The Role of IRF8 Polymorphisms in Systemic Sclerosis Development and Pathogenesis
Abstract
1. Introduction
1.1. Genetics of SSc
1.2. Manifestation of SSc and Subtypes
1.3. Type I Interferon in SSc
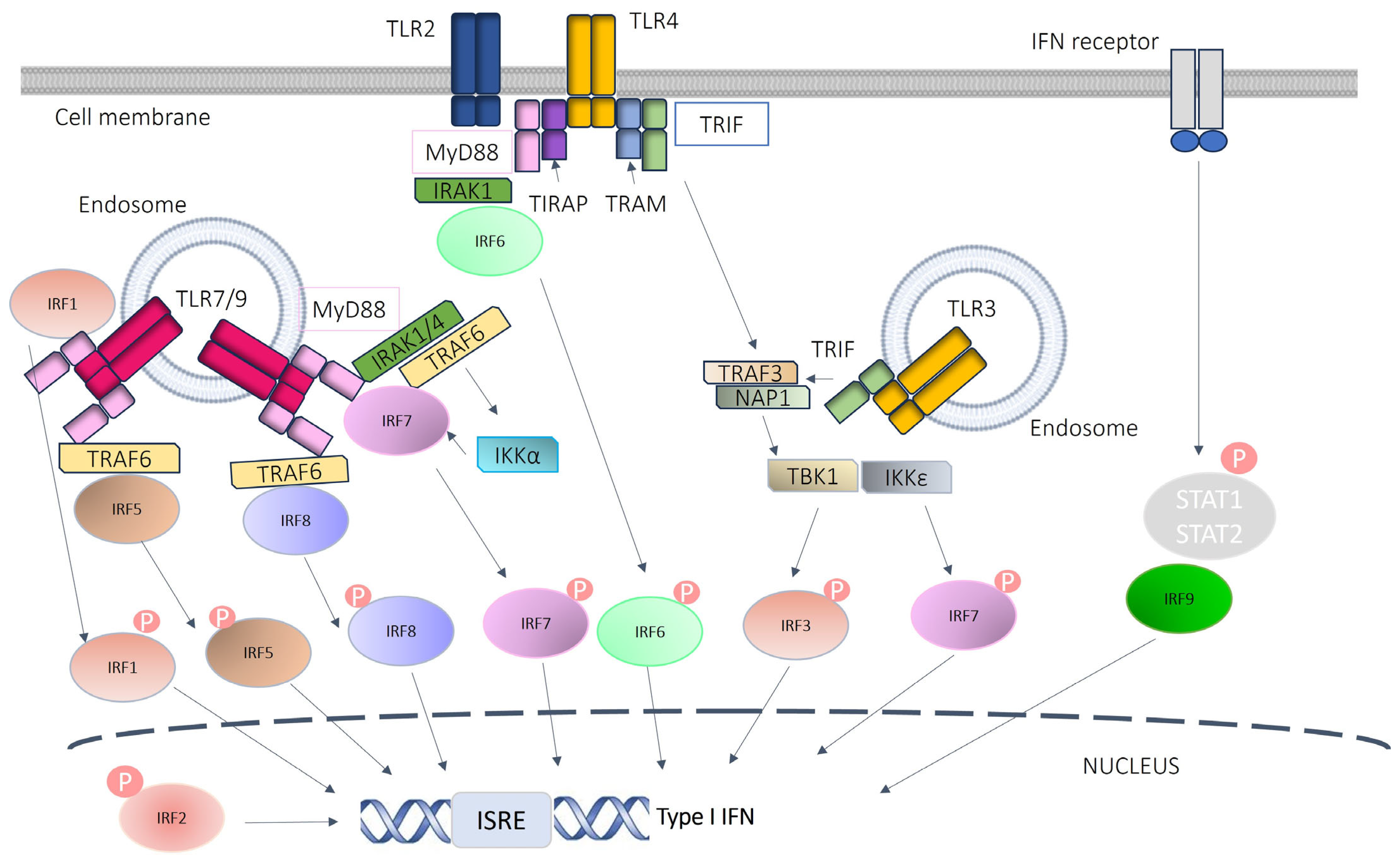
1.4. IRF Family in Humans and Mice
1.5. IRF Genes in SSc
1.6. Role of IRF8 in Autoimmune Diseases
1.7. IRF8 in SSc
2. Specific Role of IRF8 in SSc
2.1. IRF8-KLF4 (Kruppel-like Factor 4) Interaction
2.2. GWAS (Genome-Wide Association Study)
2.3. IRF8 Expression in SSc Subtypes
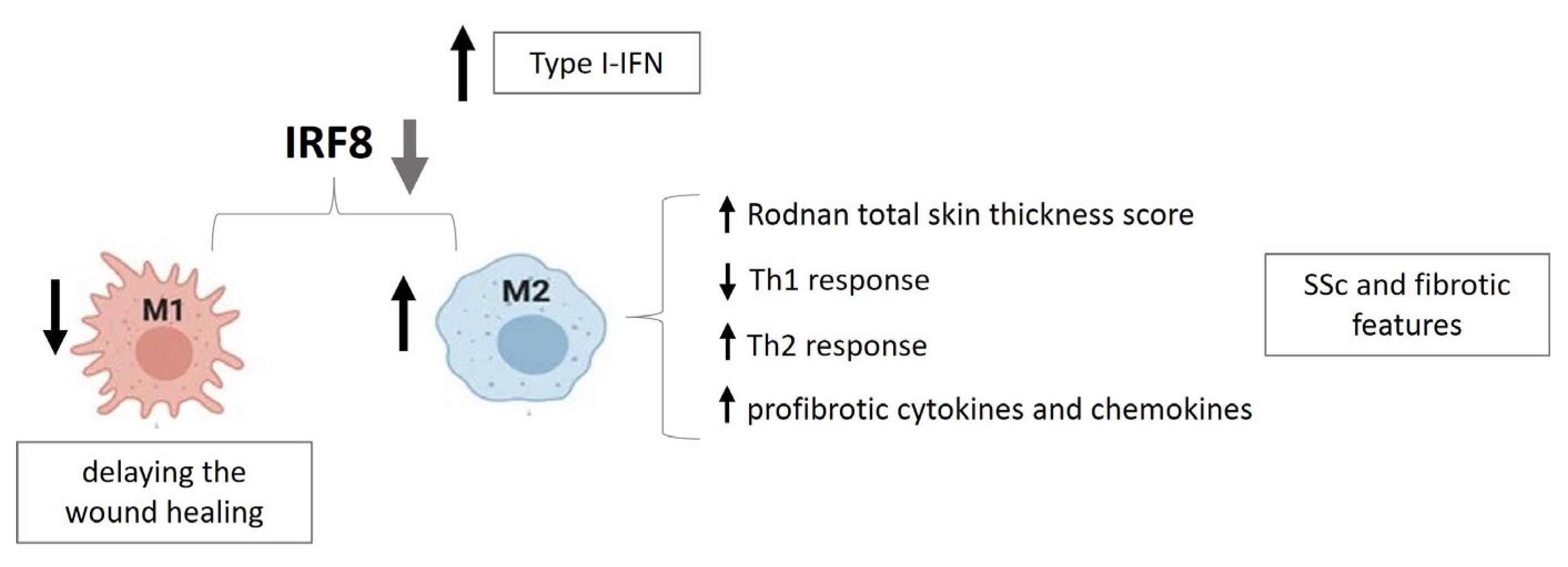
2.4. IRF8 in SSc Fibrosis
2.5. IRF8 SNPs and SSc Susceptibility
2.6. Physical Interaction with Chromatin for IRF8 Expression in Monocytes
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kucharz, E.J.; Kopeć-Mędrek, M. Systemic Sclerosis Sine Scleroderma. Adv. Clin. Exp. Med. Off. Organ. Wroc. Med. Univ. 2017, 26, 875–880. [Google Scholar] [CrossRef]
- Varga, J.; Abraham, D. Systemic Sclerosis: A Prototypic Multisystem Fibrotic Disorder. J. Clin. Investig. 2007, 117, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Furst, D.E.; Fernandes, A.W.; Iorga, S.R.; Greth, W.; Bancroft, T. Epidemiology of Systemic Sclerosis in a Large US Managed Care Population. J. Rheumatol. 2012, 39, 784–786. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Wei, J.; Varga, J. Understanding Fibrosis in Systemic Sclerosis: Shifting Paradigms, Emerging Opportunities. Nat. Rev. Rheumatol. 2011, 8, 42–54. [Google Scholar] [CrossRef]
- Mathai, S.C.; Hassoun, P.M. Pulmonary Arterial Hypertension Associated with Systemic Sclerosis. Expert Rev. Respir. Med. 2011, 5, 267–279. [Google Scholar] [CrossRef]
- Arnett, F.; Cho, M.; Chatterjee, S.; Aguilar, M.; Reveille, J.; Mayes, M. Familial Occurrence Frequencies and Relative Risks for Systemic Sclerosis (Scleroderma) in Three United States Cohorts. Arthritis Rheum. 2001, 44, 1359–1362. [Google Scholar] [CrossRef]
- Stafford, L.; Englert, H.J.; Gover, J.; Bertouch, J.V. Distribution of Macrovascular Disease in Scleroderma. Ann. Rheum. Dis. 1998, 57, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Feghali-Bostwick, C.; Medsger, T.A.; Wright, T.M. Analysis of Systemic Sclerosis in Twins Reveals Low Concordance for Disease and High Concordance for the Presence of Antinuclear Antibodies. Arthritis Rheum. 2003, 48, 1956–1963. [Google Scholar] [CrossRef] [PubMed]
- Ünlü, B.; Tursen, U.; Rajabi, Z.; Jabalameli, N.; Rajabi, F. The Immunogenetics of Systemic Sclerosis. Adv. Exp. Med. Biol. 2022, 1367, 259–298. [Google Scholar]
- Luo, Y.; Wang, Y.; Wang, Q.; Xiao, R.; Lu, Q. Systemic Sclerosis: Genetics and Epigenetics. J. Autoimmun. 2013, 41, 161–167. [Google Scholar] [CrossRef]
- Van den Hoogen, F.; Khanna, D.; Fransen, J.; Johnson, S.R.; Baron, M.; Tyndall, A.; Matucci-Cerinic, M.; Naden, R.P.; Medsger, T.A.; Carreira, P.E.; et al. 2013 Classification Criteria for Systemic Sclerosis: An American College of Rheumatology/European League against Rheumatism Collaborative Initiative. Arthritis Rheum. 2013, 65, 2737–2747. [Google Scholar] [CrossRef]
- Denton, C.P.; Khanna, D. Systemic Sclerosis. Lancet Lond. Engl. 2017, 390, 1685–1699. [Google Scholar] [CrossRef]
- Pope, J.E.; Denton, C.P.; Johnson, S.R.; Fernandez-Codina, A.; Hudson, M.; Nevskaya, T. State-of-the-Art Evidence in the Treatment of Systemic Sclerosis. Nat. Rev. Rheumatol. 2023, 19, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Avouac, J.; Fransen, J.; Walker, U.A.; Riccieri, V.; Smith, V.; Muller, C.; Miniati, I.; Tarner, I.H.; Randone, S.B.; Cutolo, M.; et al. Preliminary Criteria for the Very Early Diagnosis of Systemic Sclerosis: Results of a Delphi Consensus Study from EULAR Scleroderma Trials and Research Group. Ann. Rheum. Dis. 2011, 70, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Muangchan, C.; Baron, M.; Pope, J. The 15% Rule in Scleroderma: The Frequency of Severe Organ Complications in Systemic Sclerosis. A Systematic Review. J. Rheumatol. 2013, 40, 121380. [Google Scholar] [CrossRef] [PubMed]
- Tyndall, A.J.; Bannert, B.; Vonk, M.; Airò, P.; Cozzi, F.; Carreira, P.E.; Bancel, D.F.; Allanore, Y.; Müller-Ladner, U.; Distler, O.; et al. Causes and Risk Factors for Death in Systemic Sclerosis: A Study from the EULAR Scleroderma Trials and Research (EUSTAR) Database. Ann. Rheum. Dis. 2010, 69, 1809–1815. [Google Scholar] [CrossRef]
- Hinz, B.; Phan, S.H.; Thannickal, V.J.; Prunotto, M.; Desmoulière, A.; Varga, J.; De Wever, O.; Mareel, M.; Gabbiani, G. Recent Developments in Myofibroblast Biology: Paradigms for Connective Tissue Remodeling. Am. J. Pathol. 2012, 180, 1340–1355. [Google Scholar] [CrossRef] [PubMed]
- Rodnan, G.P.; Fennell, R.H., Jr. Progressive Systemic Sclerosis Sine Scleroderma. JAMA 1962, 180, 665–670. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, R.; Ferri, C.; Giuggioli, D.; Bajocchi, G.; Dagna, L.; Bellando-Randone, S.; Zanframundo, G.; Foti, R.; Cacciapaglia, F.; Cuomo, G.; et al. Systemic Sclerosis Sine Scleroderma: Clinical and Serological Features and Relationship with Other Cutaneous Subsets in a Large Series of Patients from the National Registry ‘SPRING’ of the Italian Society for Rheumatology. RMD Open 2023, 9, e002890. [Google Scholar] [CrossRef] [PubMed]
- Samha, R.; Ghaddar, S.A.; Raya, M.; Alhadi, S.A. Systemic Sclerosis Sine Scleroderma with Atypical Clinical Course: A Rare Case Report. Ann. Med. Surg. 2023, 85, 5656–5661. [Google Scholar] [CrossRef]
- van Leeuwen, N.M.; Boonstra, M.; Bakker, J.A.; Grummels, A.; Jordan, S.; Liem, S.; Distler, O.; Hoffmann-Vold, A.-M.; Melsens, K.; Smith, V.; et al. Anticentromere Antibody Levels and Isotypes and the Development of Systemic Sclerosis. Arthritis Rheumatol. 2021, 73, 2338–2347. [Google Scholar] [CrossRef]
- Liem, S.I.E.; Neppelenbroek, S.; Fehres, C.M.; Wevers, B.A.; Toes, R.E.M.; Allaart, C.F.; Huizinga, T.W.J.; Scherer, H.U.; De, J.K. Vries-Bouwstra Progression from Suspected to Definite Systemic Sclerosis and the Role of Anti-Topoisomerase I Antibodies. RMD Open 2023, 9, e002827. [Google Scholar] [CrossRef]
- Cavazzana, I.; Angela, C.; Paolo, A.; Stefania, Z.; Angela, T.; Franco, F. Anti-RNA Polymerase III Antibodies: A Marker of Systemic Sclerosis with Rapid Onset and Skin Thickening Progression. Autoimmun. Rev. 2009, 8, 580–584. [Google Scholar] [CrossRef]
- Tall, F.; Dechomet, M.; Riviere, S.; Cottin, V.; Ballot, E.; Tiev, K.P.; Montin, R.; Morin, C.; Chantran, Y.; Grange, C.; et al. The Clinical Relevance of Antifibrillarin (Anti-U3-RNP) Autoantibodies in Systemic Sclerosis. Scand. J. Immunol. 2017, 85, 73–79. [Google Scholar] [CrossRef]
- Assassi, S.; Mayes, M.D.; Arnett, F.C.; Gourh, P.; Agarwal, S.K.; McNearney, T.A.; Chaussabel, D.; Oommen, N.; Fischbach, M.; Shah, K.R.; et al. Systemic Sclerosis and Lupus: Points in an Interferon-mediated Continuum. Arthritis Rheum. 2010, 62, 589–598. [Google Scholar] [CrossRef]
- Brkic, Z.; van Bon, L.; Cossu, M.; van Helden-Meeuwsen, C.G.; Vonk, M.C.; Knaapen, H.; van den Berg, W.; Dalm, V.A.; Van Daele, P.L.; Severino, A.; et al. The Interferon Type I Signature Is Present in Systemic Sclerosis before Overt Fibrosis and Might Contribute to Its Pathogenesis through High BAFF Gene Expression and High Collagen Synthesis. Ann. Rheum. Dis. 2016, 75, 1567. [Google Scholar] [CrossRef]
- Solans, R.; Bosch, J.; Esteban, I.; Vilardell, M. Systemic Sclerosis Developing in Association with the Use of Interferon Alpha Therapy for Chronic Viral Hepatitis. Clin. Exp. Rheumatol. 2004, 22, 625–628. [Google Scholar]
- Kim, D.; Peck, A.; Santer, D.; Patole, P.; Schwartz, S.M.; Molitor, J.A.; Arnett, F.C.; Elkon, K.B. Induction of Interferon-α by Scleroderma Sera Containing Autoantibodies to Topoisomerase I: Association of Higher Interferon-α Activity with Lung Fibrosis. Arthritis Rheum. 2008, 58, 2163–2173. [Google Scholar] [CrossRef]
- Eloranta, M.-L.; Franck-Larsson, K.; Lövgren, T.; Kalamajski, S.; Rönnblom, A.; Rubin, K.; Alm, G.; Rönnblom, L. Type I Interferon System Activation and Association with Disease Manifestations in Systemic Sclerosis. Ann. Rheum. Dis. 2010, 69, 1396–1402. [Google Scholar] [CrossRef]
- Barrat, F.J.; Su, L. A Pathogenic Role of Plasmacytoid Dendritic Cells in Autoimmunity and Chronic Viral Infection. J. Exp. Med. 2019, 216, 1974–1985. [Google Scholar] [CrossRef]
- Pelka, K.; Shibata, T.; Miyake, K.; Latz, E. Nucleic Acid-Sensing TLRs and Autoimmunity: Novel Insights from Structural and Cell Biology. Immunol. Rev. 2016, 269, 60–75. [Google Scholar] [CrossRef]
- Ah Kioon, M.D.; Tripodo, C.; Fernandez, D.; Kirou, K.A.; Spiera, R.F.; Crow, M.K.; Gordon, J.K.; Barrat, F.J. Plasmacytoid Dendritic Cells Promote Systemic Sclerosis with a Key Role for TLR8. Sci. Transl. Med. 2018, 10, eaam8458. [Google Scholar] [CrossRef]
- Lande, R.; Lee, E.Y.; Palazzo, R.; Marinari, B.; Pietraforte, I.; Santos, G.S.; Mattenberger, Y.; Spadaro, F.; Stefanantoni, K.; Iannace, N.; et al. CXCL4 Assembles DNA into Liquid Crystalline Complexes to Amplify TLR9-Mediated Interferon-α Production in Systemic Sclerosis. Nat. Commun. 2019, 10, 1731. [Google Scholar] [CrossRef]
- Frasca, L.; Lande, R. Toll-like Receptors in Mediating Pathogenesis in Systemic Sclerosis. Clin. Exp. Immunol. 2020, 201, 14–24. [Google Scholar] [CrossRef]
- Antonczyk, A.; Krist, B.; Sajek, M.; Michalska, A.; Piaszyk-Borychowska, A.; Plens-Galaska, M.; Wesoly, J.; Bluyssen, H.A.R. Direct Inhibition of IRF-Dependent Transcriptional Regulatory Mechanisms Associated with Disease. Front. Immunol. 2019, 10, 1176. [Google Scholar] [CrossRef]
- Fujita, T.; Sakakibara, J.; Sudo, Y.; Miyamoto, M.; Kimura, Y.; Taniguchi, T. Evidence for a Nuclear Factor(s), IRF-1, Mediating Induction and Silencing Properties to Human IFN-beta Gene Regulatory Elements. EMBO J. 1988, 7, 3397–3405. [Google Scholar] [CrossRef]
- Miyamoto, M.; Fujita, T.; Kimura, Y.; Maruyama, M.; Harada, H.; Sudo, Y.; Miyata, T.; Taniguchi, T. Regulated Expression of a Gene Encoding a Nuclear Factor, IRF-1, That Specifically Binds to IFN-β Gene Regulatory Elements. Cell 1988, 54, 903–913. [Google Scholar] [CrossRef]
- Taniguchi, T.; Ogasawara, K.; Takaoka, A.; Tanaka, N. IRF Family of Transcription Factors as Regulators of Host Defense. Annu. Rev. Immunol. 2001, 19, 623–655. [Google Scholar] [CrossRef]
- Levy, D.E.; Marié, I.; Smith, E.; Prakash, A. Review: Enhancement and Diversification of IFN Induction by IRF-7-Mediated Positive Feedback. J. Interferon Cytokine Res. 2002, 22, 87–93. [Google Scholar] [CrossRef]
- Gutiérrez, P.; Delgado, M.D.; Richard, C.; Moreau-Gachelin, F.; León, J. Interferon Induces Up-Regulation of Spi-1/PU.1 in Human Leukemia K562 Cells. Biochem. Biophys. Res. Commun. 1997, 240, 862–868. [Google Scholar] [CrossRef]
- Taniguchi, T.; Takaoka, A. A Weak Signal for Strong Responses: Interferon-Alpha/Beta Revisited. Nat. Rev. Mol. Cell Biol. 2001, 2, 378–386. [Google Scholar] [CrossRef]
- Wu, M.; Assassi, S. The Role of Type 1 Interferon in Systemic Sclerosis. Front. Immunol. 2013, 4, 266. [Google Scholar] [CrossRef]
- Donn, R.P.; Barrett, J.H.; Farhan, A.; Stopford, A.; Pepper, L.; Shelley, E.; Davies, N.; Ollier, W.E.R.; Thomson, W.; British Paediatric Rheumatology Study Group. Cytokine Gene Polymorphisms and Susceptibility to Juvenile Idiopathic Arthritis. Arthritis Rheum. 2001, 44, 802–810. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kang, S.W.; Song, J.K.; Baek, H.J.; Choi, H.J.; Bae, Y.D.; Ryu, H.J.; Lee, E.Y.; Lee, E.B.; Song, Y.W. Associations between Interferon Regulatory Factor–1 Polymorphisms and Behçet’s Disease. Hum. Immunol. 2007, 68, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, G.; Calcagno, G.; Bresciamorra, V.; Salvatore, E.; Filla, A.; Capone, S.; Liguori, R.; Borelli, S.; Gentile, I.; Borrelli, F.; et al. Multiple Sclerosis and Hepatitis C Virus Infection Are Associated with Single Nucleotide Polymorphisms in Interferon Pathway Genes. J. Interferon Cytokine Res. 2008, 28, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.; Sturge, M.; Rahman, P.; Woods, M.O. Autosome-Wide Copy Number Variation Association Analysis for Rheumatoid Arthritis Using the WTCCC High-Density SNP Genotype Data. J. Rheumatol. 2011, 38, 797. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, J.; Aegerter, S.; Fevurly, R.D.; Mammoto, A.; Mammoto, T.; Sahin, M.; Mably, J.D.; Fishman, S.J.; Chan, J. RASA1 Functions in EPHB4 Signaling Pathway to Suppress Endothelial mTORC1 Activity. J. Clin. Investig. 2014, 124, 2774–2784. [Google Scholar] [CrossRef] [PubMed]
- Akahoshi, M.; Nakashima, H.; Sadanaga, A.; Miyake, K.; Obara, K.; Tamari, M.; Hirota, T.; Matsuda, A.; Shirakawa, T. Promoter Polymorphisms in the IRF3 Gene Confer Protection against Systemic Lupus Erythematosus. Lupus 2008, 17, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, S.; Nordmark, G.; Göring, H.H.H.; Lindroos, K.; Wiman, A.-C.; Sturfelt, G.; Jönsen, A.; Rantapää-Dahlqvist, S.; Möller, B.; Kere, J.; et al. Polymorphisms in the Tyrosine Kinase 2 and Interferon Regulatory Factor 5 Genes Are Associated with Systemic Lupus Erythematosus. Am. J. Hum. Genet. 2005, 76, 528–537. [Google Scholar] [CrossRef]
- Jia, X.; Hu, M.; Lin, Q.; Ren, H. Association of the IRF5 Rs2004640 Polymorphism with Rheumatoid Arthritis: A Meta-Analysis. Rheumatol. Int. 2013, 33, 2757–2761. [Google Scholar] [CrossRef] [PubMed]
- Kristjansdottir, G.; Sandling, J.K.; Bonetti, A.; Roos, I.M.; Milani, L.; Wang, C.; Gustafsdottir, S.M.; Sigurdsson, S.; Lundmark, A.; Tienari, P.J.; et al. Interferon Regulatory Factor 5 (IRF5) Gene Variants Are Associated with Multiple Sclerosis in Three Distinct Populations. J. Med. Genet. 2008, 45, 362. [Google Scholar] [CrossRef]
- Fu, Q.; Zhao, J.; Qian, X.; Wong, J.L.H.; Kaufman, K.M.; Yu, C.Y.; Hwee Siew Howe and the Tan Tock Seng Hospital Lupus Study Group; Mok, M.Y.; Harley, J.B.; Guthridge, J.M.; et al. Association of a Functional IRF7 Variant with Systemic Lupus Erythematosus. Arthritis Rheum. 2011, 63, 749–754. [Google Scholar] [CrossRef]
- Cunninghame Graham, D.S.; Morris, D.L.; Bhangale, T.R.; Criswell, L.A.; Syvänen, A.-C.; Rönnblom, L.; Behrens, T.W.; Graham, R.R.; Vyse, T.J. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with Systemic Lupus Erythematosus. PLoS Genet. 2011, 7, e1002341. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Zhang, P.; Li, H. Interferon Regulatory Factor Signalings in Cardiometabolic Diseases. Hypertension 2015, 66, 222–247. [Google Scholar] [CrossRef]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2018, 10, a028423. [Google Scholar] [CrossRef]
- Dieudé, P.; Guedj, M.; Wipff, J.; Avouac, J.; Fajardy, I.; Diot, E.; Granel, B.; Sibilia, J.; Cabane, J.; Mouthon, L.; et al. Association between the IRF5 Rs2004640 Functional Polymorphism and Systemic Sclerosis: A New Perspective for Pulmonary Fibrosis. Arthritis Rheum. 2009, 60, 225–233. [Google Scholar] [CrossRef]
- Ito, I.; Kawaguchi, Y.; Kawasaki, A.; Hasegawa, M.; Ohashi, J.; Hikami, K.; Kawamoto, M.; Fujimoto, M.; Takehara, K.; Sato, S.; et al. Association of a Functional Polymorphism in the IRF5 Region With Systemic Sclerosis in a Japanese Population. Arthritis Rheum. 2009, 60, 1845–1850. [Google Scholar] [CrossRef]
- Sharif, R.; Mayes, M.; Tan, F.; Gorlova, O.; Hummers, L.; Shah, A.; Furst, D.; Khanna, D.; Martin, J.; Bossini-Castillo, L.; et al. IRF5 Polymorphism Predicts Prognosis in Patients with Systemic Sclerosis. Ann. Rheum. Dis. 2012, 71, 1197–1202. [Google Scholar] [CrossRef]
- González-Serna, D.; Shi, C.; Kerick, M.; Hankinson, J.; Ding, J.; McGovern, A.; Tutino, M.; Villanueva Martin, G.; Ortego-Centeno, N.; Callejas, J.L.; et al. Identification of Mechanisms by Which Genetic Susceptibility Loci Influence Systemic Sclerosis Risk Using Functional Genomics in Primary T Cells and Monocytes. Arthritis Rheumatol. 2022, 75, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- López-Isac, E.; Martín, J.-E.; Assassi, S.; Simeón, C.P.; Carreira, P.; Ortego-Centeno, N.; Freire, M.; Beltrán, E.; Narváez, J.; Alegre-Sancho, J.J.; et al. Brief Report: IRF4 Newly Identified as a Common Susceptibility Locus for Systemic Sclerosis and Rheumatoid Arthritis in a Cross-Disease Meta-Analysis of Genome-Wide Association Studies. Arthritis Rheumatol. 2016, 68, 2338–2344. [Google Scholar] [CrossRef] [PubMed]
- Radstake, T.R.D.J.; Gorlova, O.; Rueda, B.; Martin, J.-E.; Alizadeh, B.Z.; Palomino-Morales, R.; Coenen, M.J.; Vonk, M.C.; Voskuyl, A.E.; Schuerwegh, A.J.; et al. Genome-Wide Association Study of Systemic Sclerosis Identifies CD247 as a New Susceptibility Locus. Nat. Genet. 2010, 42, 426–429. [Google Scholar] [CrossRef]
- Carmona-Pérez, L.; Dagenais-Lussier, X.; Mai, L.T.; Stögerer, T.; Swaminathan, S.; Isnard, S.; Rice, M.R.; Barnes, B.J.; Routy, J.-P.; van Grevenynghe, J.; et al. The TLR7/IRF-5 Axis Sensitizes Memory CD4+ T Cells to Fas-Mediated Apoptosis during HIV-1 Infection. JCI Insight 2023, 8, e167329. [Google Scholar] [CrossRef]
- Gorlova, O.; Martin, J.-E.; Rueda, B.; Koeleman, B.P.C.; Ying, J.; Teruel, M.; Diaz-Gallo, L.-M.; Broen, J.C.; Vonk, M.C.; Simeon, C.P.; et al. Identification of Novel Genetic Markers Associated with Clinical Phenotypes of Systemic Sclerosis through a Genome-Wide Association Strategy. PLoS Genet. 2011, 7, e1002178. [Google Scholar] [CrossRef]
- Arismendi, M.; Giraud, M.; Ruzehaji, N.; Dieudé, P.; Koumakis, E.; Ruiz, B.; Airo, P.; Cusi, D.; Matucci-Cerinic, M.; Salvi, E.; et al. Identification of NF-κB and PLCL2 as New Susceptibility Genes and Highlights on a Potential Role of IRF8 through Interferon Signature Modulation in Systemic Sclerosis. Arthritis Res. Ther. 2015, 17, 71. [Google Scholar] [CrossRef]
- Tan, F.K.; Zhou, X.; Mayes, M.D.; Gourh, P.; Guo, X.; Marcum, C.; Jin, L.; Arnett, F.C., Jr. Signatures of Differentially Regulated Interferon Gene Expression and Vasculotrophism in the Peripheral Blood Cells of Systemic Sclerosis Patients. Rheumatology 2006, 45, 694–702. [Google Scholar] [CrossRef]
- De Oliveira, D.B.; de Freitas Almeida, G.M.; Guedes, A.C.M.; Santos, F.P.S.T.; Bonjardim, C.A.; Ferreira, P.C.P.; Kroon, E.G. Basal Activation of Type I Interferons (Alpha2 and Beta) and OAS Genes: Insights into Differential Expression Profiles of Interferon System Components in Systemic Sclerosis. Int. J. Rheumatol. 2011, 2011, 275617. [Google Scholar] [CrossRef]
- Kanno, Y.; Levi, B.-Z.; Tamura, T.; Ozato, K. Immune Cell-Specific Amplification of Interferon Signaling by the IRF-4/8-PU.1 Complex. J. Interferon Cytokine Res. 2005, 25, 770–779. [Google Scholar] [CrossRef]
- Moorman, H.R.; Reategui, Y.; Poschel, D.B.; Liu, K. IRF8: Mechanism of Action and Health Implications. Cells 2022, 11, 2630. [Google Scholar] [CrossRef]
- Driggers, P.H.; Ennist, D.L.; Gleason, S.L.; Mak, W.H.; Marks, M.S.; Levi, B.Z.; Flanagan, J.R.; Appella, E.; Ozato, K. An Interferon Gamma-Regulated Protein That Binds the Interferon-Inducible Enhancer Element of Major Histocompatibility Complex Class I Genes. Proc. Natl. Acad. Sci. USA 1990, 87, 3743–3747. [Google Scholar] [CrossRef]
- Saito, E.; Suzuki, D.; Kurotaki, D.; Mochizuki, A.; Manome, Y.; Suzawa, T.; Toyoshima, Y.; Ichikawa, T.; Funatsu, T.; Inoue, T.; et al. Down-Regulation of Irf8 by Lyz2-Cre/loxP Accelerates Osteoclast Differentiation in Vitro. Cytotechnology 2017, 69, 443–450. [Google Scholar] [CrossRef]
- Salem, S.; Salem, D.; Gros, P. Role of IRF8 in Immune Cells Functions, Protection against Infections, and Susceptibility to Inflammatory Diseases. Hum. Genet. 2020, 139, 707–721. [Google Scholar] [CrossRef]
- Chrabot, B.S.; Kariuki, S.N.; Zervou, M.I.; Feng, X.; Arrington, J.; Jolly, M.; Boumpas, D.T.; Reder, A.T.; Goulielmos, G.N.; Niewold, T.B. Genetic Variation near IRF8 Is Associated with Serologic and Cytokine Profiles in Systemic Lupus Erythematosus and Multiple Sclerosis. Genes Immun. 2013, 14, 471–478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, Y.; Feng, D.; Jiang, W.; Zhang, T.; Lu, Q.; Zhao, M. 3D Genome Organization and Epigenetic Regulation in Autoimmune Diseases. Front. Immunol. 2023, 14, 1196123. [Google Scholar] [CrossRef] [PubMed]
- Sjöstrand, M.; Johansson, A.; Aqrawi, L.; Olsson, T.; Wahren-Herlenius, M.; Espinosa, A. The Expression of BAFF Is Controlled by IRF Transcription Factors. J. Immunol. Baltim. Md 2016, 196, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Hambleton, S.; Salem, S.; Bustamante, J.; Bigley, V.; Boisson-Dupuis, S.; Azevedo, J.; Fortin, A.; Haniffa, M.; Ceron-Gutierrez, L.; Bacon, C.M.; et al. IRF8 Mutations and Human Dendritic-Cell Immunodeficiency. N. Engl. J. Med. 2011, 365, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Kong, H.J.; Li, H.; Huang, B.; Yang, M.; Zhu, C.; Bogunovic, M.; Zheng, F.; Mayer, L.; Ozato, K.; et al. IRF-8/Interferon (IFN) Consensus Sequence-Binding Protein Is Involved in Toll-like Receptor (TLR) Signaling and Contributes to the Cross-Talk between TLR and IFN-Gamma Signaling Pathways. J. Biol. Chem. 2006, 281, 10073–10080. [Google Scholar] [CrossRef] [PubMed]
- Farh, K.K.-H.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhu, X.; Ye, Z.; Wang, Y.-F.; Yao, C.; Xu, N.; Zhou, M.; Ma, J.; Qin, Y.; Shen, Y.; et al. Lupus Enhancer Risk Variant Causes Dysregulation of IRF8 through Cooperative lncRNA and DNA Methylation Machinery. Nat. Commun. 2022, 13, 1855. [Google Scholar] [CrossRef] [PubMed]
- Geiman, D.E.; Ton-That, H.; Johnson, J.M.; Yang, V.W. Transactivation and Growth Suppression by the Gut-Enriched Krüppel-like Factor (Krüppel-like Factor 4) Are Dependent on Acidic Amino Acid Residues and Protein–Protein Interaction. Nucleic Acids Res. 2000, 28, 1106–1113. [Google Scholar] [CrossRef]
- Kurotaki, D.; Yamamoto, M.; Nishiyama, A.; Uno, K.; Ban, T.; Ichino, M.; Sasaki, H.; Matsunaga, S.; Yoshinari, M.; Ryo, A.; et al. IRF8 Inhibits C/EBPα Activity to Restrain Mononuclear Phagocyte Progenitors from Differentiating into Neutrophils. Nat. Commun. 2014, 5, 4978. [Google Scholar] [CrossRef]
- López-Isac, E.; Acosta-Herrera, M.; Kerick, M.; Assassi, S.; Satpathy, A.T.; Granja, J.; Mumbach, M.R.; Beretta, L.; Simeón, C.P.; Carreira, P.; et al. GWAS for Systemic Sclerosis Identifies Multiple Risk Loci and Highlights Fibrotic and Vasculopathy Pathways. Nat. Commun. 2019, 10, 4955. [Google Scholar] [CrossRef]
- Nelson, M.R.; Tipney, H.; Painter, J.L.; Shen, J.; Nicoletti, P.; Shen, Y.; Floratos, A.; Sham, P.C.; Li, M.J.; Wang, J.; et al. The Support of Human Genetic Evidence for Approved Drug Indications. Nat. Genet. 2015, 47, 856–860. [Google Scholar] [CrossRef]
- Terao, C.; Ohmura, K.; Kawaguchi, Y.; Nishimoto, T.; Kawasaki, A.; Takehara, K.; Furukawa, H.; Kochi, Y.; Ota, Y.; Ikari, K.; et al. PLD4 as a Novel Susceptibility Gene for Systemic Sclerosis in a Japanese Population. Arthritis Rheum. 2013, 65, 472–480. [Google Scholar] [CrossRef]
- Nihtyanova, S.I.; Tang, E.C.; Coghlan, J.G.; Wells, A.U.; Black, C.M.; Denton, C.P. Improved Survival in Systemic Sclerosis Is Associated with Better Ascertainment of Internal Organ Disease: A Retrospective Cohort Study. QJM Mon. J. Assoc. Physicians 2010, 103, 109–115. [Google Scholar] [CrossRef]
- Steen, V.; Mayes, M.; Merkel, P. Assessment of Kidney Involvement. Clin. Exp. Rheumatol. 2003, 21, S29–S31. [Google Scholar]
- Guo, Y.; Yang, Z.; Wu, S.; Xu, P.; Peng, Y.; Yao, M. Inhibition of IRF8 Negatively Regulates Macrophage Function and Impairs Cutaneous Wound Healing. Inflammation 2017, 40, 68–78. [Google Scholar] [CrossRef]
- Ototake, Y.; Yamaguchi, Y.; Asami, M.; Komitsu, N.; Akita, A.; Watanabe, T.; Kanaoka, M.; Kurotaki, D.; Tamura, T.; Aihara, M. Downregulated IRF8 in Monocytes and Macrophages of Patients with Systemic Sclerosis May Aggravate the Fibrotic Phenotype. J. Investag. Dermatol. 2021, 141, 1954–1963. [Google Scholar] [CrossRef]
- Elvira, D.; Masri, R. Current Update on the Role of Inflammation in the Pathogenesis of SSc. In Systemic Sclerosis—Recent Advances and New Perspectives; IntechOpen: London, UK, 2023. [Google Scholar]
- Trombetta, A.C.; Soldano, S.; Contini, P.; Tomatis, V.; Ruaro, B.; Paolino, S.; Brizzolara, R.; Montagna, P.; Sulli, A.; Pizzorni, C.; et al. A Circulating Cell Population Showing Both M1 and M2 Monocyte/Macrophage Surface Markers Characterizes Systemic Sclerosis Patients with Lung Involvement. Respir. Res. 2018, 19, 186. [Google Scholar] [CrossRef] [PubMed]
- Higashi-Kuwata, N.; Jinnin, M.; Makino, T.; Fukushima, S.; Inoue, Y.; Muchemwa, F.C.; Yonemura, Y.; Komohara, Y.; Takeya, M.; Mitsuya, H.; et al. Characterization of Monocyte/Macrophage Subsets in the Skin and Peripheral Blood Derived from Patients with Systemic Sclerosis. Arthritis Res. Ther. 2010, 12, R128. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, J.; Taroni, J.; Martyanov, V.; Wood, T.; Greene, C.; Pioli, P.; Hinchcliff, M.; Whitfield, M. Systems Level Analysis of Systemic Sclerosis Shows a Network of Immune and Profibrotic Pathways Connected with Genetic Polymorphisms. PLoS Comput. Biol. 2015, 11, e1004005. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Sato, S.; Takehara, K. Augmented Production of Chemokines (Monocyte Chemotactic Protein-1 (MCP-1), Macrophage Inflammatory Protein-1α (MIP-1α) and MIP-1β) in Patients with Systemic Sclerosis: MCP-1 and MIP-1α May Be Involved in the Development of Pulmonary Fibrosis. Clin. Exp. Immunol. 1999, 117, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Chauhan, K.S.; Kumar, H.; Tailor, P. Mutation in Irf8 Gene (Irf8R294C) Impairs Type I IFN-Mediated Antiviral Immune Response by Murine pDCs. Front. Immunol. 2021, 12, 758190. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Yu, H.; Zhu, Y.; Ye, Z.; Deng, J.; Su, W.; Cao, Q.; Yuan, G.; Kijlstra, A.; Yang, P. Hypermethylation of Interferon Regulatory Factor 8 (IRF8) Confers Risk to Vogt-Koyanagi-Harada Disease. Sci. Rep. 2017, 7, 1007. [Google Scholar] [CrossRef] [PubMed]
- Bigley, V.; Maisuria, S.; Cytlak, U.; Jardine, L.; Care, M.A.; Green, K.; Gunawan, M.; Milne, P.; Dickinson, R.; Wiscombe, S.; et al. Biallelic Interferon Regulatory Factor 8 Mutation: A Complex Immunodeficiency Syndrome with Dendritic Cell Deficiency, Monocytopenia, and Immune Dysregulation. J. Allergy Clin. Immunol. 2018, 141, 2234–2248. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kim, H.S.; Baek, S.Y.; Lee, G.R. Transcription factor IRF8 controls Th1-like regulatory T-cell function. Cell Mol. Immunol. 2018, 13, 785–794. [Google Scholar] [CrossRef]
- Frantz, C.; Auffray, C.; Avouac, J.; Allanore, Y. Regulatory T Cells in Systemic Sclerosis. Front. Immunol. 2018, 15, 2356. [Google Scholar] [CrossRef]
- Jin, W.; Zheng, Y.; Zhu, P. T cell abnormalities in systemic sclerosis. Autoimmun. Rev. 2022, 21, 103185. [Google Scholar] [CrossRef]
- Liang, K.L.; Laurenti, E.; Taghon, T. Circulating IRF8-expressing CD123+CD127+ lymphoid progenitors: Key players in human hematopoiesis. Trends Immunol. 2023, 44, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Shikhagaie, M.; Germar, K.; Bal, S.; Ros, X.R.; Spits, H. Innate lymphoid cells in autoimmunity: Emerging regulators in rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 164–173. [Google Scholar] [CrossRef] [PubMed]
| Authors | Years of Publication | Methods | Results |
|---|---|---|---|
| Gorlova et al. [63] | 2011 | Genome-wide association study | Association between the IRF8 gene and the lcSSc subtype and the ACA positive subgroup |
| Kurotaki et al. [80] | 2013 | Chromatin immunoprecipitation sequencing; gene expression | IRF8-KLF4 in monocyte differentiation |
| Terao et al. [83] | 2013 | Genome-wide association study | IRF8 as susceptibility gene for SSc in a Japanese population |
| Mahoney et al. [91] | 2015 | Microarray | Connection between inflammatory- and fibroproliferative-specific genes |
| Arismendi et al. [64] | 2015 | Gene expression, genotyping | IRF8 rs11117432 SNP and SSc susceptibility in lcSSc subtype and the ACA positive subgroup |
| Qiu et al. [94] | 2017 | Gene expression, quantitative methylation analysis | IRF8 demethylation as suppressor of Th1 response |
| Lòpez-Isac et al. [81] | 2019 | Genome-wide association study | IRF8 rs11117420 SNP interaction and regulation of its promoter |
| Ototake et al. [87] | 2021 | Gene expression, immunoblotting, Irf8 knockout mice | Involvement of altered IRF8 regulation in monocytes and macrophages in lcSSc |
| González et al. [59] | 2023 | Promoter capture Hi-C | IRF8 rs11117420 variant as new hypothetical SNP for SSc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mennella, A.; Ocone, G.; Stefanantoni, K.; Frasca, L. The Role of IRF8 Polymorphisms in Systemic Sclerosis Development and Pathogenesis. J. Mol. Pathol. 2024, 5, 120-132. https://doi.org/10.3390/jmp5010008
Mennella A, Ocone G, Stefanantoni K, Frasca L. The Role of IRF8 Polymorphisms in Systemic Sclerosis Development and Pathogenesis. Journal of Molecular Pathology. 2024; 5(1):120-132. https://doi.org/10.3390/jmp5010008
Chicago/Turabian StyleMennella, Anna, Giuseppe Ocone, Katia Stefanantoni, and Loredana Frasca. 2024. "The Role of IRF8 Polymorphisms in Systemic Sclerosis Development and Pathogenesis" Journal of Molecular Pathology 5, no. 1: 120-132. https://doi.org/10.3390/jmp5010008
APA StyleMennella, A., Ocone, G., Stefanantoni, K., & Frasca, L. (2024). The Role of IRF8 Polymorphisms in Systemic Sclerosis Development and Pathogenesis. Journal of Molecular Pathology, 5(1), 120-132. https://doi.org/10.3390/jmp5010008







