A Longitudinal Study of Bacteriophages as Indicators of Norovirus Contamination of Mussels (Mytilus edulis) and Their Overlying Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Site
2.2. Sample Collection
2.3. Sample Processing
2.3.1. Preparation of Mussel Homogenate and Overlying Water Samples for Phage Analysis
2.3.2. Preparation of Mussel Homogenate for Bacterial Analysis
2.4. Detection and Enumeration of Bacterial Indicators
2.5. Detection and Enumeration of Phage-Based Indicators
2.6. Detection and Enumeration of NoV
2.7. Statistical Analysis
3. Results
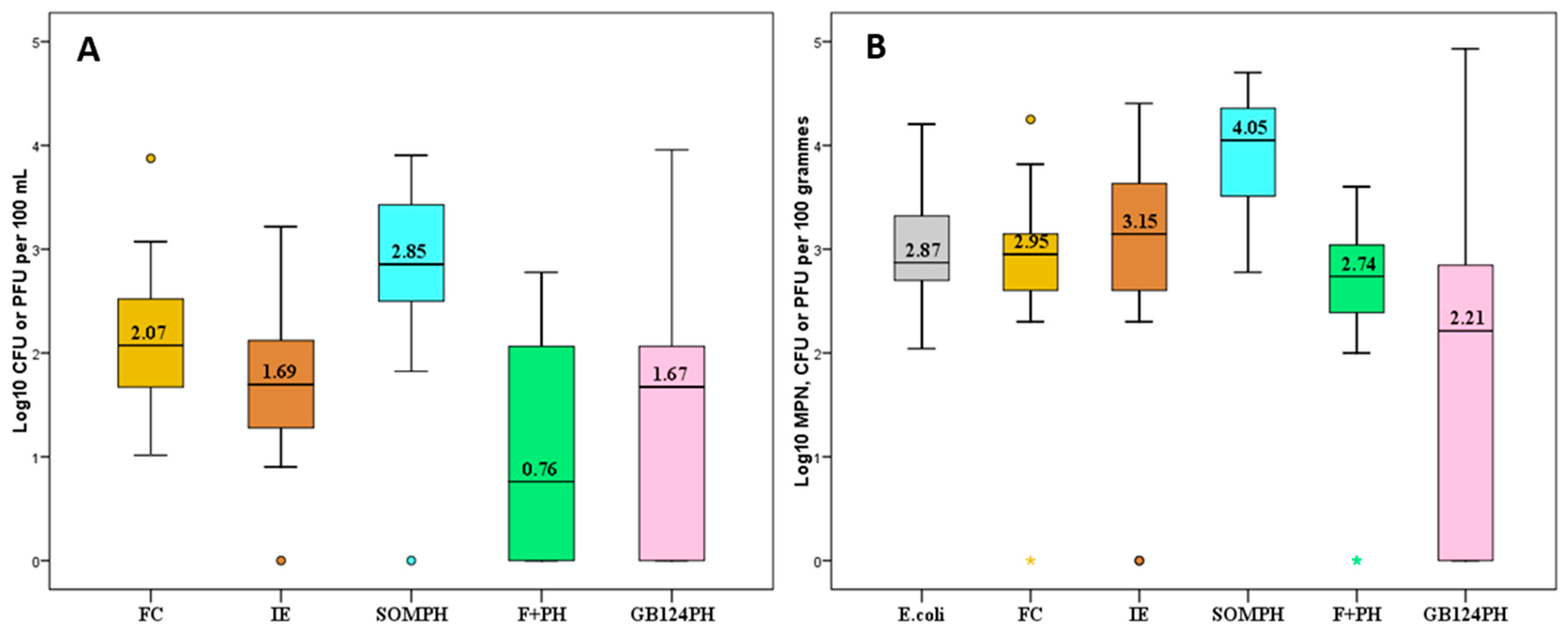
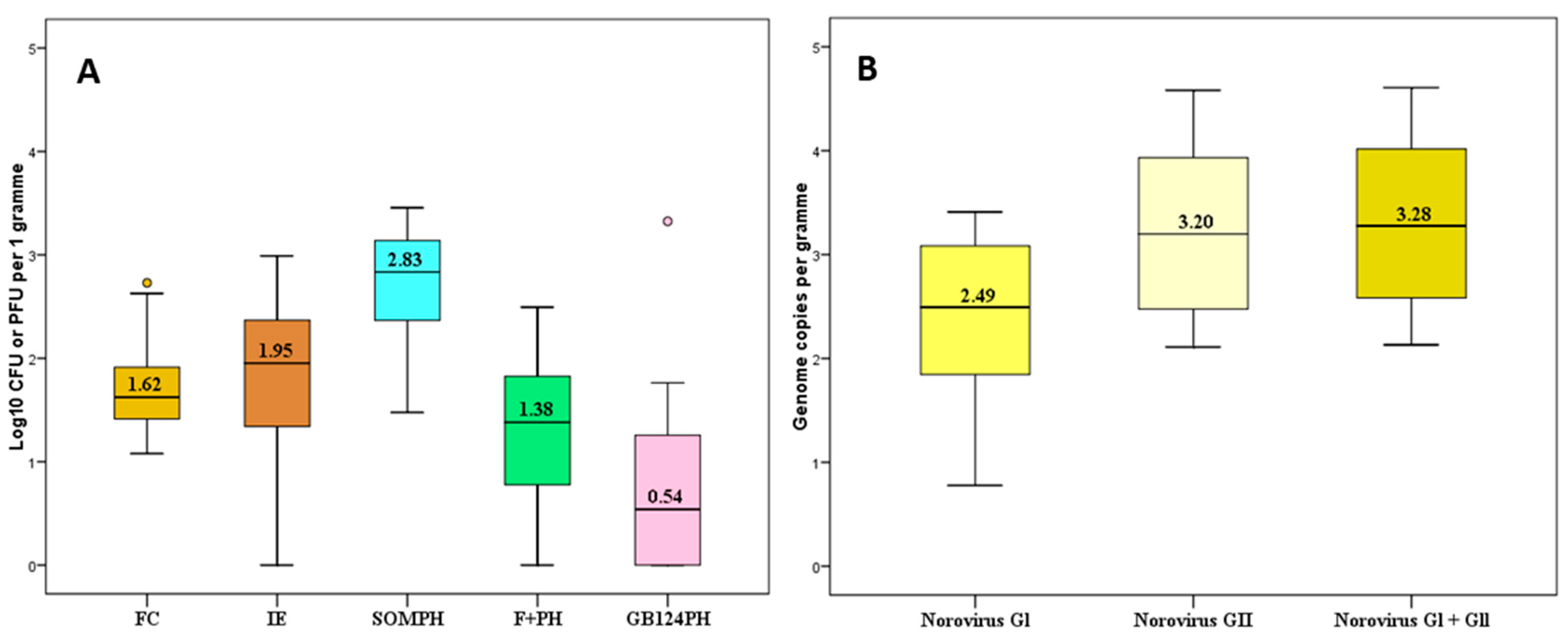
3.1. Levels of Faecal Pollution Indicators and Norovirus (NoV)
3.2. Relationship between Faecal Indicators and NoV in Mussels
4. Discussion
4.1. Compliance with EU Shellfish Classification Criteria
4.2. Effect of Seasonality
4.3. Faecal Pollution Indicators and NoV in Mussel Matrices
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CFU | Colony-Forming Units |
| CSO | Combined sewer overflow |
| FC | Faecal coliforms |
| F + PH | F-specific RNA phages |
| G I/II | Norovirus genogroup I and II |
| GB124PH | Phages infecting Bacteroides fragilis strain GB-124 |
| IE | Intestinal enterococci |
| NoV | Norovirus |
| NSSP | National Shellfish Sanitation Program |
| PFU | Plaque-forming units |
| RT-PCR | Real-time polymerase chain reaction |
| SOMPH | Somatic coliphages |
| WWTW | Wastewater treatment works |
References
- Crane, S.; Moore, J. A Management Strategy to Reduce Bacterial Pollution in Shellfish Areas: A Case Study. Environ. Manag. 1986, 10, 41–51. [Google Scholar] [CrossRef]
- Schaeffer, J.; Treguier, C.; Piquet, J.; Gachelin, S.; Cochennec-Laureau, N.; Le Saux, J.; Garry, P.; Le Guyader, F. Improving the Efficacy of Sewage Treatment Decreases Norovirus Contamination in Oysters. Int. J. Food Microbiol. 2018, 286, 1–5. [Google Scholar] [CrossRef]
- Martinez-Albores, A.; Lopez-Santamarina, A.; Rodriguez, J.; Ibarra, I.; Mondragón, A.; Miranda, J.; Lamas, A.; Cepeda, A. Complementary Methods to Improve the Depuration of Bivalves: A Review. Foods 2020, 9, 129. [Google Scholar] [CrossRef]
- Ang, L.H. An outbreak of viral gastroenteritis associated with eating raw oysters. Commun. Dis. Public Health 1998, 1, 38–40. [Google Scholar]
- Gerba, C.; Wallis, C.; Melnick, J. Viruses in Water: The problem, some solutions. Environ. Sci. Technol. 1975, 9, 1122–1126. [Google Scholar] [CrossRef]
- McMinn, B.; Ashbolt, N.; Korajkic, A. Bacteriophages as Indicators of Faecal Pollution and Enteric Virus Removal. Lett. Appl. Microbiol. 2017, 65, 11–26. [Google Scholar] [CrossRef]
- Love, D.C.; Lovelace, G.L.; Sobsey, M.D. Removal of Escherichia coli, Enterococcus fecalis, coliphage MS-2, poliovirus, and hepatitis A virus from oysters (Crassostrea virginica) and hard-shell clams (Mercinaria mercinaria) by depuration. Int. J. Food Microbiol. 2010, 143, 211–217. [Google Scholar] [CrossRef]
- Iwamoto, M.; Ayers, T.; Mahon, B.; Swerdlow, D. Epidemiology of Seafood-Associated Infections in the United States. Clinic. Microbiol. Rev. 2010, 23, 399–411. [Google Scholar] [CrossRef]
- Bellou, M.; Kokkinos, P.; Vantarakis, A. Shellfish-Borne Viral Outbreaks: A Systematic Review. Food Environ. Virol. 2012, 5, 13–23. [Google Scholar] [CrossRef]
- Wu, J.; Long, S.C.; Das, D.; Dorner, S.M. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health 2011, 9, 265–278. [Google Scholar] [CrossRef]
- Simpson, J.M.; Santo Domingo, J.W.; Reasoner, D.J. Microbial Source Tracking: State of the Science. Environ. Sci. Technol. 2002, 36, 5279–5288. [Google Scholar] [CrossRef]
- Lees, D.; CEN WG6 TAG4. International standardisation of a method for detection of human pathogenic viruses in molluscan shellfish. Food Environ. Virol. 2010, 2, 146–155. [Google Scholar] [CrossRef]
- Anon. ISO 15216-1:2017; Microbiology of the Food Chain—Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 1: Method for Quantification. International Organization for Standardization: Geneva, Switzerland, 2017.
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Corrêa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef]
- Lowther, J.; Avant, J.; Gizynski, K.; Rangdale, R.; Lees, D. Comparison between Quantitative Real-Time Reverse Transcription PCR Results for Norovirus in Oysters and Self-Reported Gastroenteric Illness in Restaurant Customers. J. Food Prot. 2010, 73, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, K.; Torok, V.; Turnbull, A. Bacteriophages as Enteric Viral Indicators in Bivalve Mollusc Management. Food Microbiol. 2017, 65, 284–293. [Google Scholar] [CrossRef]
- Formiga-Cruz, M.; Allard, A.K.; Conden-Hansson, A.C.; Henshilwood, K.; Hernroth, B.E.; Jofre, J.; Lees, D.N.; Lucena, F.; Papapetropoulou, M.; Rangdale, R.E.; et al. Evaluation of potential indicators of viral contamination in shellfish and their applicability to diverse geographical areas. Appl. Environ. Microbiol. 2003, 69, 1556–1563. [Google Scholar] [CrossRef]
- Doré, W.J.; Henshilwood, K.; Lees, D.N. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl. Environ. Microbiol. 2000, 66, 1280–1285. [Google Scholar] [CrossRef]
- Lucena, F.; Lasobras, J.; Mcintosh, D.; Forcadell, M.; Jofre, J. Effect of distance from the polluting focus on relative concentrations of Bacteroides fragilis phages and coliphages in mussels. Appl. Environ. Microbiol. 1994, 60, 2272–2277. [Google Scholar] [CrossRef]
- Da Silva, D.T.G. Bacteriophages as Indicators of Human Enteric Viruses in Mussels. Ph.D. Thesis, University of Brighton, Brighton, UK, 2013. [Google Scholar]
- Olalemi, A.; Purnell, S.; Caplin, J.; Ebdon, J.; Taylor, H. The application of phage-based faecal pollution markers to predict the concentration of adenoviruses in mussels (Mytilus edulis) and their overlying waters. J. Appl. Microbiol. 2016, 121, 1152–1162. [Google Scholar] [CrossRef]
- Potasman, I.; Paz, A.; Odeh, M. Infectious outbreaks associated with bivalve shellfish consumption: A worldwide perspective. Clin. Infect. Dis. 2002, 35, 921–928. [Google Scholar] [CrossRef]
- Ashbolt, N.; Fujioka, R.; Glymph, T.; McGee, C.; Schaub, S.; Sobsey, M.; Toranzos, G. Pathogens, pathogen indicators, and indicators of fecal contamination. In Report of the Experts Scientific Workshop on Critical Research Needs for the Development of New or Revised Recreational Water Quality Criteria; EPA 823-R-07-006; Environmental Protection Agency Office of Water Office of Research and Development: Warrenton, VA, USA, 2007; Volume 147, pp. 35–52. [Google Scholar]
- Ebdon, J.E.; Sellwood, J.; Shore, J.; Taylor, H. Phages of Bacteroides (GB-124): A novel tool for viral waterborne disease control? Environm. Sci. Technol. 2012, 46, 1163–1169. [Google Scholar] [CrossRef]
- McMinn, B.R.; Korajkic, A.; Ashbolt, H.J. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated fecal indicators in the United States. Lett. Appl. Microbiol. 2014, 59, 115–121. [Google Scholar] [CrossRef]
- USEPA Office of Water. Review of Coliphages as Possible Indicators of Fecal Contamination for Ambient Water Quality; 820-R-15-098; United States Environment Protection Agency, Office of Science and Technology, Office of Water: Washington, DC, USA, 2015. Available online: http://water.epa.gov/scitech/swguidance/standards/criteria/health/microbial/upload/coliphages-literature-review-report-2015.pdf (accessed on 25 July 2021).
- OART. The Sussex River Ouse Corridor. Available online: https://oart.org.uk/rivers-new/river-ouse/ (accessed on 2 November 2021).
- Anon. ISO/TS 16649-3; Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Enumeration of Beta-Glucoronidase-positive Escherichia coli. Part 3: Most Probable Number Technique Using 5-Bromo-4-Chloro-3-Indolyl-Beta-D-Glucuro-Nide. International Organization for Standardization: Geneva, Switzerland, 2005.
- Anon. ISO 9308-1:2000; Water Quality. Detection and Enumeration of Escherichia coli and Coliform Bacteria Part 1: Membrane Filtration Method. International Organization for Standardization: Geneva, Switzerland, 2000.
- Anon. ISO 7899-2:2000; Water Quality: Detection and Enumeration of Intestinal Enterococci Part 2: Membrane Filtration Method. International Organization for Standardization: Geneva, Switzerland, 2000.
- Anon. ISO 10705-2; Water Quality—Detection and Enumeration of Bacteriophages—Part 2: Enumeration of Somatic Coliphages. International Organization for Standardization: Geneva, Switzerland, 2001.
- Anon. ISO 10705-1; Water Quality—Detection and Enumeration of Bacteriophages—Part 1: Enumeration of F-Specific RNA Bacteriophages. International Organisation for Standardization: Geneva, Switzerland, 2001.
- Anon. ISO 10705-4; Water Quality—Detection and Enumeration of Bacteriophages—Part 4: Enumeration of Bacteriophages Infecting Bacteroides fragilis. International Organisation for Standardization: Geneva, Switzerland, 2001.
- Helsel, D.R. Advantages of nonparametric procedures for analysis of water-quality data. Hydrol. Sci. J. 1987, 32, 179–190. [Google Scholar] [CrossRef]
- Lowther, J.; Gustar, N.; Powell, A.; Hartnell, R.; Lees, D. Two-Year Systematic Study to Assess Norovirus Contamination in Oysters from Commercial Harvesting Areas in the United Kingdom. Appl. Environ. Microbiol. 2012, 78, 5812–5817. [Google Scholar] [CrossRef]
- Muniain-Mujika, I.; Calvo, M.; Lucena, F.; Girones, R. Comparative analysis of viral pathogens and potential indicators in shellfish. Int. J. Food Microbiol. 2003, 83, 75–85. [Google Scholar] [CrossRef]
- Flannery, J.; Keaveney, S.; Rajko-Nenow, P.; O’Flaherty, V.; Dore, W. NoV and FRNA bacteriophage determined by RT-qPCR and infectious FRNA bacteriophage in wastewater and oysters. Water Res. 2013, 47, 5222–5231. [Google Scholar] [CrossRef]
- EC Regulation No 854/2004 of the European Parliament and of the Council of 29 April 2004 Laying Down Specific Rules for the Organization of Official Controls on Products of Animal Origin Intended for Human Consumption. Off. J. Eur. Union 2004, L226, 83–127.
- Food Standards Agency (FSA). Shellfish Harvesting Classifications England and Wales: 2020–2021. Available online: https://www.food.gov.uk/sites/default/files/media/document/shellfish-classifications-2020-2021-enw.pdf (accessed on 17 January 2021).
- NSSP. National Shellfish Sanitation Program. In Guide for the Control of Molluscan Shellfish: 2013 Revision. Available online: http://wayback.archive-it.org/7993/20180126093259/https://www.fda.gov/downloads/Food/GuidanceRegulation/FederalStateFoodPrograms/UCM415522.pdf (accessed on 5 March 2015).
- NSSP. National Shellfish Sanitation Program. In Guide for the Control of Molluscan Shellfish: 2019 Revision. Available online: https://www.fda.gov/media/143238/download (accessed on 22 November 2021).
- Contreras-Coll, N.; Lucena, F.; Mooijman, K.; Havelaar, A.; Pierzo, V.; Boque, M.; Gawler, A.; Holler, C.; Lambiri, M.; Mirolo, G.; et al. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Res. 2002, 36, 4963–4974. [Google Scholar] [CrossRef]
- Wiggins, B.A.; Alexander, M. Minimum bacterial density for bacteriophage replication—Implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 1985, 49, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J. Is the replication of somatic coliphages in water environments significant? J. Appl. Microbiol. 2009, 106, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Sinton, L.W.; Finlay, R.K.; Lynch, P.A. Sunlight inactivation of fecal bacteriophages and bacteria in sewage-polluted seawater. Appl. Environ. Microbiol. 1999, 65, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Mocé-Llivina, L.; Lucena, F.; Jofre, J. Enteroviruses and Bacteriophages in Bathing Waters. Appl. Environ. Microbiol. 2005, 71, 6838–6844. [Google Scholar] [CrossRef]
- Leclerc, H.; Edberg, S.; Pierzo, V.; Delattre, J.M. Bacteriophages as indicators of enteric viruses and public health risk in groundwaters. J. Appl. Microbiol. 2000, 88, 5–21. [Google Scholar] [CrossRef]
- Grabow, W.O.K. Bacteriophages: Update on application as models for viruses in water. Water SA 2001, 27, 251–268. [Google Scholar] [CrossRef]
- Payan, A.; Ebdon, J.; Taylor, H.; Gantzer, C.; Ottoson, J.; Papageorgiou, G.T.; Blanch, A.R.; Lucena, F.; Jofre, J.; Muniesa, M. Method for isolation of Bacteroides bacteriophage host strains suitable for tracking sources of fecal pollution in water. Appl. Environ. Microbiol. 2005, 71, 5659–5662. [Google Scholar] [CrossRef]
- Flannery, J.; Keaveney, S.; Dore, W. Use of F-RNA bacteriophages to indicate the risk of NoV contamination in Irish oysters. J. Food Prot. 2009, 72, 2358–2362. [Google Scholar] [CrossRef] [PubMed]
- Hartard, C.; Banas, S.; Loutreul, J.; Rincé, A.; Benoit, F.; Boudaud, N.; Gantzer, C. Relevance of F-Specific RNA Bacteriophages in Assessing Human Norovirus Risk in Shellfish and Environmental Waters. Appl. Environ. Microbiol. 2016, 82, 5709–5719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gyawali, P.; Devane, M.; Scholes, P.; Hewitt, J. Application of Crassphage, F-RNA Phage and Pepper Mild Mottle Virus as Indicators of Human Faecal and Norovirus Contamination in Shellfish. Sci. Total Environ. 2021, 783, 146848. [Google Scholar] [CrossRef] [PubMed]
- Westrell, T.; Dusch, V.; Ethelberg, S.; Harris, J.; Hjertqvist, M.; Jourdan-Da Silva, N.; Koller, A.; Lenglet, A.; Lisby, M.; Vold, L. NoV outbreaks linked to oyster consumption in the United Kingdom, Norway, France, Sweden and Denmark, 2010. Eurosurveillance 2010, 15, 8–11. [Google Scholar] [CrossRef]
- Ballesté, E.; Blanch, A.; Mendez, J.; Sala-Comorera, L.; Maunula, L.; Monteiro, S.; Farnleitner, A.; Tiehm, A.; Jofre, J.; García-Aljaro, C. Bacteriophages Are Good Estimators of Human Viruses Present in Water. Front. Microbiol. 2021, 12, 973. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; .Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinjé, J.; et al. Enteric bacteria promote human and mouse NoV infection of B cells. Science 2014, 6210, 755–759. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ko, G. Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 2012, 55, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.; Choi, M.; Kim, J.; Ha, K.; Kwon, J.; Jeong, S.; Lee, H.; Jung, Y.; Ha, J.; Park, S. Characterizing the Effects of Thermal Treatment on Human Norovirus GII.4 Viability Using Propidium Monoazide Combined with RT-Qpcr and Quality Assessments in Mussels. Food Control 2020, 109, 106954. [Google Scholar] [CrossRef]
- Quijada, N.; Fongaro, G.; Barardi, C.; Hernández, M.; Rodríguez-Lázaro, D. Propidium Monoazide Integrated with Qpcr Enables the Detection and Enumeration of Infectious Enteric RNA and DNA Viruses in Clam and Fermented Sausages. Front. Microbiol. 2016, 7, 2008. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lowther, J.; Cross, L.; Stapleton, T.; Gustar, N.; Walker, D.; Sills, M.; Treagus, S.; Pollington, V.; Lees, D. Use of F-Specific RNA Bacteriophage to Estimate Infectious Norovirus Levels in Oysters. Food Environ. Virol. 2019, 11, 247–258. [Google Scholar] [CrossRef]
- Flannery, J.; Rajko-Nenow, P.; Winterbourn, J.B.; Malham, S.K.; Jones, D.L. Effectiveness of cooking to reduce Norovirus and infectious F-specific RNA bacteriophage concentrations in Mytilus edulis. J. Appl. Microbiol. 2014, 117, 564–571. [Google Scholar] [CrossRef]
- Polo, D.; Alvarez, C.; Diez, J.; Darriba, S.; Longa, A.; Romalde, J.L. Viral elimination during commercial depuration of shellfish. Food Control 2014, 43, 206–212. [Google Scholar] [CrossRef]
| Indicator | Sample Matrix | (n = 42) | |
|---|---|---|---|
| n | % | ||
| Overlying waters | 0 | 0 | |
| Faecal coliforms (FC) | Mussel flesh | 2 | 4.8 |
| Mussel gland | 0 | 0 | |
| Overlying waters | 2 | 4.8 | |
| Intestinal enterococci (IE) | Mussel flesh | 3 | 7.1 |
| Mussel gland | 2 | 4.8 | |
| Overlying waters | 1 | 2.4 | |
| Somatic coliphages (SOMPH) | Mussel flesh | 0 | 0 |
| Mussel gland | 0 | 0 | |
| Overlying waters | 13 | 31 | |
| F-specific RNA phages (F + PH) | Mussel flesh | 5 | 11.9 |
| Mussel gland | 3 | 9.1 | |
| Overlying waters | 23 | 54.8 | |
| Bacteroides fragilis phages (GB124PH) | Mussel flesh | 18 | 42.9 |
| Mussel gland | 18 | 42.9 | |
| Mean (%) Undetected Samples Per Matrix | |||
| Overlying waters | 7.8 | 18.5 | |
| Mussel flesh | 5.6 | 13.3 | |
| Mussel gland | 4.6 | 10.9 | |
| FC | IE | SOMPH | F + PH | GB124PH | NoV | |
|---|---|---|---|---|---|---|
| Mussel overlying Waters | ||||||
| FC | 1.000 | 0.848 ** | 0.601 ** | 0.417 * | 0.532 ** | 0.577 ** |
| IE | 1.000 | 0.506 * | 0.481 * | 0.439 * | 0.615 ** | |
| SOMPH | 1.000 | 0.577 ** | 0.720 ** | 0.859 ** | ||
| F + PH | 1.000 | 0.471 * | 0.601 ** | |||
| GB124PH | 1.000 | 0.596 ** | ||||
| Mussel flesh and intravalvular liquid | ||||||
| E. coli | 0.600 ** | 0.843 ** | 0.723 ** | 0.678 ** | 0.725 ** | 0.646 ** |
| FC | 1.000 | 0.601 ** | 0.518 ** | 0.267 | 0.461 * | 0.386 |
| IE | 1.000 | 0.753 ** | 0.699 ** | 0.699 ** | 0.656 ** | |
| SOMPH | 1.000 | 0.706 ** | 0.821 ** | 0.761 ** | ||
| F + PH | 1.000 | 0.718 ** | 0.658 ** | |||
| GB124PH | 1.000 | 0.661 ** | ||||
| Mussel digestive gland | ||||||
| FC | 1.000 | 0.603 ** | 0.529 ** | 0.534 ** | 0.545 ** | 0.466 ** |
| IE | 1.000 | 0.694 ** | 0.755 ** | 0.778 ** | 0.689 ** | |
| SOMPH | 1.000 | 0.755 ** | 0.764 ** | 0.684 ** | ||
| F + PH | 1.000 | 0.780 ** | 0.879 ** | |||
| GB124PH | 1.000 | 0.734 ** | ||||
| FC | IE | SOMPH | F + PH | GB124PH | NoV | |
|---|---|---|---|---|---|---|
| Mussel Overlying Waters | ||||||
| FC | 1.000 | 0.591 | 0.415 | 0.335 | 0.256 | 0.336 |
| IE | 1.000 | 0.027 | 0.156 | −0.135 | 0.418 | |
| SOMPH | 1.000 | 0.359 | 0.081 | 0.506 | ||
| F + PH | 1.000 | 0.189 | 0.480 | |||
| GB124PH | 1.000 | −0.243 | ||||
| Mussel Flesh and Intravalvular Liquid | ||||||
| E. coli | 0.167 | 0.499 | 0.115 | 0.485 | 0.703 * | −0.238 |
| FC | 1.000 | 0.526 | 0.639 * | 0.262 | 0.672 * | 0.415 |
| IE | 1.000 | 0.437 | 0.470 | 0.608 * | 0.056 | |
| SOMPH | 1.000 | 0.314 | 0.620 * | 0.610 * | ||
| F + PH | 1.000 | 0.428 | 0.182 | |||
| GB124PH | 1.000 | 0.035 | ||||
| Mussel Digestive Gland | ||||||
| FC | 1.000 | 0.327 | 0.492 | 0.550 | 0.426 | 0.395 |
| IE | 1.000 | 0.629 * | 0.372 | 0.450 | 0.009 | |
| SOMPH | 1.000 | 0.656 * | 0.336 | 0.683 * | ||
| F + PH | 1.000 | 0.160 | 0.723 * | |||
| GB124PH | 1.000 | 0.035 | ||||
| FC | IE | SOMPH | F + PH | GB124PH | NoV | |
|---|---|---|---|---|---|---|
| Mussel Overlying Waters | ||||||
| FC | 1.000 | 0.786 ** | 0.264 | 0.240 | 0.408 | 0.236 |
| IE | 1.000 | 0.335 | 0.612 * | 0.425 | 0.451 | |
| SOMPH | 1.000 | 0.657 * | 0.764 ** | 0.929 ** | ||
| F + PH | 1.000 | 0.517 | 0.746 ** | |||
| GB124PH | 1.000 | 0.708 ** | ||||
| Mussel Flesh and Intravalvular Liquid | ||||||
| E. coli | 0.768 ** | 0.719 ** | 0.598 * | 0.575 * | 0.563 * | 0.733 * |
| FC | 1.000 | 0.623 * | 0.501 | 0.201 | 0.470 | 0.476 |
| IE | 1.000 | 0.415 | 0.656 * | 0.553 * | 0.641 * | |
| SOMPH | 1.000 | 0.629 * | 0.684 ** | 0.764 ** | ||
| F + PH | 1.000 | 0.599 ** | 0.770 * | |||
| GB124PH | 1.000 | 0.759 ** | ||||
| Mussel Digestive Gland | ||||||
| FC | 1.000 | 0.617 * | 0.310 | 0.249 | 0.466 | 0.340 |
| IE | 1.000 | 0.253 | 0.629 * | 0.769 ** | 0.725 ** | |
| SOMPH | 1.000 | 0.441 | 0.501 | 0.495 | ||
| F + PH | 1.000 | 0.744 ** | 0.767 ** | |||
| GB124PH | 1.000 | 0.840 ** | ||||
| Period | Sample Matrices | ||
|---|---|---|---|
| Overlying Water | Mussel Flesh | Mussel Gland | |
| Annual | SOMPH | SOMPH | F + PH |
| (rho = 0.859) | (rho = 0.761) | (rho = 0.879) | |
| Spring/Summer | None | SOMPH | F+PH |
| (rho = 0.610) | (rho = 0.723) | ||
| None | SOMPH | F + PH | GB124PH |
| (rho = 0.929) | (rho = 0.770) | (rho = 0.840) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, D.T.G.; Ebdon, J.; Dancer, D.; Baker-Austin, C.; Taylor, H. A Longitudinal Study of Bacteriophages as Indicators of Norovirus Contamination of Mussels (Mytilus edulis) and Their Overlying Waters. Pollutants 2022, 2, 66-81. https://doi.org/10.3390/pollutants2010008
da Silva DTG, Ebdon J, Dancer D, Baker-Austin C, Taylor H. A Longitudinal Study of Bacteriophages as Indicators of Norovirus Contamination of Mussels (Mytilus edulis) and Their Overlying Waters. Pollutants. 2022; 2(1):66-81. https://doi.org/10.3390/pollutants2010008
Chicago/Turabian Styleda Silva, Diogo Trajano Gomes, James Ebdon, Daniel Dancer, Craig Baker-Austin, and Huw Taylor. 2022. "A Longitudinal Study of Bacteriophages as Indicators of Norovirus Contamination of Mussels (Mytilus edulis) and Their Overlying Waters" Pollutants 2, no. 1: 66-81. https://doi.org/10.3390/pollutants2010008
APA Styleda Silva, D. T. G., Ebdon, J., Dancer, D., Baker-Austin, C., & Taylor, H. (2022). A Longitudinal Study of Bacteriophages as Indicators of Norovirus Contamination of Mussels (Mytilus edulis) and Their Overlying Waters. Pollutants, 2(1), 66-81. https://doi.org/10.3390/pollutants2010008






