pMyc and pMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptide Design and Synthesis
2.2. Electrophoretic Mobility Shift Assay
2.3. Nanosystem Construction
2.4. Hemolysis Test
2.5. Cell Viability Assays
2.6. Statistical Analyzes
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Conacci-Sorrell, M.; McFerrin, L.; Eisenman, R.N. An Overview of MYC and Its Interactome. Cold Spring Harb. Perspect. Med. 2014, 4, a014357. [Google Scholar] [CrossRef] [PubMed]
- Allevato, M.; Bolotin, E.; Grossman, M.; Mane-Padros, D.; Sladek, F.M.; Martinez, E. Sequence-Specific DNA Binding by MYC/MAX to Low-Affinity Non-E-Box Motifs. PLoS ONE 2017, 12, e0180147. [Google Scholar] [CrossRef] [PubMed]
- Barsouk, A.; Padala, S.A.; Vakiti, A.; Mohammed, A.; Saginala, K.; Thandra, K.C.; Rawla, P.; Barsouk, A. Epidemiology, Staging and Management of Prostate Cancer. Med. Sci. 2020, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Leni, R.; Bray, F.; Fleshner, N.; Freedland, S.J.; Kibel, A.; Stattin, P.; Van Poppel, H.; La Vecchia, C. Epidemiology and Prevention of Prostate Cancer. Eur. Urol. Oncol. 2021, 4, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, H.; Qing, G. Targeting Oncogenic Myc as a Strategy for Cancer Treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Wang, Y.; Geer, L.Y.; Chappey, C.; Kans, J.A.; Bryant, S.H. Cn3D: Sequence and Structure Views for Entrez. Trends Biochem. Sci. 2000, 25, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Kwasigroch, J.M.; Rooman, M. SCooP: An Accurate and Fast Predictor of Protein Stability Curves as a Function of Temperature. Bioinformatics 2017, 33, 3415–3422. [Google Scholar] [CrossRef] [PubMed]
- Jabir, M.S.; Taha, A.A.; Sahib, U.I.; Taqi, Z.J.; Al-Shammari, A.M.; Salman, A.S. Novel of Nano Delivery System for Linalool Loaded on Gold Nanoparticles Conjugated with CALNN Peptide for Application in Drug Uptake and Induction of Cell Death on Breast Cancer Cell Line. Mater. Sci. Eng. C 2019, 94, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Verimli, N.; Demiral, A.; Yılmaz, H.; Çulha, M.; Erdem, S.S. Design of Dense Brush Conformation Bearing Gold Nanoparticles as Theranostic Agent for Cancer. Appl. Biochem. Biotechnol. 2019, 189, 709–728. [Google Scholar] [CrossRef] [PubMed]
- Carabet, L.A.; Lallous, N.; Leblanc, E.; Ban, F.; Morin, H.; Lawn, S.; Ghaidi, F.; Lee, J.; Mills, I.G.; Gleave, M.E.; et al. Computer-Aided Drug Discovery of Myc-Max Inhibitors as Potential Therapeutics for Prostate Cancer. Eur. J. Med. Chem. 2018, 160, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.G.; Parker, J.B.; Sagar, V.; Truica, M.I.; Soni, P.N.; Han, H.; Schiltz, G.E.; Abdulkadir, S.A.; Chakravarti, D. A MYC Inhibitor Selectively Alters the MYC and MAX Cistromes and Modulates the Epigenomic Landscape to Regulate Target Gene Expression. Sci. Adv. 2022, 8, eabh3635. [Google Scholar] [CrossRef] [PubMed]
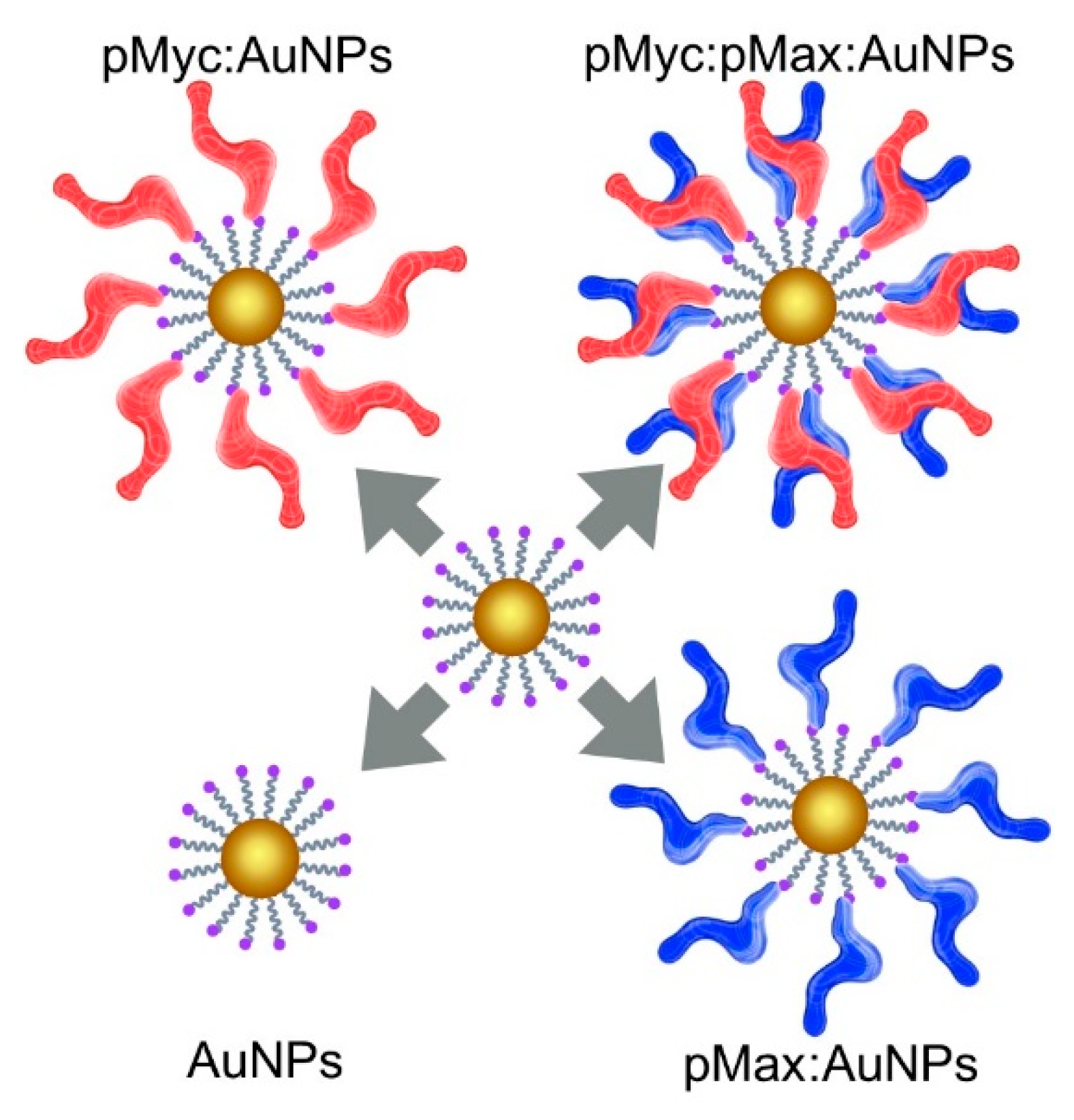
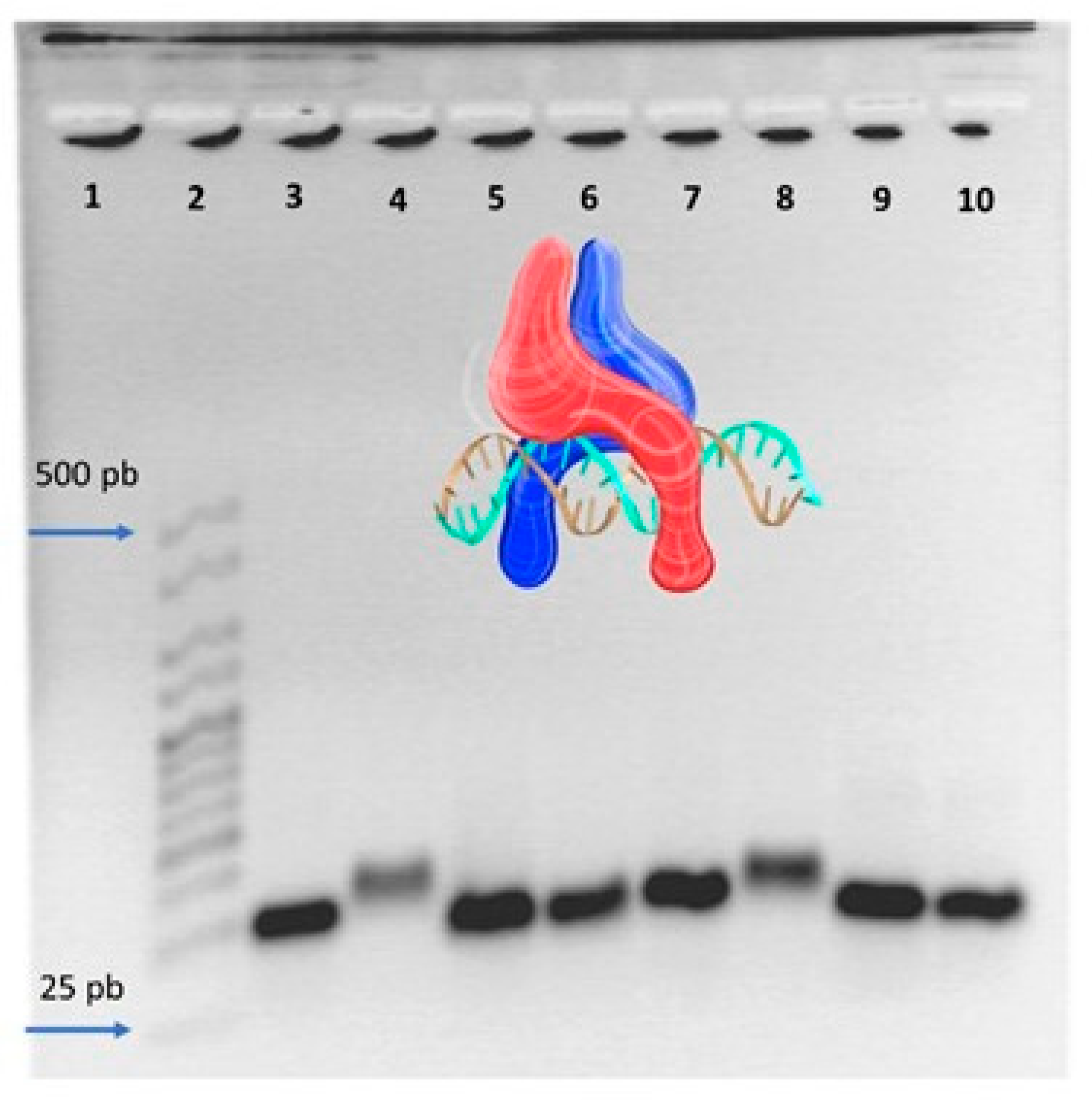

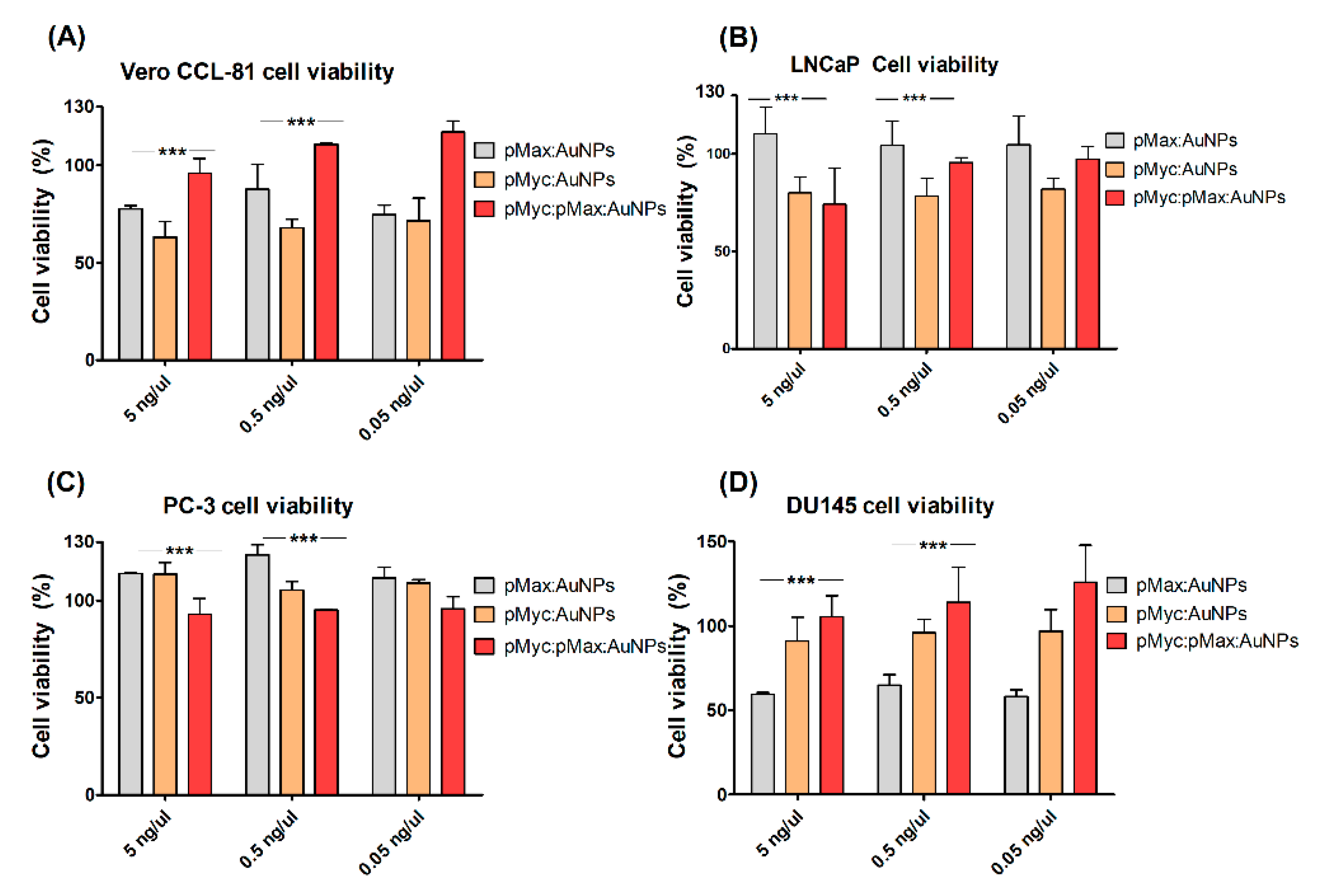
| IC | Sequence |
|---|---|
| CME Allevato F | 5′ CCG GCC ACG TGC ACG TGT TAA TAG CTC AGA CTA CTG TGT CGA CG 3′ |
| CME Allevato R | 5′ CGT CGA CAC AGT AGT CTG AGC TAT TAA CAC GTG CAC GTG GCC GG 3′ |
| CME F | 5′ AGA TCT CGA GCT GCA TGC TGT ACA CGT GAT GTC GTA CGT CGA GCT CTA GT 3′ |
| CME R | 5′ ACT AGA GCT CGA CGT ACG ACA TCA CGT GTA CAG CAT GCA GCT CGA GAT CT 3′ |
| NE F | 5′ AGA TCT CGA GCT GCA TGC TGT AAA CGT TAT GTC GTA CGT CGA GCT CTA GT 3′ |
| NE R | 5′ ACT AGA GCT CGA CGT ACG ACA TAA CGT TTA CAG CAT GCA GCT CGA GAT CT 3′ |
| CTRL F | 5′ AGA TCT CGA GCT GCA TGC TGT ATT AGC AAT GTC GTT ATC AGA GCT CTA GT 3′ |
| CTRL R | 5′ ACT AGA GCT CTG ATA ACG ACA TTG CTA ATA CAG CAT GCA GCT CGA GAT CT 3′ |
| Peptide | Tm (°C) | ΔG (kcal/mol) |
|---|---|---|
| pMyc | 80.1 | −8.4 |
| pMax | 81.4 | −6 |
| pMyc:pMax dimer | 76.6 | −4.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Longoria-García, S.; Sánchez-Domínguez, C.N.; Sánchez-Domínguez, M.; Delgado-Balderas, J.R.; Gallardo-Blanco, H.L. pMyc and pMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays. Mater. Proc. 2023, 14, 46. https://doi.org/10.3390/IOCN2023-14501
Longoria-García S, Sánchez-Domínguez CN, Sánchez-Domínguez M, Delgado-Balderas JR, Gallardo-Blanco HL. pMyc and pMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays. Materials Proceedings. 2023; 14(1):46. https://doi.org/10.3390/IOCN2023-14501
Chicago/Turabian StyleLongoria-García, Samuel, Celia N. Sánchez-Domínguez, Margarita Sánchez-Domínguez, Jesús R. Delgado-Balderas, and Hugo L. Gallardo-Blanco. 2023. "pMyc and pMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays" Materials Proceedings 14, no. 1: 46. https://doi.org/10.3390/IOCN2023-14501
APA StyleLongoria-García, S., Sánchez-Domínguez, C. N., Sánchez-Domínguez, M., Delgado-Balderas, J. R., & Gallardo-Blanco, H. L. (2023). pMyc and pMax Peptides Nanosystems and the Potential Treatment of Prostate Cancer, In Vitro Assays. Materials Proceedings, 14(1), 46. https://doi.org/10.3390/IOCN2023-14501









