars, respectively. Gene expression levels of the metabolic enzymes are expressed as log fold change (logFC). (B) Regulation of the interferon-stimulated metabolic gene signature and transcription factors by IFN-α treatment for 8 h in A549 and HTBE cells.
3.6. Visualization of the Protein Interaction Network of the Metabolic Gene Signature in the STRING Database
Protein interactions mediate post-translational modifications such as phosphorylation and ubiquitination that activate or inhibit signaling and provide novel insights into biological functions [37]. Protein interactions play a critical role in all aspects of type I IFN signaling, including the interactions between type I interferons and their receptors, activation of the Jak–STAT pathway, and interaction between STAT proteins and other transcription factors to regulate ISG expression [28]. These interactions can be visualized as a graph using the information in several bioinformatic databases such as BIOGRID and STRING [37]. The protein interactions of the genes in the metabolic gene signature were interrogated in the STRING database, which revealed that SAMHD1, EIF2AK2, and IDO1 extensively interact with multiple major hubs in the antiviral subnetwork such as STAT1, STAT3, IRF7, and IRF1. In contrast, NAMPT and CH25H interact specifically with STAT3 and IRF1, respectively (Figure 7). These interactions may play a major role in the regulation of interferon signaling in response to multiple respiratory viruses [55]. In addition, IDO1 also interacts with suppressors of cytokine signaling 1 (SOCS1) and suppressors of cytokine signaling 3 (SOCS3) proteins, which are involved in the negative regulation of cytokine signaling. Respiratory viruses can also interfere with protein interactions that regulate interferon-mediated gene expression. For example, the influenza virus NS1 protein can bind to STAT1 and inhibit its transcriptional activity. This prevents the expression of ISGs and allows the virus to replicate more efficiently [50,51]. The development of therapeutic strategies to target protein interactions that regulate gene expression in response to respiratory viruses is a promising area of research [53,56].
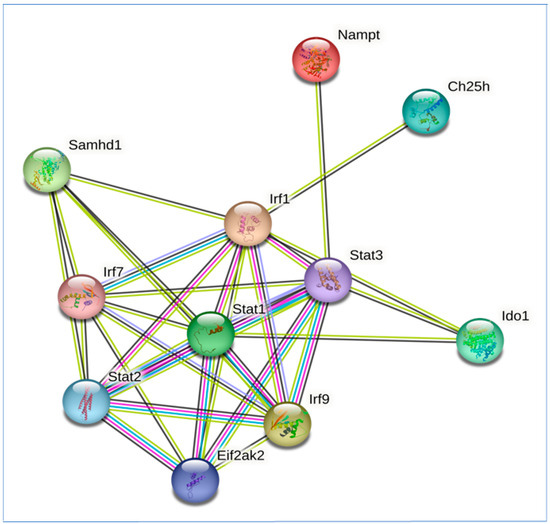
Figure 7.
Protein–protein interaction (PPI) network of genes in metabolism gene signature induced by respiratory viruses and the transcription factor network of type I interferon signaling. Protein interactions in the signaling network were obtained from the STRING database. Interacting proteins are indicated by ovals and protein interactions are indicated by edges.
3.7. Lipopolysaccharide (LPS) Regulation of Metabolic Gene Signature in Acute Lung Injury (ALI)
Secondary bacterial infections are a common complication of respiratory virus infections in up to 30% of patients, and they are a leading cause of morbidity and mortality [57]. LPS is a component of the outer membrane of Gram-negative bacteria and a potent activator of the innate immune system. It can induce inflammatory responses, including fever, pain, and inflammation [58]. LPS binds to Toll-like receptor 4 (TLR4) on immune cells, such as macrophages and dendritic cells [58,59]. This activation of TLR4 triggers a cascade of events that leads to the release of type I interferons and proinflammatory cytokines, such as tumor necrosis factor-alpha (TNF-α) and interleukin-1β (IL-1β). These cytokines can damage the lungs by increasing vascular permeability, promoting neutrophil infiltration, and stimulating the production of reactive oxygen species. This can lead to a number of complications, including acute lung injury (ALI), acute respiratory distress syndrome (ARDS), and sepsis [59].
LPS stimulation induces PKR mRNA expression through the activation of the TLR4 signaling pathway, which is essential for the activation of interferon regulatory factor 3 (IRF3) and the production of type I interferons [60]. Furthermore, LPS activation of transcription factors such as IRF3, IRF7, and NF-κB regulates the expression of CH25H, SAMHD1, NAMPT, and IDO1 in macrophages and dendritic cells [61,62,63]. Consistent with in vitro results in human immune cells, the metabolic gene signature was also induced within 8–24 h after intratracheal administration of LPS in mice (Figure 8). Bpifa1 (BPI fold-containing group A member 1) is a mouse airway-secreted protein with immunomodulatory functions that plays a major role in the lung epithelial barrier [64]. The deletion of Bpifa1 in mice results in the abrogation of LPS-induced cytokine (interferon) and chemokine (Cxcl9 and Cxcl10) production and the recruitment of neutrophils as well as acute lung injury [35]. Furthermore, induction of the metabolic gene signature was significantly abrogated at both 8 and 24 h after the intratracheal administration of LPS in Bpifa1-KO mice. These results indicate that Bpifa1 regulates type I interferon induction and the sustained activation of the metabolic gene signature in response to bacterial infection [35,64].
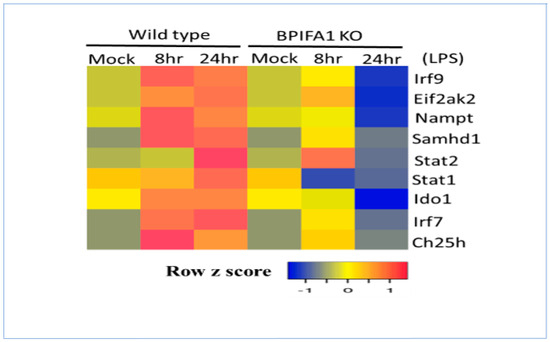
Figure 8.
LPS regulation of the interferon-stimulated metabolic gene signature and transcription factors in wild-type and BPIFA1-knockout (KO) mice. Wild-type or BPIFA1-KO mice were mock-treated or treated with LPS for 8 or 24 h. RNA expression values were normalized and are shown as a heatmap.
3.8. Role of Metabolic Gene Signature in Integrated Stress Response (ISR)
ISR is an evolutionarily conserved intracellular signaling pathway in mammalian cells that is activated in response to a variety of stress conditions, including nutrient deprivation, the unfolded protein response (UPR), viral infection, and oxidative stress [65]. The core event in ISR is the phosphorylation of eukaryotic translation initiation factor 2 alpha (eIF2α) by one of the four members of the eIF2α kinase family. This phosphorylation decreases global protein synthesis and induces selected genes, including the transcription factor ATF4. ATF4 then activates several genes that promote cellular recovery, such as those encoding chaperones, proteases, and heat shock proteins [65]. EIF2AK2 is one of four members of the eIF2α kinase family. It is activated by a variety of stress signals, including unfolded protein folding stress (UPR), nutrient deprivation, and viral infection. EIF2AK2 phosphorylates eIF2α, which leads to a decrease in global protein synthesis and the induction of the ISR. IDO1 is activated by a variety of stress signals, including inflammation, infection, and tumor growth [15]. IDO1 can induce the expression of genes encoding transcription factors and solute transporters as well as genes involved in redox regulation [45]. There is strong evidence for the activation of the unfolded protein response by SARS-CoV-2 [29,66]. Consistent with previous results, SARS-CoV-2 infection of Calu-3 cells induced markers of the UPR, such as ATF4, XBP1, IRE1A, DDIT3, DDIT4, and NFE2L2, as well as IDO1-regulated genes such as ATF3, ATF4, NFE2L2, MTHFD2, SESN, ALDH1L2, and SLC3A2 (Figure 9A,B). Furthermore, a protein interaction analysis using the STRING database revealed strong interactions between unfolded protein response (UPR) and IDO1-regulated proteins that were activated specifically in response to SARS-CoV-2 infection as well as with transcription factors of the antiviral subnetwork and the metabolic gene signature to form a gene regulatory network or GRN (Figure 10). These results indicate that IDO1 is a critical regulator at the intersection of metabolism, inflammation, and cancer [67].
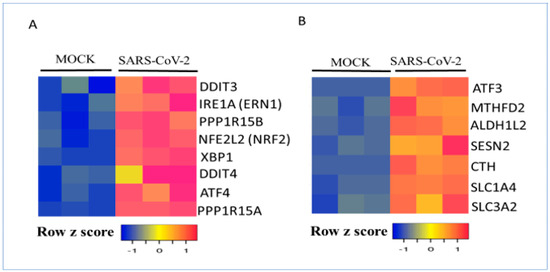
Figure 9.
Regulation of the unfolded protein response (UPR) and IDO1 target genes by SARS-CoV-2 in Calu-3 human lung cells. (A) Calu-3 cells were mock-infected or infected with SARS-CoV-2 for 24 h. UPR target gene expressions are shown. (B) Calu-3 cells were mock-infected or infected with SARS-CoV-2 for 24 h. IDO1 target gene expression is shown. RNA expression values were normalized and are shown as a heatmap.
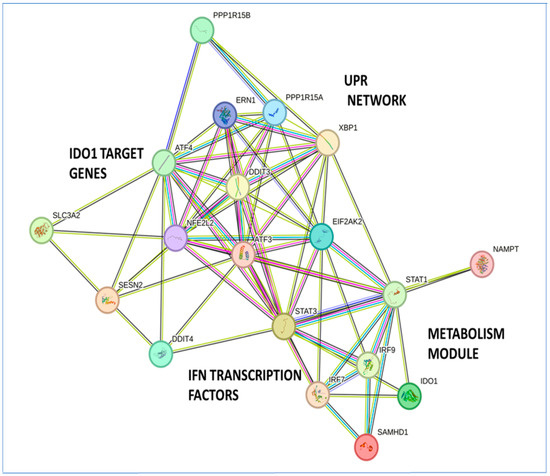
Figure 10.
Visualization of the protein–protein interaction (PPI) network of the metabolic gene signature with the unfolded protein response (UPR), type I interferon signaling, and IDO1 target genes. Protein interactions in the signaling network were obtained from the STRING database. Interacting proteins are indicated by ovals and protein interactions are indicated by edges.
3.9. Regulation of the Metabolic Gene Signature in Healthy and COVID-19 Patients
SARS-CoV-2 infection can cause various metabolic alterations in patients, both during acute infection and in the long term. These alterations can affect all aspects of metabolism, including glucose metabolism, lipid metabolism, amino acid metabolism, and energy metabolism [68,69]. Some of the most common metabolic changes observed in COVID-19 patients include hyperglycemia, increased triglycerides, increased levels of aromatic amino acids such as tryptophan and tyrosine, and decreased levels of arginine leading to defects in immune function and wound healing as well as muscle wasting [68,69,70]. Interrogation of the gene expression datasets of healthy and COVID-19 patients revealed that metabolic markers such as EIF2AK2 and IDO1 were upregulated in the lung tissue samples of COVID-19 patients compared with those of healthy patients. This upregulation was also correlated with an increase in interferon-inducible transcription factors such as STAT1, STAT2, and IRF7 in the lung tissue samples of COVID-19 patients (Figure 11A). In contrast, the metabolic markers CH25H and SAMHD1, and inducible transcription factors STAT1, STAT2, and IRF8 were significantly increased in the peripheral blood (PBMC) samples of COVID-19 patients compared with those of healthy individuals (Figure 11B). A transcriptomic analysis of a different dataset revealed that the interferon-induced transcription factors STAT1, STAT2, and IRF7 and the metabolic gene signature genes EIF2AK2, IDO1, and NAMPT were significantly upregulated in the lung tissue samples of COVID-19 patients compared with those of healthy patients (Figure 11C). The innate immune response to SARS-CoV-2 infection must be precisely controlled, and any imbalance in the cytokine response may result in lung injury and death [5,6,8,10]. A mix of antiviral proteins, proinflammatory cytokines such as IL-α, and IL-1β, and chemokines such as CXCL10, CXCL11, and CCL8 were upregulated in the lung tissue samples of COVID-19 patients compared with those of healthy controls (Figure 11C).
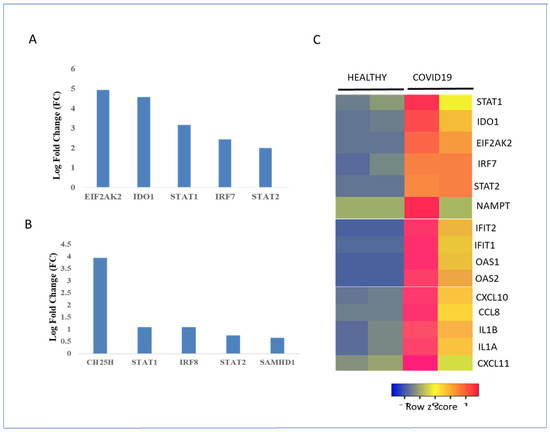
Figure 11.
Regulation of interferon-stimulated metabolic gene signature and transcription factors in healthy controls and COVID-19 patients. (A) Regulation of metabolic enzyme and transcription factor gene expression in COVID-19 lung samples. Gene expression levels are expressed as log fold change or logFC. (B) Regulation of metabolic enzyme and transcription factor gene expression in the peripheral blood (PBMC) of COVID-19 patients. Gene expression levels are expressed as logFC. (C) RNA expression levels of metabolic enzymes, chemokines, and transcription factors in healthy and COVID-19 lung samples. Low and high gene expression in the healthy and COVID-19 lung biopsies are represented by blue and red, respectively. RNA expression values were normalized by the DEseq2 method and are shown as a heatmap. Data from two samples for each condition are shown.
4. Discussion
In this study, transcriptome profiling revealed a common metabolic gene signature mediated by type I interferon signaling in the host response to several respiratory viruses. The same metabolic signature was observed in response to double-stranded RNA, bacterial LPS, and the UPR. This metabolic signature is part of a protein interaction network with transcription factors of type I interferon signaling. Furthermore, EIF2AK2/PKR was identified as a key node in the metabolic signature as suggested by the attenuation of the metabolic gene signature in Eif2ak2-KO mice in response to RSV infection. Pathogen molecular pattern recognition by lung resident immune cells such as dendritic cells and macrophages and the production of type I interferons play an important role in innate immunity to respiratory viruses [1]. The type I interferon effects are pleiotropic with antiviral, antiproliferative, immunomodulatory, and metabolic effects through the modulation of gene expression [2]. Previous bioinformatic studies have revealed an antiviral module composed of transcription factors from the STAT, IRF, and SP families and an inflammatory module composed of STAT1, STAT3, JUN, RELA, and BRCA1 in innate immunity [27,28]. In this study, the profiling of interferon-regulated metabolic enzymes in the host response to respiratory viruses led to the identification of a common five-gene signature consisting of EIF2AK2, IDO1, CH25H, NAMPT, and SAMHD1. The induction of these metabolic enzymes by type I interferon depletes metabolic building blocks such as essential amino acids, lipids, and nucleotide pools to restrict respiratory virus replication and multiplication [16,19]. The interferon-stimulated metabolic gene signature was induced by both low-pathogenicity (RSV and HPIV3) and highly pathogenic respiratory viruses (1918 H1N1 and SARS-CoV-2). Furthermore, several components of the gene signature were upregulated in the PBMC and lung tissue samples of COVID-19 patients compared with those of healthy individuals [31]. Both host-encoded and viral factors are involved in the induction of the metabolic gene signature in response to respiratory viruses. Double-stranded RNA and EIF2AK2 were required for the induction of the metabolic gene signature and correlated with the induction of IFNB1 and IFN-induced transcription factor expression. Virally encoded proteins, such as influenza NS1, and host cellular proteins involved in viral entry, such as ACE2, in lung epithelial cells are required for the activation of the metabolic gene signature [46,50,51].
Genetic variability in different individuals leads to different clinical courses and treatment responses to COVID-19. Genetic analyses revealed that about 10% of critically ill COVID-19 patients have alterations in the components of type I interferon signaling or autoantibodies to interferon that inactivate antiviral signaling [71,72]. Furthermore, the gene expression profiling of nasal swabs revealed that patients with mild infections or those that have recovered from COVID-19 have a more robust type I interferon response in the nasopharynx than critically ill patients [73]. These results suggest that type I interferon signaling and the metabolic gene signature are important for innate immunity to respiratory virus infection and of diagnostic value for predicting disease progression. The identification of a metabolic gene signature in the lung epithelial response to respiratory viruses indicates that a comparative transcriptome analysis for metabolic gene signatures in a variety of lung diseases such as asthma, acute respiratory distress syndrome (ARDS), and septic shock is warranted. Further investigations are required to test the role of EIF2AK2/PKR inhibitors in response to respiratory viruses and inflammatory diseases.
The interferon-stimulated metabolic signature was also activated by bacterial LPS, which is commonly found in opportunistic secondary bacterial infections following respiratory virus infection [57]. LPS was detected in the peripheral blood samples of COVID-19 patients [74]. The protein interaction network analysis revealed that these five metabolic enzymes form extensive connections with the core transcription factors of the type I interferon signaling pathway, IDO1 target genes, and members of the integrated stress response [45,65,66]. These results suggest that the interferon-stimulated metabolic gene signature activated in response to respiratory viruses may participate in the feedback regulation of type I interferon signaling and in the cross-talk with LPS and proinflammatory cytokine signaling as well as the integrated stress response. An imbalance in the antiviral and proinflammatory gene expression in lung cells in response to respiratory viruses may be a causal factor in the severity of respiratory disease [5,75]. Understanding the role of the metabolic gene signature in the feedback regulation of interferon and proinflammatory cytokine regulation of gene expression in response to respiratory viruses may have therapeutic implications in vaccine development and drug design.
EIF2AK2 (PKR) is a serine kinase that promotes the activation of transcription factors such as STAT1, NF-κB, and IRF1; the production of proinflammatory cytokines; and the inhibition of protein synthesis [34,42,53,60]. Induction of the metabolic gene signature and acute lung injury were attenuated in Eif2ak2-KO mice in response to RSV infection, demonstrating that EIF2AK2 is a critical node in metabolism and inflammation in innate immunity [27]. The activation of PKR by immunostimulatory double-stranded RNA and bacterial RNA leads to the activation of transcription factors and proinflammatory cytokine and chemokine gene expression [52,76,77]. Furthermore, synthetic RNA containing modified uridine bases failed to activate PKR or an innate immune response, leading to the development of mRNA vaccine technology [78,79]. NAMPT and SAMHD1 regulate NAD+ metabolism and type I interferon signaling [43,44,63]. SAMHD1 inhibits IRF7 and NF-κB activation [63]. IDO1 was recently shown to be a regulator of stress-induced transcription factors involved in the regulation of solute transporter gene expression [45]. CH25H is a critical enzyme in lipid metabolism, the production of 25-hydroxycholesterol (25-HC), which is involved in the activation of transcription factors; and the production of proinflammatory cytokines [41,61,80,81]. Previous studies have shown that CH25H plays a central role in lipid metabolism and is regulated by interferons, TLR ligands such as LPS and poly I:C, and a high-fat diet [80,81,82,83]. The induction of CH25H and the production of 25-HC by TLR ligands are mediated by type I interferon signaling [82]. The transcriptional effect of 25-HC includes activation of the transcriptional factors involved in lipid metabolism such as liver X receptor (LXR α/β), retinoic-related orphan receptor (ROR α/β/χ), sterol response element-binding protein (SREBP), and the AP1 family [80]. CH25H and 25-HC may have proinflammatory or anti-inflammatory effects depending on the cell type and under different experimental conditions such as obesity, diabetes, and viral infections [83]. NAMPT and the NAD+-dependent enzyme Sirturin 2 (SIRT2) mediate the inhibition of p53 transactivation, whereas NAMPT and SIRT 1-activate RelB-mediated NF-κB transactivation [84,85]. These studies revealed that IFN-regulated metabolic enzymes play a major role in transcriptional regulation and coordinate cross-talk between multiple signal transduction pathways involved in the antiviral response, inflammation, and cell growth. Many cancers express high levels of IDO1, depleting the essential amino acid tryptophan and enabling them to escape immune surveillance. IDO1 inhibitors are in clinical trials either as a monotherapy or in combination with other therapies for cancer treatment [16,86]. Similarly, NAMPT inhibitors were developed to target pancreatic cancers [87]. The utility of these metabolic inhibitors to block viral replication and inflammation and for treating respiratory viral diseases remains to be determined. These studies suggest that the inhibition of the metabolic module is a potential therapeutic target for treating respiratory viruses. A major limitation of this study is the lack of direct experimental evidence for a particular metabolic state induced by type I interferon in cells. However, the activation of several transcription factors known to be involved in redox regulation, such as NF-κB, Stat1, Stat3, p53, ATF4, and NRF2, by the identified metabolic module strongly suggests that redox regulation may play a major role [45,87].
5. Conclusions
Identifying a lung epithelial metabolic gene signature regulated by type I interferon signaling in the host response to respiratory viruses holds significant promise for the following applications:
- This metabolic gene signature could potentially serve as a biomarker for the early detection of respiratory pathogens.
- By tracking changes in the metabolic gene signature by RT-PCR, clinicians could monitor respiratory disease severity and the response to therapy.
- Screening for chemical inhibitors of the metabolic gene signature could lead to novel therapeutic drugs. Further research is required to test the presence of similar metabolic gene signatures in other bacterial, viral, and inflammatory diseases where type I interferon signaling plays a critical role.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The author alone is responsible for the content and writing of this article.
Acknowledgments
The author thanks his colleagues for feedback on the preprint version of the manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Kohlmeier, J.E.; Woodland, D.L. Immunity to respiratory viruses. Annu. Rev. Immunol. 2009, 27, 61–82. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Kerr, I.M.; Williams, B.R.; Silverman, R.H.; Schreiber, R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998, 67, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.Y.; Danielson, M.L.; Town, M.; Derado, G.; Greenlund, K.J.; Kirley, P.D.; Alden, N.B.; Yousey-Hindes, K.; Anderson, E.J.; Ryan, P.A.; et al. COVID-NET Surveillance Team. Risk Factors for Coronavirus Disease 2019 (COVID-19)-Associated Hospitalization: COVID-19-Associated Hospitalization Surveillance Network and Behavioral Risk Factor Surveillance System. Clin. Infect. Dis. 2021, 72, e695–e703. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Tonetti, T.; Protti, A.; Langer, T.; Girardis, M.; Bellani, G.; Laffey, J.; Carrafiello, G.; Carsana, L.; Rizzuto, C.; et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: A multicentre prospective observational study. Lancet Respir. Med. 2020, 8, 1201–1208. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Jiang, L.; Dua, K.; Hansbro, P.M.; Liu, G. SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir. Res. 2020, 21, 182. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef]
- O’Meara, M.J.; Guo, J.Z.; Swaney, D.L.; Tummino, T.A.; Hüttenhain, R. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ziegler, C.G.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef]
- Delorey, T.M.; Ziegler, C.G.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, Å.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Lionetto, L.; Ulivieri, M.; Capi, M.; De Bernardini, D.; Fazio, F.; Petrucca, A.; Pomes, L.M.; De Luca, O.; Gentile, G.; Casolla, B.; et al. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: An observational cohort study. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166042. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Malachowski, W.J.; Mondal, A.; Scherle, P.; Muller, A.J. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int. Rev. Cell Mol. Biol. 2018, 336, 175–203. [Google Scholar] [PubMed]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef]
- Rudiansyah, M.; Jasim, S.A.; Mohammad Pour, Z.G.; Athar, S.S.; Jeda, A.S.; Doewes, R.I.; Jalil, A.T.; Bokov, D.O.; Mustafa, Y.F.; Noroozbeygi, M.; et al. Coronavirus disease 2019 (COVID-19) update: From metabolic reprogramming to immunometabolism. J. Med. Virol. 2022, 94, 4611–4627. [Google Scholar] [CrossRef]
- Ryu, K.W.; Nandu, T.; Kim, J.; Challa, S.; DeBerardinis, R.J.; Kraus, W.L. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 2018, 360, aa5780. [Google Scholar] [CrossRef]
- Raniga, K.; Liang, C. Interferons: Reprogramming the Metabolic Network against Viral Infection. Viruses 2018, 10, 36. [Google Scholar] [CrossRef]
- Phipps, S.; Lam, C.E.; Mahalingam, S.; Newhouse, M.; Ramirez, R.; Rosenberg, H.F.; Foster, P.S.; Matthaei, K.I. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 2007, 110, 1578–1586. [Google Scholar] [CrossRef]
- Bajwa, G.; DeBerardinis, R.J.; Shao, B.; Hall, B.; Farrar, J.D.; Gill, M.A. Cutting Edge: Critical Role of Glycolysis in Human Plasmacytoid Dendritic Cell Antiviral Responses. J. Immunol. 2016, 196, 2004–2009. [Google Scholar] [CrossRef]
- Wu, D.; Sanin, D.E.; Everts, B.; Chen, Q.; Qiu, J.; Buck, M.D.; Patterson, A.; Smith, A.M.; Chang, C.H.; Liu, Z.; et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity 2016, 44, 1325–1336. [Google Scholar] [CrossRef]
- Smith, T.; Rohaim, M.A.; Munir, M. Mapping Molecular Gene Signatures Mediated by SARS-COV-2 and Large-Scale and Genome-Wide Transcriptomics Comparative Analysis among Respiratory Viruses of Medical Importance. Mol. Cell. Probes 2022, 64, 101820. [Google Scholar] [CrossRef]
- Bucasas, K.L.; Mian, A.I.; Demmler-Harrison, G.J.; Caviness, A.C.; Piedra, P.A.; Franco, L.M.; Shaw, C.A.; Zhai, Y.; Wang, X.; Bray, M.S.; et al. Global gene expression profiling in infants with acute respiratory syncytial virus bronchiolitis demonstrates systemic activation of interferon signaling networks. Pediatr. Infect. Dis. J. 2013, 32, e68–e76. [Google Scholar] [CrossRef]
- Liu, H.L.; Yeh, I.J.; Phan, N.N.; Wu, Y.H.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Jiang, J.Z.; et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect. Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Scalambra, L.; Triboli, L.; Ray, F.; Giorgi, F.M. Gene regulatory network inference resources: A practical overview. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194430. [Google Scholar] [CrossRef] [PubMed]
- Ramana, C.V. Insights into functional connectivity in mammalian signal transduction pathways by pairwise comparison of protein interaction partners of critical signaling hubs. Biomol. Concepts 2022, 13, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Ramana, C.V.; Das, B. Profiling transcription factor sub-networks in type I interferon signaling and in response to SARS-CoV-2 infection. Comput. Math. Biophys 2021, 9, 273–288. [Google Scholar] [CrossRef]
- Nguyen, L.C.; Renner, D.M.; Silva, D.; Yang, D.; Parenti, N.; Medina, K.M.; Nicolaescu, V.; Gula, H.; Drayman, N.; Valdespino, A.; et al. SARS-CoV2 diverges from other betacoronaviruses in only partially activating the IRE1a/XBP1 ER stress pathway in human lung-derive cells. mBio 2022, e0241522. [Google Scholar] [CrossRef] [PubMed]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef] [PubMed]
- Daamen, A.R.; Bachali, P.; Owen, K.A.; Kingsmore, K.M.; Hubbard, E.L.; Labonte, A.C.; Robl, R.; Shrotri, S.; Grammer, A.C.; Lipsky, P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021, 11, 7052. [Google Scholar] [CrossRef]
- Walters, K.A.; D’Agnillo, F.; Sheng, Z.M.; Kindrachuk, J.; Schwartzman, L.M.; Kuestner, R.E.; Chertow, D.S.; Golding, B.T.; Taubenberger, J.K.; Kash, J.C. 1918 pandemic influenza virus and Streptococcus pneumoniae co-infection results in activation of coagulation and widespread pulmonary thrombosis in mice and humans. J. Pathol. 2016, 238, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.; Sridhar, S.; Peng, R.; Phillips, J.E.; Cohn, R.G.; Burns, L.; Woods, J.; Ramanujam, M.; Loubeau, M.; Tyagi, G.; et al. Double-stranded RNA induces molecular and inflammatory signatures that are directly relevant to COPD. Mucosal. Immunol. 2013, 6, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Minor, R.A.; Linnmon, G.V.; Miller-DeGraff, L.; Dixon, D.; Andrews, D.M.; Kaufman, R.J.; Imani, F. Double-stranded RNA-activated protein kinase regulates early innate immune responses during respiratory syncytial virus infection. J. Interferon Cytokine Res. 2010, 30, 263–272. [Google Scholar] [CrossRef]
- Britto, C.J.; Niu, N.; Khanal, S.; Huleihel, L.; Herazo-Maya, J.D.; Thompson, A.; Sauler, M.; Slade, M.D.; Sharma, L.; Dela Cruz, C.S.; et al. BPIFA1 regulates lung neutrophil recruitment and interferon signaling during acute inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 2019, 316, L321–L333. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web- enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017. Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349. [Google Scholar] [CrossRef]
- Henrickson, K.J. Parainfluenza viruses. Clin. Microbiol. Rev. 2003, 16, 242–264. [Google Scholar] [CrossRef]
- Blanc, M.; Hsieh, W.Y.; Robertson, K.A.; Kropp, K.A.; Forster, T.; Shui, G.; Lacaze, P.; Watterson, S.; Griffiths, S.J.; Spann, N.J.; et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 2013, 38, 106–118. [Google Scholar] [CrossRef]
- Garcia-Ortega, M.B.; Lopez, G.J.; Jimenez, G.; Garcia-Garcia, J.A.; Conde, V.; Boulaiz, H.; Carrillo, E.; Perán, M.; Marchal, J.A.; Garcia, M.A. Clinical and therapeutic potential of protein kinase PKR in cancer and metabolism. Expert. Rev. Mol. Med. 2017, 19, e9. [Google Scholar] [CrossRef]
- Deutschmann, J.; Gramberg, T. SAMHD1 … and Viral Ways around It. Viruses 2021, 13, 395. [Google Scholar] [CrossRef]
- Dantoft, W.; Robertson, K.A.; Watkins, W.J.; Strobl, B.; Ghazal, P. Metabolic Regulators Nampt and Sirt6 Serially Participate in the Macrophage Interferon Antiviral Cascade. Front. Microbiol. 2019, 10, 355. [Google Scholar] [CrossRef]
- Fiore, A.; Zeitler, L.; Russier, M.; Groß, A.; Hiller, M.K.; Parker, J.L.; Stier, L.; Köcher, T.; Newstead, S.; Murray, P.J. Kynurenine importation by SLC7A11 propagates anti-ferroptotic signaling. Mol. Cell 2022, 82, 920–932.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Chiou, J.; Poirion, O.B.; Buchanan, J.; Valdez, M.J.; Verheyden, J.M.; Hou, X.; Kudtarkar, P.; Narendra, S.; Newsome, J.M.; et al. Single-cell multi omic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife 2020, 9, e62522. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, V.; Ravà, M.; Marotta, D.; Di Lucia, P.; Laura, C.; Sala, E.; Grillo, M.; Bono, E.; Giustini, L.; Perucchini, C.; et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 2022, 7, eabl9929. [Google Scholar] [CrossRef] [PubMed]
- Michael, P.; Brabant, D.; Bleiblo, F.; Ramana, C.V.; Rutherford, M.; Khurana, S.; Tai, T.C.; Kumar, A.; Kumar, A. Influenza A induced cellular signal transduction pathways. J. Thorac. Dis. 2013, 5 (Suppl. 2), S132–S141. [Google Scholar] [PubMed]
- Kash, J.C.; Tumpey, T.M.; Proll, S.C.; Carter, V.; Perwitasari, O.; Thomas, M.J.; Basler, C.F.; Palese, P.; Taubenberger, J.K.; García-Sastre, A.; et al. Genomic analysis of increased host immune response and cell death responses induced by 1918 influenza virus. Nature 2008, 443, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Han, C.W.; Jeong, M.S.; Jang, S.B. Structure and Function of the Influenza A Virus Non-Structural Protein 1. J. Microbiol. Biotechnol. 2019, 29, 1184–1192. [Google Scholar] [CrossRef]
- Bleiblo, F.; Michael, P.; Brabant, D.; Ramana, C.V.; Tai, T.; Saleh, M.; Parrillo, J.E.; Kumar, A.; Kumar, A. JAK kinases are required for the bacterial RNA and poly I:C induced tyrosine phosphorylation of PKR. Int. J. Clin. Exp. Med. 2013, 6, 16–25. [Google Scholar]
- Dalet, A.; Gatti, E.; Pierre, P. Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Lett. 2015, 589, 1539–1545. [Google Scholar] [CrossRef]
- Samuel, C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Kerr, C.H.; Skinnider, M.A.; Andrews, D.D.T.; Madero, A.M.; Chan, Q.W.T.; Stacey, R.G.; Stoynov, N.; Jan, E.; Foster, L.J. Dynamic rewiring of the human interactome by interferon signaling. Genome Biol. 2020, 21, 140. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Sharma, S.D.; Kumar, A.; Ende, Z.; Mishina, M.; Wang, Y.; Falls, Z.; Samudrala, R.; Pohl, J.; Knight, P.R.; et al. Antiviral Approaches against Influenza Virus. Clin. Microbiol. Rev. 2023, 36, e0004022. [Google Scholar] [CrossRef] [PubMed]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef] [PubMed]
- Zamyatina, A.; Heine, H. Lipopolysaccharide Recognition in the Crossroads of TLR4 and Caspase-4/11 Mediated Inflammatory Pathways. Front. Immunol. 2020, 11, 585146. [Google Scholar] [CrossRef] [PubMed]
- Beutler, B. LPS in microbial pathogenesis: Promise and fulfillment. J. Endotoxin Res. 2002, 8, 329–335. [Google Scholar] [CrossRef]
- Czerkies, M.; Korwek, Z.; Prus, W.; Kochańczyk, M.; Jaruszewicz-Błońska, J.; Tudelska, K.; Błoński, S.; Kimmel, M.; Brasier, A.R.; Lipniacki, T. Cell fate in antiviral response arises in the crosstalk of IRF, NF-κB and JAK/STAT pathways. Nat. Commun. 2018, 9, 493. [Google Scholar] [CrossRef]
- Liu, S.Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013, 38, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Valverde-Estrella, L.; López-Serrat, M.; Sánchez-Sànchez, G.; Vico, T.; Lloberas, J.; Celada, A. Induction of Samhd1 by interferon gamma and lipopolysaccharide in murine macrophages requires IRF1. Eur. J. Immunol. 2020, 50, 1321–1334. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bonifati, S.; Qin, Z.; St Gelais, C.; Kodigepalli, K.M.; Barrett, B.S.; Kim, S.H.; Antonucci, J.M.; Ladner, K.J.; Buzovetsky, O.; et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways. Proc. Natl. Acad. Sci. USA 2018, 115, E3798–E3807. [Google Scholar] [CrossRef]
- Tsou, Y.A.; Tung, M.C.; Alexander, K.A.; Chang, W.D.; Tsai, M.H.; Chen, H.L.; Chen, C.M. The Role of BPIFA1 in Upper Airway Microbial Infections and Correlated Diseases. BioMed Res. Int. 2018, 2018, 2021890. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, Z.; Li, Y.; Li, Y. The Regulation of Integrated Stress Response Signaling Pathway on Viral Infection and Viral Antagonism. Front. Microbiol. 2022, 12, 814635. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.C.; Yang, D.; Nicolaescu, V.; Best, T.J.; Gula, H.; Saxena, D.; Gabbard, J.D.; Chen, S.N.; Ohtsuki, T.; Friesen, J.B.; et al. Cannabidiol inhibits SARS-CoV-2 replication through induction of the host ER stress and innate immune responses. Sci. Adv. 2022, 8, eabi6110. [Google Scholar] [CrossRef]
- Stone, T.W.; Williams, R.O. Interactions of IDO and the Kynurenine Pathway with Cell Transduction Systems and Metabolism at the Inflammation-Cancer Interface. Cancers 2023, 15, 2895. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Wang, T. Metabolic Reprogramming in COVID-19. Int. J. Mol. Sci. 2021, 22, 11475. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Wu, Q.; Wang, X.; Qin, Z.; Wang, Y.; He, Y.; Wu, Q.; Li, L.; Chen, H. Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 2022, 13, 235. [Google Scholar] [CrossRef]
- Costantini, S.; Madonna, G.; Di Gennaro, E.; Capone, F.; Bagnara, P.; Capone, M.; Sale, S.; Nicastro, C.; Atripaldi, L.; Fiorentino, G.; et al. New Insights into the Identification of Metabolites and Cytokines Predictive of Outcome for Patients with Severe SARS-CoV-2 Infection Showed Similarity with Cancer. Int. J. Mol. Sci. 2023, 24, 4922. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity tomild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Miao, V.N.; Owings, A.H.; Navia, A.W.; Tang, Y.; Bromley, J.D.; Lotfy, P.; Sloan, M.; Laird, H.; Williams, H.B.; et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 2021, 184, 4713–4733.e22. [Google Scholar] [CrossRef]
- Kumar, A.; Yang, Y.L.; Flati, V.; Der, S.; Kadereit, S.; Deb, A.; Haque, J.; Reis, L.; Weissmann, C.; Williams, B.R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: Role of IRF-1 and NF-kappaB. EMBO J. 1997, 16, 406–416. [Google Scholar] [CrossRef]
- Bleiblo, F.; Michael, P.; Brabant, D.; Ramana, C.V.; Tai, T.; Saleh, M.; Parrillo, J.E.; Kumar, A.; Kumar, A. Bacterial RNA induces myocyte cellular dysfunction through the activation of PKR. J. Thorac. Dis. 2012, 4, 114–125. [Google Scholar]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Hull, C.M.; Bevilacqua, P.C. Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR. Acc. Chem. Res. 2016, 49, 1242–1249. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, Z.; Xiong, Y.; Zhong, Z.; Ye, Q. Multiple Roles of 25-Hydroxycholesterol in Lipid Metabolism, Antivirus Process, Inflammatory Response, and Cell Survival. Oxid. Med. Cell Longev. 2020, 2020, 8893305. [Google Scholar] [CrossRef] [PubMed]
- Gold, E.S.; Diercks, A.H.; Podolsky, I.; Podyminogin, R.L.; Askovich, P.S.; Treuting, P.M.; Aderem, A. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 10666–10671. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Scott, A.L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 2010, 88, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.; Muir, L.; Geletka, L.; Delproposto, J.; Baker, N.; Flesher, C.; O’Rourke, R.; Lumeng, C.N. Cholesterol 25-hydroxylase (CH25H) as a promoter of adipose tissue inflammation in obesity and diabetes. Mol. Metab. 2020, 39, 100983. [Google Scholar] [CrossRef]
- Xu, Y.; Nasri, M.; Dannenmann, B.; Mir, P.; Zahabi, A.; Welte, K.; Morishima, T.; Skokowa, J. NAMPT/SIRT2-mediated inhibition of the p53-p21 signaling pathway is indispensable for maintenance and hematopoietic differentiation of human iPS cells. Stem Cell Res. Ther. 2021, 12, 112. [Google Scholar] [CrossRef]
- Liu, T.F.; Yoza, B.K.; El Gazzar, M.; Vachharajani, V.T.; McCall, C.E. NAD+-dependent SIRT1 deacetylase participates in epigenetic reprogramming during endotoxin tolerance. J. Biol. Chem. 2011, 286, 9856–9864. [Google Scholar] [CrossRef]
- Zhai, L.; Ladomersky, E.; Lenzen, A.; Nguyen, B.; Patel, R.; Lauing, K.L.; Wu, M.; Wainwright, D.A. IDO1 in cancer: A Gemini of immune checkpoints. Cell Mol. Immunol. 2018, 15, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.M.; Zhou, L.; Cui, J.; Li, L.; Wu, N.; Yu, A.; Poddar, S.; Liang, K.; Abt, E.R.; Kim, S.; et al. NAD+ depletion by type I interferon signaling sensitizes pancreatic cancer cells to NAMPT inhibition. Proc. Natl. Acad. Sci. USA 2021, 118, e2012469118. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).