Analytical Validation and Clinical Utilization of the Oncomine Comprehensive Assay Plus Panel for Comprehensive Genomic Profiling in Solid Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. Nucleic Acid Isolation
2.3. Library Preparation and Next-Generation Sequencing
2.4. Bioinformatics Pipeline and Statistical Analyses
3. Results
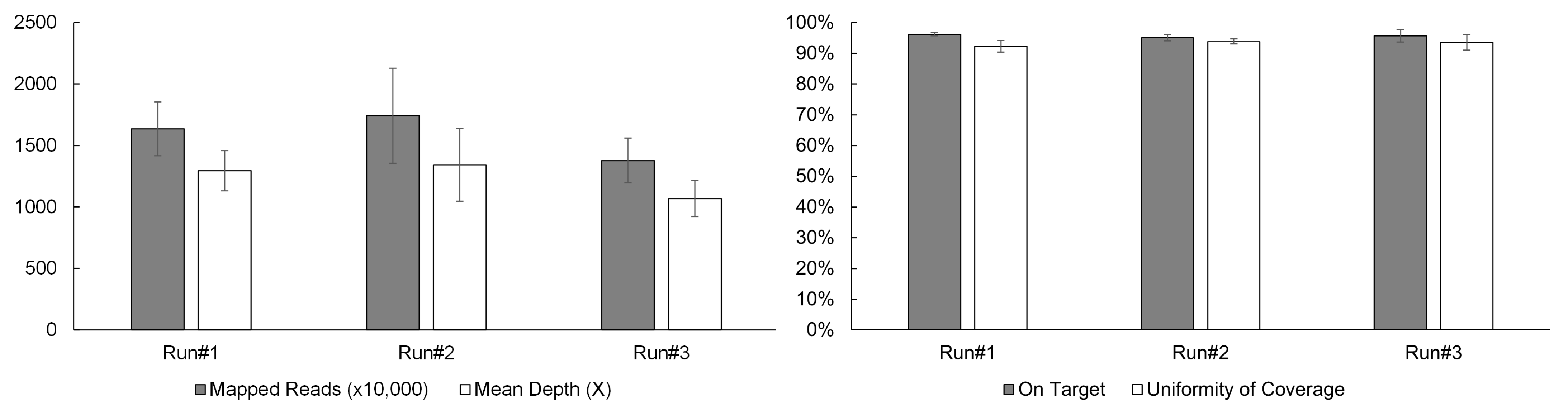
3.1. Sequencing Performance
3.2. Accuracy
3.2.1. SNVs and Indels
3.2.2. Gene Fusions
3.2.3. CNVs
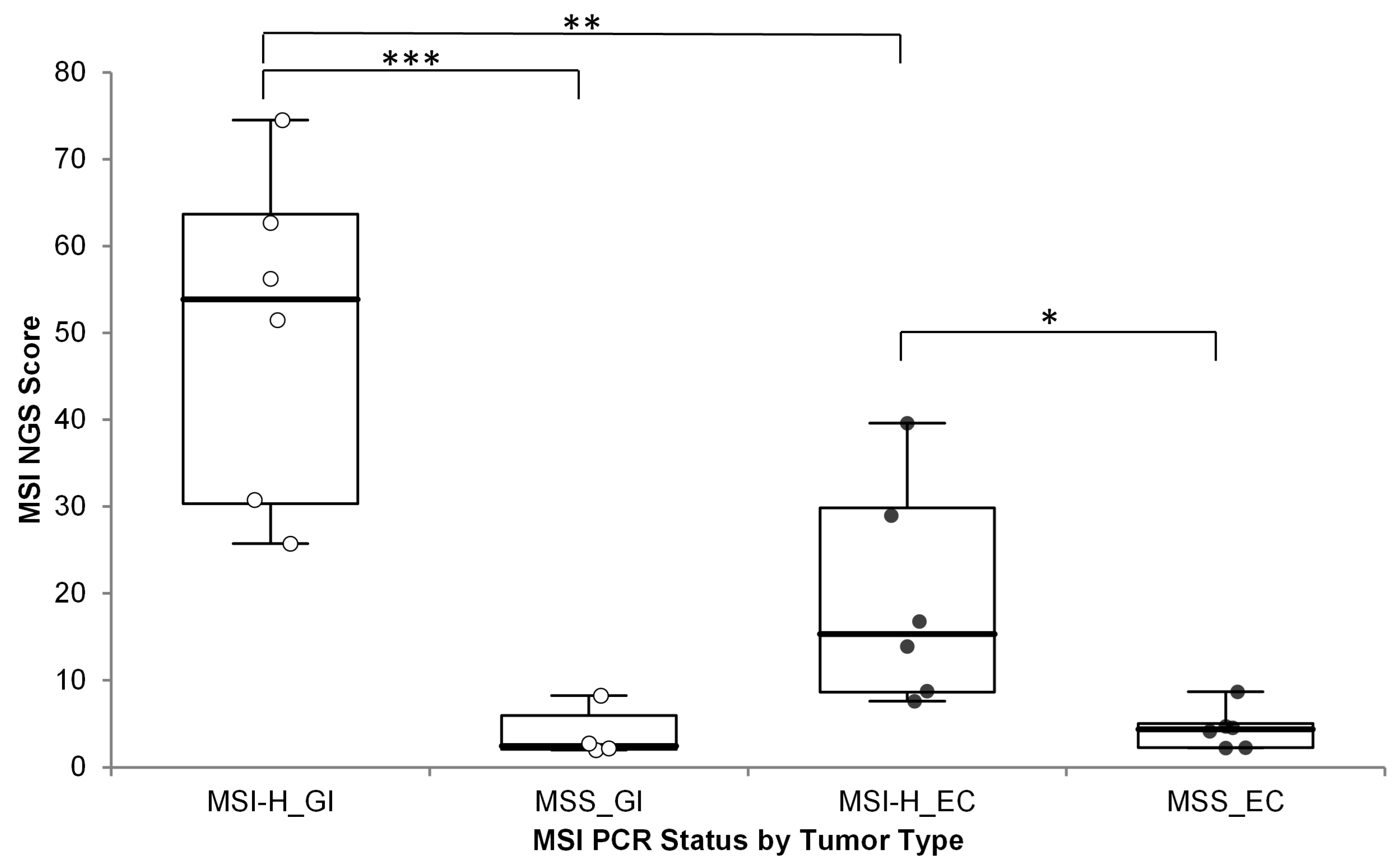
3.2.4. MSI
3.2.5. TMB and HRD
3.3. Limit of Detection
3.3.1. SNVs and Indels
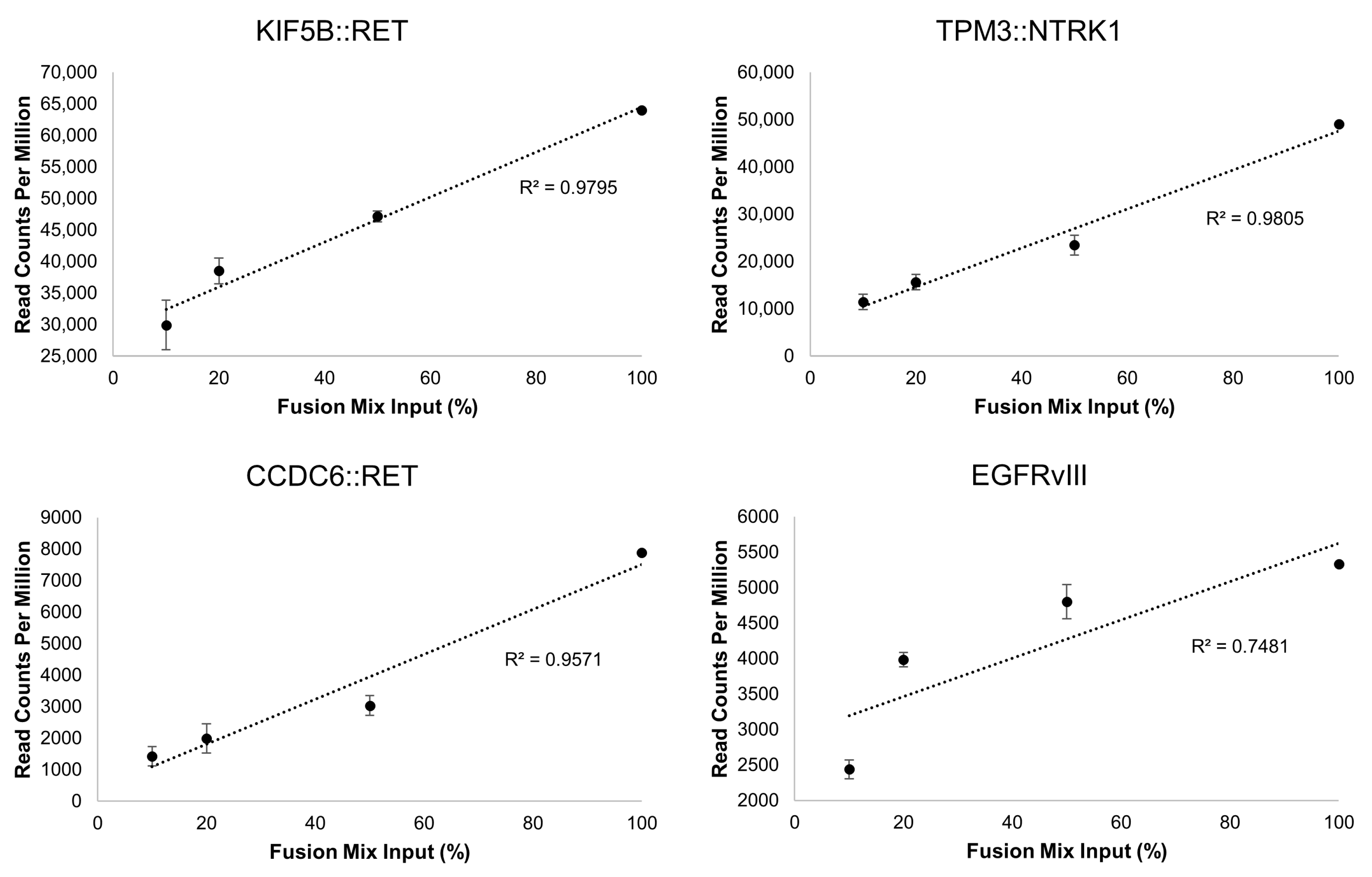
3.3.2. Gene Fusions
3.4. Precision
3.4.1. SNVs and Indels
3.4.2. Gene Fusions
3.4.3. CNVs and Genomic Signatures
3.5. Performance on Clinical Specimens
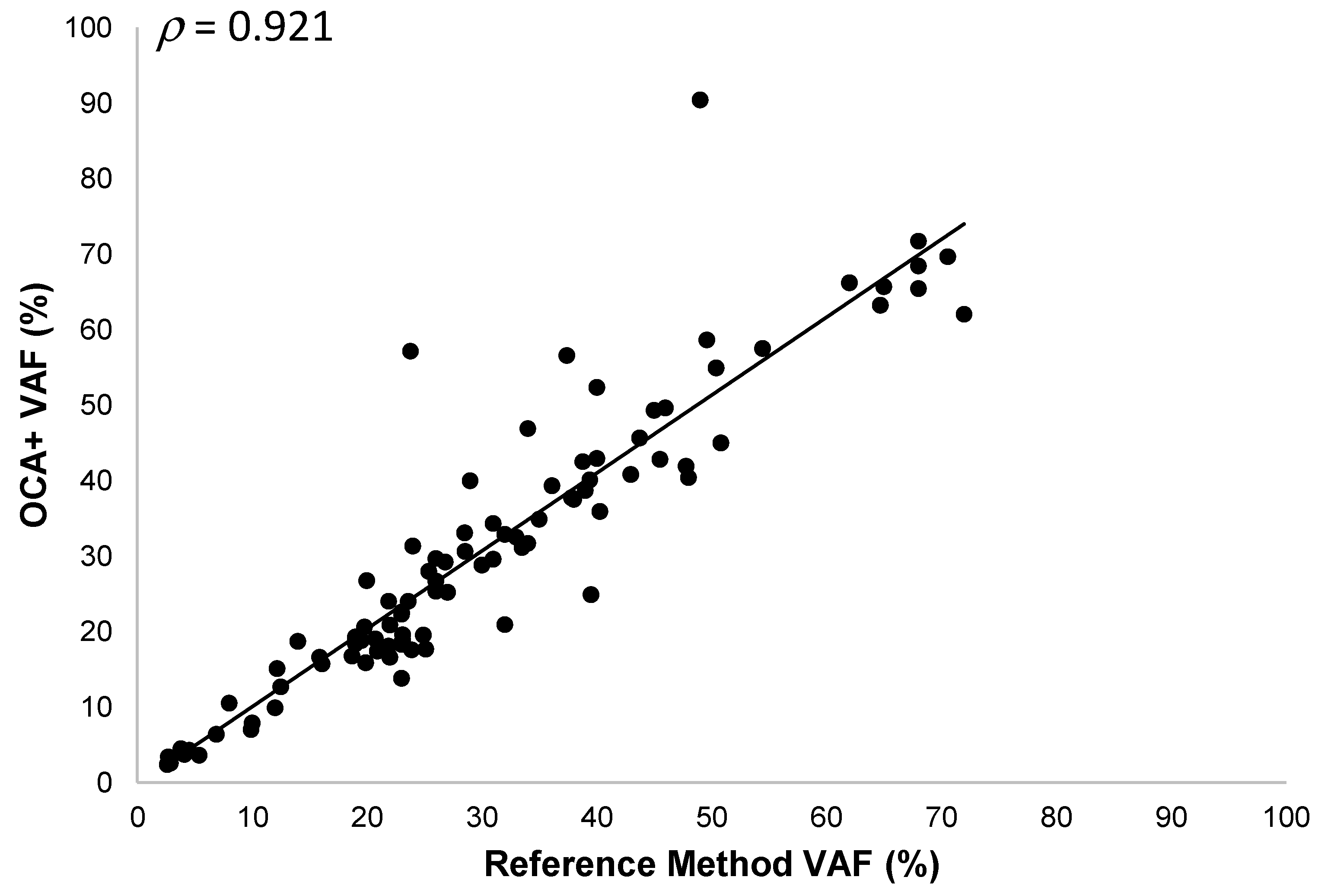
3.5.1. SNVs and Indels
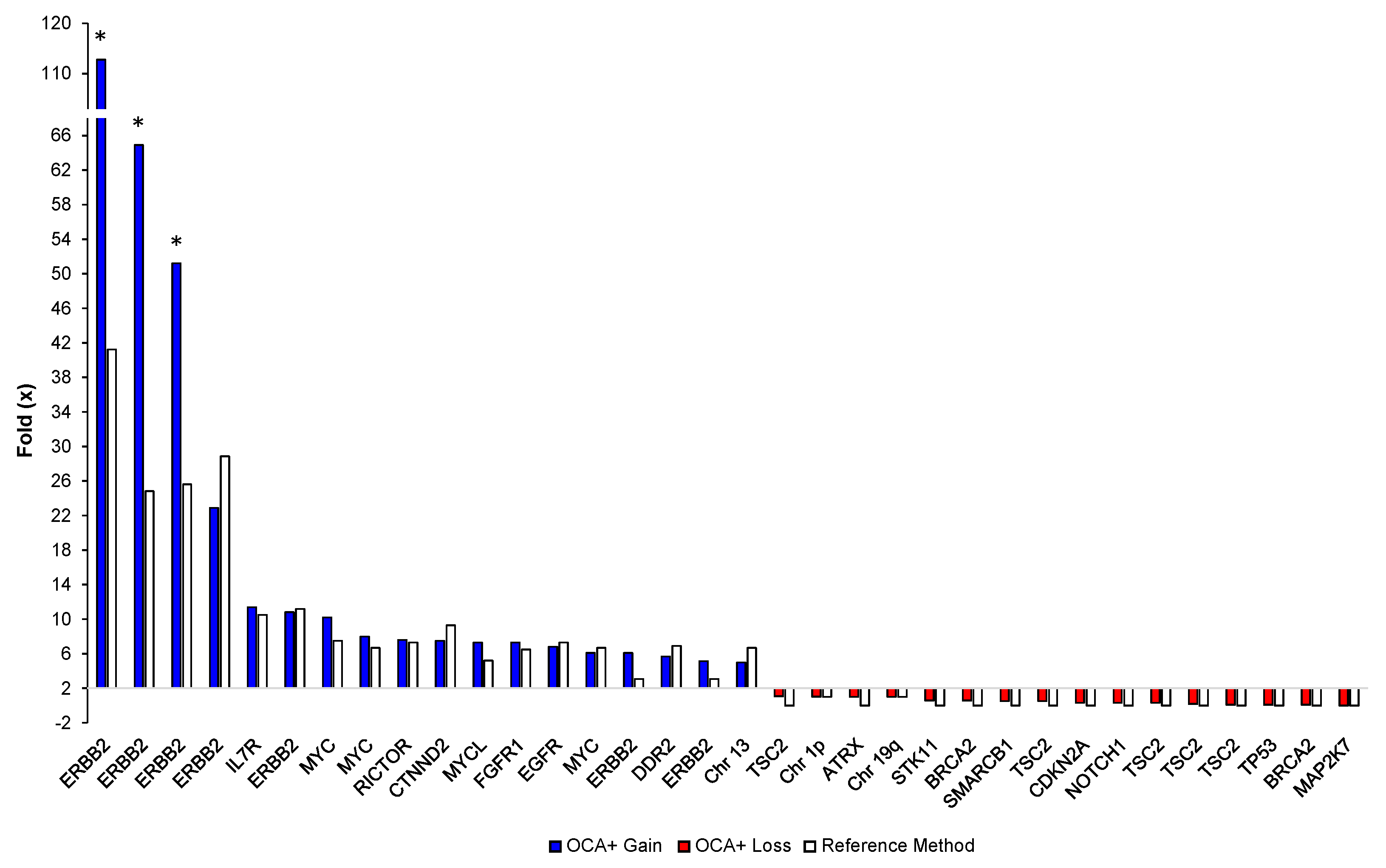
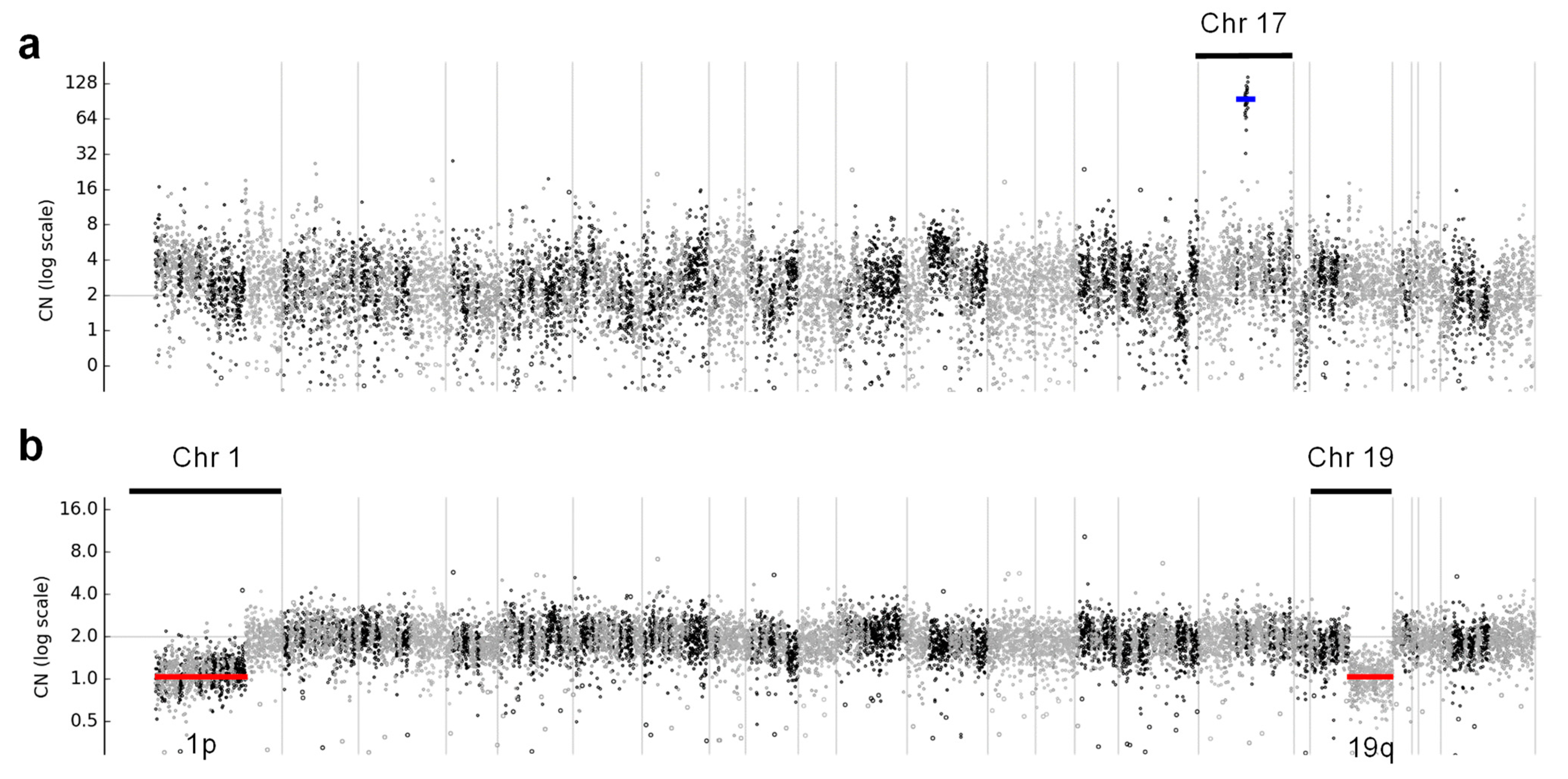
3.5.2. CNVs
3.5.3. Gene Fusions
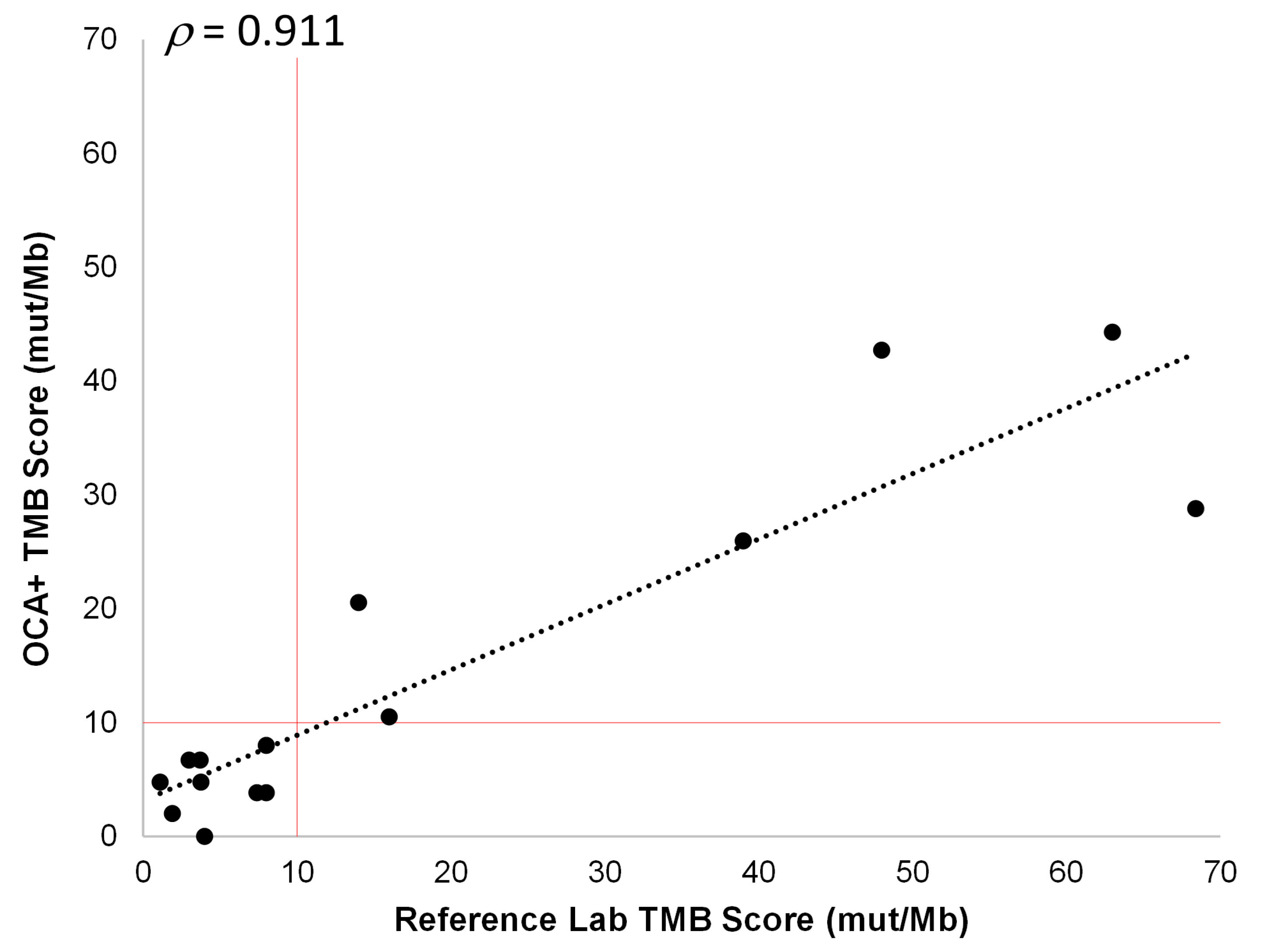
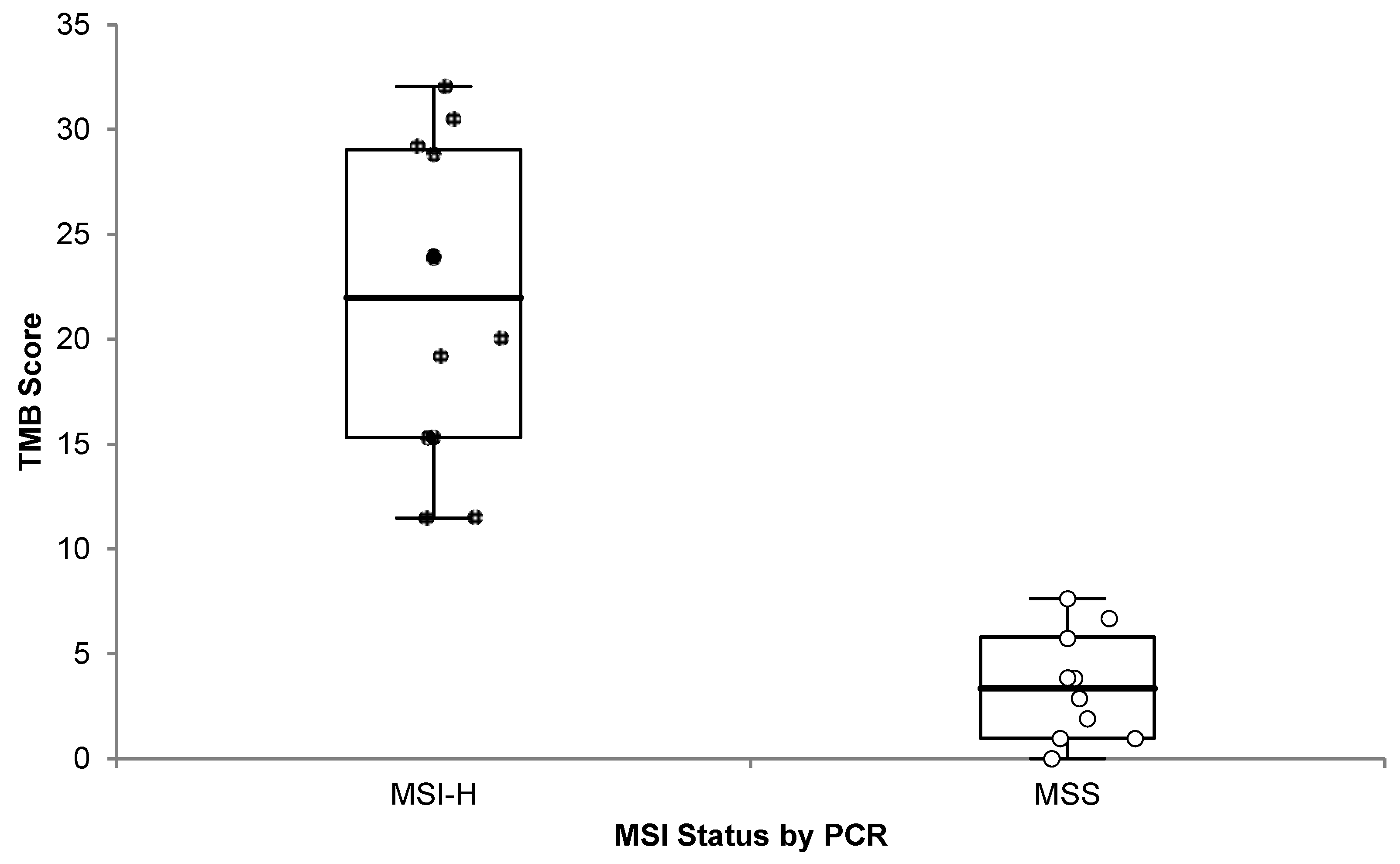
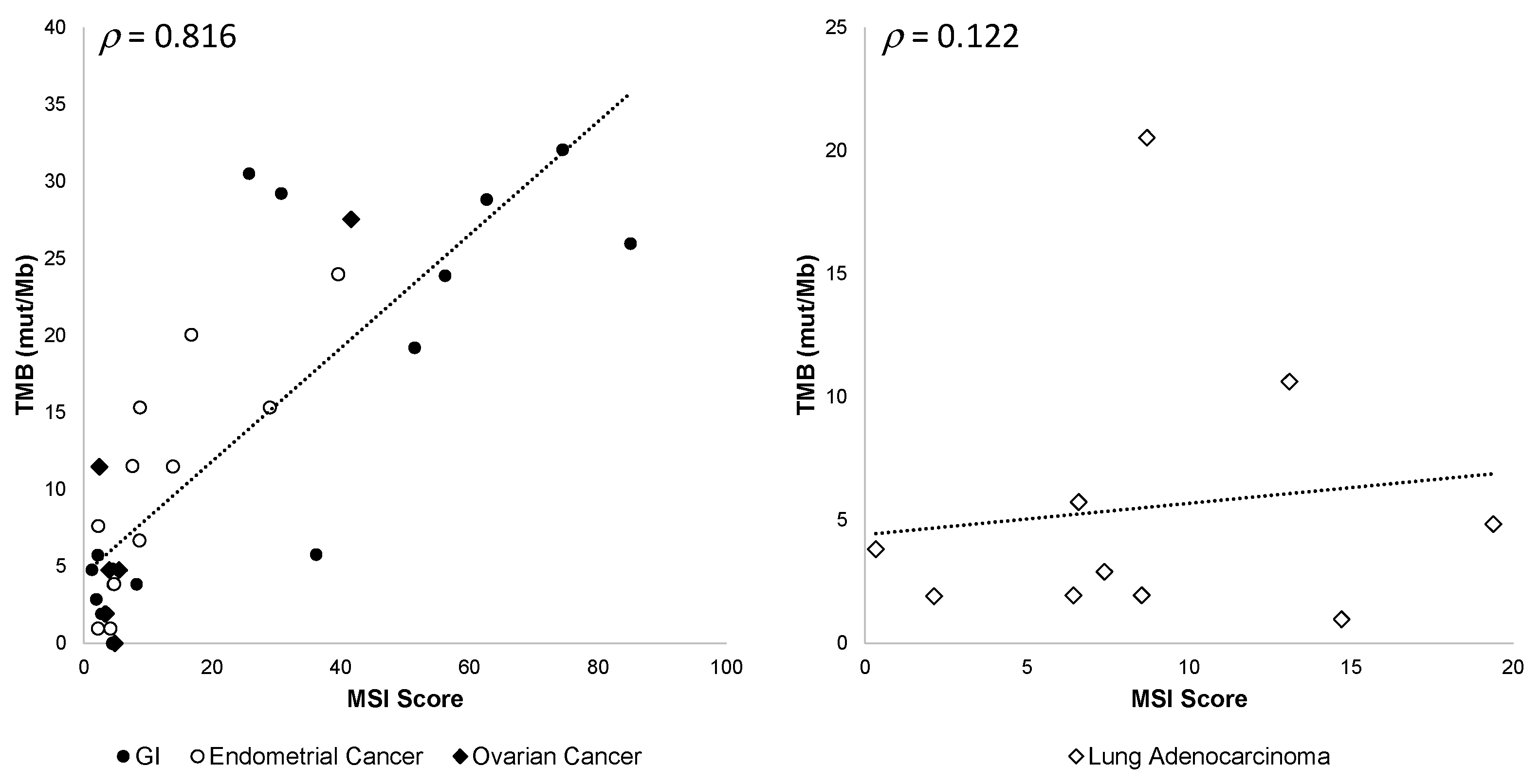
3.5.4. TMB and MSI
3.5.5. HRD
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.-I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and Trametinib in Patients With Tumors With BRAF V600E Mutations: Results of the NCI-MATCH Trial Subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Solit, D.B. Clinical cancer genomic profiling. Nat. Rev. Genet. 2021, 22, 483–501. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Mateo, J.; Porta, N.; Bianchini, D.; Ursula McGovern, U.; Elliott, T.; Jones, R.; Syndikus, I.; Ralph, C.; Jain, S.; Varughese, M. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020, 21, 162–174. [Google Scholar] [CrossRef]
- Hodgson, D.R.; Dougherty, B.A.; Lai, Z.; Fielding, A.; Grinsted, L.; Spencer, S.; O’Connor, M.J.; Ho, T.W.; Robertson, J.D.; Lanchbury, J.S.; et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 2018, 119, 1401–1409. [Google Scholar] [CrossRef]
- Subbiah, V.; Wolf, J.; Konda, B.; Kang, H.; Spira, A.; Weiss, J.; Takeda, M.; Ohe, Y.; Khan, S.; Ohashi, K.; et al. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): A phase 1/2, open-label, basket trial. Lancet Oncol. 2022, 23, 1261–1273. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef] [PubMed]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Roma, G.; Saddar, S.; Zheng, Q.; Pepe, F.; Bruzzese, D.; Vigliar, E.; Bellevicine, C.; Luthra, R.; et al. Consistency and reproducibility of next-generation sequencing in cytopathology: A second worldwide ring trial study on improved cytological molecular reference specimens. Cancer Cytopathol. 2019, 127, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Pepe, F.; Pisapia, P.; Altimari, A.; Bellevicine, C.; Brunnström, H.; Bruno, R.; Büttner, R.; Cirnes, L.; De Andrea, C.E.; et al. Reference standards for gene fusion molecular assays on cytological samples: An international validation study. J. Clin. Pathol. 2023, 76, 47–52. [Google Scholar] [CrossRef]
- Redegalli, M.; Grassini, G.; Magliacane, G.; Pecciarini, L.; Schiavo Lena, M.; Smart, C.E.; Johnston, R.L.; Waddell, N.; Maestro, R. Routine Molecular Profiling in Both Resectable and Unresectable Pancreatic Adenocarcinoma: Relevance of Cytologic Samples. Clin. Gastroenterol. Hepatol. 2022. S1542-3565(22)01003-5. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Yip, S.; Butterfield, Y.S.; Morozova, O.; Chittaranjan, S.; Blough, M.D.; An, J.; Birol, I.; Chesnelong, C.; Chiu, R.; Chuah, E.; et al. Concurrent CIC mutations, IDH mutations and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012, 226, 7–16. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Telli, M.L.; Timms, K.M.; Reid, J.; Hennessy, B.; Mills, G.B.; Jensen, K.C.; Szallasi, Z.; Barry, W.T.; Winer, E.P.; Tung, N.M.; et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancers. Clin. Cancer Res. 2016, 22, 3764–3773. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012, 107, 1776–1782. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Johnson, A.; Jeffrey Sklar, J.; Lindeman, N.I.; Moore, K.; Ganesan, S.; Lovly, C.M.; Perlmutter, J.; Gray, S.W.; Hwang, J.; et al. Somatic Genomic Testing in Patients With Metastatic or Advanced Cancer: ASCO Provisional Clinical Opinion. J. Clin. Oncol. 2022, 40, 1231–1258. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.D.; Lomboy, A.; Lawrence, C.A.; Yourshaw, M.; Bocsi, G.T.; Camidge, D.R.; Aisner, D.L. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J. Thorac. Oncol. 2019, 14, 737–741. [Google Scholar] [CrossRef]
- Parilla, M.; Ritterhouse, L.L. Beyond the Variants: Mutational Patterns in Next-Generation Sequencing Data for Cancer Precision Medicine. Front Cell Dev. Biol. 2020, 8, 370. [Google Scholar] [CrossRef]
- Vestergaard, L.K.; Oliveira, D.N.P.; Poulsen, T.S.; Høgdall, C.K.; Høgdall, E.V. Oncomine™ Comprehensive Assay v3 vs. Oncomine™Comprehensive Assay Plus. Cancers 2021, 13, 5230. [Google Scholar] [CrossRef]
- Mantripragada, K.C.; Olszewski, A.J.; Schumacher, A.; Perez, K.; Birnbaum, A.; Reagan, J.L.; Mega, A.; Khurshid, H.; Bartley, C.; Lombardo, A.; et al. Clinical Trial Accrual Targeting Genomic Alterations After Next-Generation Sequencing at a Non-National Cancer Institute-Designated Cancer Program. J. Oncol. Pract. 2016, 12, e396–e404. [Google Scholar] [CrossRef]
- Hagemann, I.S.; Devarakonda, S.; Lockwood, C.M.; Spencer, D.H.; Guebert, K.; Bredemeyer, A.J.; Al-Kateb, H.; Nguyen, T.T.; Duncavage, E.J.; Cottrell, C.E.; et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015, 121, 631–639. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Wu, Z.; Zeng, F.; Song, B.; Zhang, Y.; Li, J.; Lui, S.; Wu, M. Tumor Mutational Burden Predicting the Efficacy of Immune Checkpoint Inhibitors in Colorectal Cancer: A Systematic Review and Meta-Analysis. Front. Immunol. 2021, 12, 751407. [Google Scholar] [CrossRef]
- Gjoerup, O.; Brown, C.A.; Ross, J.S.; Huang, R.S.P.; Schrock, A.; Creeden, J.; Fabrizio, D.; Tolba, K. Identification and Utilization of Biomarkers to Predict Response to Immune Checkpoint Inhibitors. AAPS J. 2020, 22, 132. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, W.; Zhang, X.; Ge, M.; Song, C. Correlation between TMB and MSI in patients with solid tumors. J. Clin. Oncol. 2020, 38, e15169. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, C.; Eisenberg, R.; Vnencak-Jones, C.L. Differences in Microsatellite Instability Profiles between Endometrioid and Colorectal Cancers: A Potential Cause for False-Negative Results? J. Mol. Diagn. 2017, 19, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Buhard, O.; Cattaneo, F.; Wong, Y.F.; Yim, S.F.; Friedman, E.; Flejou, J.F.; Duval, A.; Hamelin, R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J. Clin. Oncol. 2006, 24, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Chun, S.-M.; Sung, C.O.; Kim, S.Y.; Kim, T.W.; Jang, S.J.; Kim, J. Clinical Utility of a Fully Automated Microsatellite Instability Test with Minimal Hands-on Time. J. Pathol. Transl. Med. 2019, 53, 386–392. [Google Scholar] [CrossRef]
- Dedeurwaerdere, F.; Claes, K.B.; Van Dorpe, J.; Rottiers, I.; Van der Meulen, J.; Breyne, J.; Swaerts, K.; Martens, G. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef]
| Gene (Variant) | Expected VAF (%) | OCA+ VAF (%) |
|---|---|---|
| EGFR (p.E746_A750delELREA) | 25.0 ± 1.6 | 23.8 ± 4.7 |
| KRAS (p.A146T) | 4.3 ± 0.6 | 3.4 ± 0.7 |
| KRAS (p.G12C) | 6.2 ± 0.8 | 6.7 ± 1.4 |
| PIK3CA (p.E542K) | 4.1 ± 0.7 | 3.8 ± 0.5 |
| Gene Fusion | OCA+ (Read Counts) |
|---|---|
| KIF5B::RET | 64,018 |
| ETV6::NTRK3 | 49,745 |
| TPM3::NTRK1 | 49,052 |
| EML4::ALK | 46,932 |
| TMPRSS2::ERG | 42,573 |
| LMNA::NTRK1 | 35,393 |
| PAX8::PPARG | 34,650 |
| TFG::NTRK1 | 38,610 |
| SLC34A2::ROS1 | 28,861 |
| CD74::ROS1 | 25,762 |
| MET Exon 14 Skipping | 29,684 |
| FGFR3::TACC3 | 29,571 |
| NCOA4::RET | 17,646 |
| EGFR::SEPT14 | 14,389 |
| SLC45A3::BRAF | 15,719 |
| FGFR3::BAIAP2L1 | 15,502 |
| CCDC6::RET | 7894 |
| EGFRvIII | 5336 |
| Sample ID | FISH Results | OCA+ Result | |
|---|---|---|---|
| Average HER2 CN 1 per Cell | HER2/chr 17 Ratio | ERBB2 CNV (×) | |
| BHL_F13 | 5.38 | 1.31 | 6.1 |
| BHL_F15 | 23.70 | 6.58 | 22.9 |
| BHL_F16 | 15.70 | 10.83 | 10.8 |
| Seraseq® RM | Expected Score | OCA+ Score |
|---|---|---|
| gDNA TMB Mix Score 7 | 7.2 ± 0.2 | 7.7 ± 0.0 |
| gDNA TMB Mix Score 26 | 25.8 ± 0.5 | 20.3 ± 0.4 |
| FFPE HRD Negative | 31 ± 2 | 35 ± 6 |
| FFPE HRD Low-Positive | 54 ± 2 | 54 ± 2 |
| FFPE HRD High-Positive | 72 ± 3 | 68 ± 4 |
| Gene | Variant Type | Mutation | Target VAF (%) | OCA+ VAF (%) |
|---|---|---|---|---|
| MPL | SNV | p.W515L | 10.0 | 6.8 |
| AKT1 | SNV | p.E17K | 10.0 | 9.8 |
| APC | SNV | p.R1450* | 10.0 | 10.3 |
| GNA11 | SNV | p.Q209L | 10.0 | 10.4 |
| GNAQ | SNV in HP 3N | p.Q209P | 10.0 | 11.4 |
| KIT | SNV | p.D816V | 10.0 | 11.4 |
| PIK3CA | SNV | p.E545K | 10.0 | 12.1 |
| PDGFRA | SNV | p.D842V | 10.0 | 13.6 |
| EGFR | DEL | p.E746_A750delELREA | 10.0 | 4.5 |
| EGFR | INS | p.D770_N771insG | 10.0 | 4.5 |
| SMAD4 | INS | p.A466fs*28 | 10.0 | 11.7 |
| APC | INS in HP 7N | p.T1556fs*3 | 10.0 | 6.3 |
| ERBB2 | INS | p.A775_G776insYVMA | 10.0 | 9.9 |
| JAK2 | SNV in HP 3N | p.V617F | 7.0 | 4.1 |
| TP53 | SNV | p.R248Q | 7.0 | 7.0 |
| EGFR | SNV | p.L858R | 7.0 | 7.1 |
| TP53 | SNV | p.R175H | 7.0 | 7.3 |
| TP53 | SNV | p.R273H | 7.0 | 7.6 |
| KRAS | SNV | p.G12D | 7.0 | 7.7 |
| CTNNB1 | SNV | p.T41A | 7.0 | 7.8 |
| NRAS | SNV | p.Q61R | 7.0 | 7.9 |
| GNAS | SNV | p.R201C | 7.0 | 8.9 |
| PTEN | DEL 6N > 5N | p.K267fs*9 | 7.0 | Not Called |
| TP53 | DEL | p.C242fs*5 | 7.0 | 8.0 |
| PTEN | INS | p.P248fs*5 | 7.0 | 7.6 |
| RET | SNV | p.M918T | 4.0 | 3.9 |
| EGFR | SNV | p.T790M | 4.0 | 4.2 |
| IDH1 | SNV | p.R132C | 4.0 | 4.2 |
| FOXL2 | SNV | p.C134W | 4.0 | 4.5 |
| BRAF | SNV | p.V600E | 4.0 | 4.8 |
| FLT3 | SNV | p.D835Y | 4.0 | 5.5 |
| PIK3CA | SNV | p.H1047R | 4.0 | 5.9 |
| FGFR3 | SNV | p.S249C | 4.0 | 6.7 |
| ATM | DEL | p.C353fs*5 | 4.0 | 9.6 |
| TP53 | DEL 5N > 4N | p.S90fs*33 | 4.0 | Not Called |
| PDGFRA | INS | p.S566fs*6 | 4.0 | 6.1 |
| Gene (Variant) | Library Prep. | Day 1 | Day 2 | Day 3 | Intra-Run | Inter-Run | ||
|---|---|---|---|---|---|---|---|---|
| Expected VAF (%) | Chip 1.1 | Chip 1.2 | Chip 1.3 | Chip 2 | Chip 3 | CV (%) 1 | CV (%) | |
| EGFR (p.E746_ A750delELREA) | 25.0 | 19.7 | 20.9 | 23.0 | 23.9 | 31.6 | 7.9 | 19.5 |
| KRAS (p.A146T) | 4.3 | 4.2 | 3.5 | 3.9 | 2.6 | 2.6 | 9.1 | 21.9 |
| KRAS (p.G12C) | 6.2 | 5.4 | 7.7 | 7.5 | 7.8 | 5.0 | 18.6 | 20.4 |
| MET (p.T1010I) | 6.2 | 7.1 | 7.0 | 7.2 | 7.0 | 8.2 | 1.4 | 7.0 |
| PIK3CA (p.E542K) | 4.1 | 4.0 | 4.2 | 4.0 | 3.0 | 4.0 | 2.8 | 12.4 |
| Sample ID | Library Prep. | Day 1 (Tech. 1) | Day 2 | Day 3 | Intra-Run | Inter-Run | ||
|---|---|---|---|---|---|---|---|---|
| Gene/Signature (Unit) | Replicate 1 | Replicate 2 | Replicate 3 | Tech. 2 | Tech. 3 | CV (%) 1 | CV (%) | |
| BHL_C03 | IL7R Gain (×) | 10.8 | 10.0 | 9.6 | 11.4 | 7.3 | 6.0 | 16.0 |
| BHL_C61 | MDM2 Gain (×) | 18.3 | 13.4 | 12.70 | 16.0 | 17.9 | 20.6 | 16.3 |
| BHL_F03 | ERBB2 Gain (×) | 64.9 | 58.7 | 61.1 | 62.9 | 33.4 | 5.1 | 23.0 |
| BHL_C11 | MSS (MSI Score) | 8.2 | 8.1 | 8.5 | 5.2 | 4.3 | 2.3 | 28.2 |
| BHL_C40 | MSI-H (MSI Score) | 25.3 | 18.7 | 18.25 | 16.7 | 31.4 | 19.3 | 28.1 |
| BHL_C66 | MSI-H (MSI Score) | 78.7 | 73.3 | 81.1 | 48.6 | 84.8 | 5.1 | 19.6 |
| BHL_C03 | TMB-L (mut/Mb) 2 | 4.8 | 4.8 | 4.8 | 6.7 | 4.8 | 0.5 | 16.3 |
| BHL_C40 | TMB-H (mut/Mb) | 17.1 | 19.0 | 17.1 | 20.1 | 19.2 | 6.2 | 7.1 |
| BHL_C66 | TMB-H (mut/Mb) | 40.5 | 37.7 | 40.3 | 39.9 | 41.6 | 4.0 | 3.6 |
| BHL_C05 | gLOH (%) | 4.9 | 1.8 | 5.5 | 4.2 | 4.1 | 48.8 | 34.2 |
| BHL_C10 | gLOH (%) | 13.9 | 14.5 | 16.9 | 9.6 | 14.2 | 10.5 | 19.1 |
| BHL_C55 | gLOH (%) | 25.5 | 26.0 | 25.7 | 26.5 | 26.5 | 1.0 | 1.8 |
| Sample ID | Tumor Type | Tumor Content (%) | Expected Fusion (Exon Junctions) | OCA+ Result (Exon Junctions) |
|---|---|---|---|---|
| BHL_C09 | Prostate Cancer | 25 | TMPRSS2::ERG 1 | TMPRSS2::ERG (T1E2) |
| BHL_C13 | MASC | 85 | ETV6-Fusion 1 | ETV6::NTRK3 (E5N15) |
| BHL_C14 | NSCLC | 30 | FGFR3::TACC3 (F17T8) | FGFR3::TACC3 (F17T8) |
| BHL_C15 | Prostate Cancer | 30 | TMPRSS2::ERG (T1E4) | TMPRSS2::ERG (T1E4) |
| BHL_C16 | NSCLC | 50 | FGFR3::TACC3 (F17T8) | FGFR3::TACC3 (F17T8) |
| BHL_C17 | NSCLC | 70 | METex14 (M13M15) 2 | METex14 (M13M15) |
| BHL_C18 | NSCLC | 70 | KIF5B::RET (K15R12) | KIF5B::RET (K15R12) |
| BHL_C19 | NSCLC | 70 | ETV6::NTRK3 (E5N15) | ETV6::NTRK3 (E5N15) |
| BHL_C20 | NSCLC | 5 | EML4::ALK (E6A19) | EML4::ALK (E6A19) |
| BHL_C21 | NSCLC | 50 | CCDC6::RET (C1R12) | CCDC6::RET (C1R12) |
| BHL_C22 | NSCLC | 20 | EML4::ALK (E13A20) | EML4::ALK (E13A20) |
| BHL_C23 | NSCLC | 50 | EML4::ALK (E20A20) | EML4::ALK (E20A20) |
| BHL_C24 | NSCLC | 5 | CD74::ROS1 (C6R34) | CD74::ROS1 (C6R34) |
| BHL_C25 | Prostate Cancer | 50 | TMPRSS2::ERG (T2E4) | TMPRSS2::ERG (T2E4) |
| BHL_C26 | NSCLC | 30 | EML4::ALK (E2A20) | EML4::ALK (E2A20) |
| BHL_C27 | NSCLC | 10 | METex14 (M13M15) | METex14 (M13M15) |
| BHL_C28 | NSCLC | 40 | KIF5B::RET (K15R12) | KIF5B::RET (K15R12) |
| BHL_C29 | NSCLC | 10 | METex14 (M13M15) | METex14 (M13M15) |
| BHL_C30 | NSCLC | 10 | METex14 (M13M15) | METex14 (M13M15) |
| BHL_C31 | NSCLC | 50 | EML4::ALK (E6ALK20) | EML4::ALK (E6ALK20) |
| Sample ID | HRD Class | HRR Mutated Gene | Reference Score Type | Reference Score | OCA+ Score Type | OCA+ Score |
|---|---|---|---|---|---|---|
| BHL_C04 | HRD-NEG | ATM | %LOH | 1 | nLOH | 6 |
| BHL_C12 | HRD-NEG | FANCG | %LOH | 0 | nLOH | 1 |
| BHL_C67 | HRD-NEG | None | %LOH | 9 | nLOH | 5 |
| BHL_C73 | HRD-NEG | None | %LOH | 0 | nLOH | 2 |
| BHL_C68 | HRD-NEG | None | GIS | 1 | HRD | 2 |
| BHL_C69 | HRD-NEG | None | GIS | 12 | HRD | 24 |
| BHL_C70 | HRD-NEG | None | GIS | 3 | HRD | 7 |
| BHL_C71 | HRD-NEG | None | GIS | 19 | HRD | 29 |
| BHL_C72 | HRD-POS | BRCA2 | GIS | 64 | HRD | 53 |
| BHL_C74 | HRD-POS | BRCA1 | GIS | 75 | HRD | 58 |
| BHL_C06 | HRD-POS | BRCA2 | GIS | + 1 | HRD | 71 |
| Sample ID | HRD Class | HRD Score (v5.18) | OCA+ GIM (v5.20) |
|---|---|---|---|
| BHL_C08 | HRD-NEG | 1 | 0 |
| BHL_C73 | HRD-NEG | 2 | 0 |
| BHL_C68 | HRD-NEG | 2 | 0 |
| BHL_C09 | HRD-NEG | 6 | 0 |
| BHL_C70 | HRD-NEG | 7 | 0 |
| BHL_C12 | HRD-NEG | 12 | 0 |
| BHL_C11 | HRD-NEG | 13 | 0 |
| BHL_C05 | HRD-NEG | 17 | 3 |
| BHL_C04 | HRD-NEG | 20 | 1 |
| BHL_C67 | HRD-NEG | 23 | 4 |
| BHL_C69 | HRD-NEG | 24 | 7 |
| BHL_C03 | HRD-NEG | 26 | 5 |
| BHL_C71 | HRD-NEG | 29 | 4 |
| BHL_C07 | HRD-NEG | 32 | 15 |
| BHL_C01 | HRD-NEG | 38 | 10 |
| BHL_C02 | HRD-NEG | 40 | 13 |
| BHL_C72 | HRD-POS | 53 | 16 |
| BHL_C10 | HRD-POS | 56 | 16 |
| BHL_C74 | HRD-POS | 58 | 19 |
| BHL_C06 | HRD-POS | 71 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumur, C.I.; Krishnan, R.; Almenara, J.A.; Brown, K.E.; Dugan, K.R.; Farni, C.; Ibrahim, F.Z.; Sanchez, N.A.; Rathore, S.; Pradhan, D.; et al. Analytical Validation and Clinical Utilization of the Oncomine Comprehensive Assay Plus Panel for Comprehensive Genomic Profiling in Solid Tumors. J. Mol. Pathol. 2023, 4, 109-127. https://doi.org/10.3390/jmp4020012
Dumur CI, Krishnan R, Almenara JA, Brown KE, Dugan KR, Farni C, Ibrahim FZ, Sanchez NA, Rathore S, Pradhan D, et al. Analytical Validation and Clinical Utilization of the Oncomine Comprehensive Assay Plus Panel for Comprehensive Genomic Profiling in Solid Tumors. Journal of Molecular Pathology. 2023; 4(2):109-127. https://doi.org/10.3390/jmp4020012
Chicago/Turabian StyleDumur, Catherine I., Ramakrishnan Krishnan, Jorge A. Almenara, Kathleen E. Brown, Kailyn R. Dugan, Christiana Farni, Fatima Z. Ibrahim, Naomi A. Sanchez, Sumra Rathore, Dinesh Pradhan, and et al. 2023. "Analytical Validation and Clinical Utilization of the Oncomine Comprehensive Assay Plus Panel for Comprehensive Genomic Profiling in Solid Tumors" Journal of Molecular Pathology 4, no. 2: 109-127. https://doi.org/10.3390/jmp4020012
APA StyleDumur, C. I., Krishnan, R., Almenara, J. A., Brown, K. E., Dugan, K. R., Farni, C., Ibrahim, F. Z., Sanchez, N. A., Rathore, S., Pradhan, D., & Hughes, J. H. (2023). Analytical Validation and Clinical Utilization of the Oncomine Comprehensive Assay Plus Panel for Comprehensive Genomic Profiling in Solid Tumors. Journal of Molecular Pathology, 4(2), 109-127. https://doi.org/10.3390/jmp4020012








