1. Introduction
The Brain–Computer Interface (BCI) uses a single modality to acquire brain signals performed during a task and convert them into signals that can be actuated on other devices. However, this method has become old with the current research on Hybrid BCI, which can overcome the limitations of traditional BCI by combining two acquisition modalities. Non-invasive methods like electroencephalogram (EEG), functional near-infrared spectroscopy (fNIRS), and functional magnetic resonance imaging (fMRI) were more prominently combined for signal acquisition. EEG acquires the electrical activity of the cortex, while fNIRS and fMRI capture the changes in the Blood Oxygen Level Dependent (BOLD) signal due to changes in the hemodynamic activity of the cortex during a mental activation task. Although fMRI is proven to provide better information, fNIRS attracts better attention than fMRI due to its portability and cost efficiency [
1]. EEG-fNIRS is a common form of Hybrid BCI since the former has a good temporal resolution while the latter has a good spatial resolution.
Motor tasks are among the common BCI activation signals, which can either be motor imagery signals or motor execution signals. A µ wave (8–13 Hz) is generated in the motor cortex of the brain during imagined or executed motor tasks [
2]. These signals are primarily used to improve or supplement BCI applications. Hence, the choice of the task for obtaining motor imagery/execution signals is carefully chosen. Researchers have studied both the two-class and multi-class classification of these signals. The common choice for two-class classification was right/left hand, while for multi-class (besides right/left hand), the other choices were either foot/tongue or others like mental arithmetic [
3,
4]. From these, we can conclude that the spatial activation of the brain signal is important for classification, besides the temporal characteristics; thereby, only contralateral mental tasks were initially chosen. Nevertheless, some works have been carried out with both contralateral and ipsilateral activations, like right/left hand and right/left arm [
5]. The accuracy attained by these works is low, proving the spatial co-occurrence of features. Another author suggested a channel selection method for obtaining improved spatial features [
6]. However, this may lead to the loss of spatial features preserved in the rejected channels.
This calls for the utilization of deep learning models to obtain a complex feature extraction and classification method that can overlook the spatial co-occurrence of the features. Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) are found to have improved memory, sequential information, and spatio-temporal information encoding [
7]. In our previous work, we showed the use of CNNs to attain a good classification accuracy without omitting any channel [
8]. This also showed that every channel has spatial information that cannot be omitted. In addition to this, the current work investigates the use of a hybrid CNN, which is a combination of a CNN and Bidirectional long short-term memory (Bi-LSTM), in the performance of half the total number of channels to understand if the acquisition system can be operated on a switched mode.
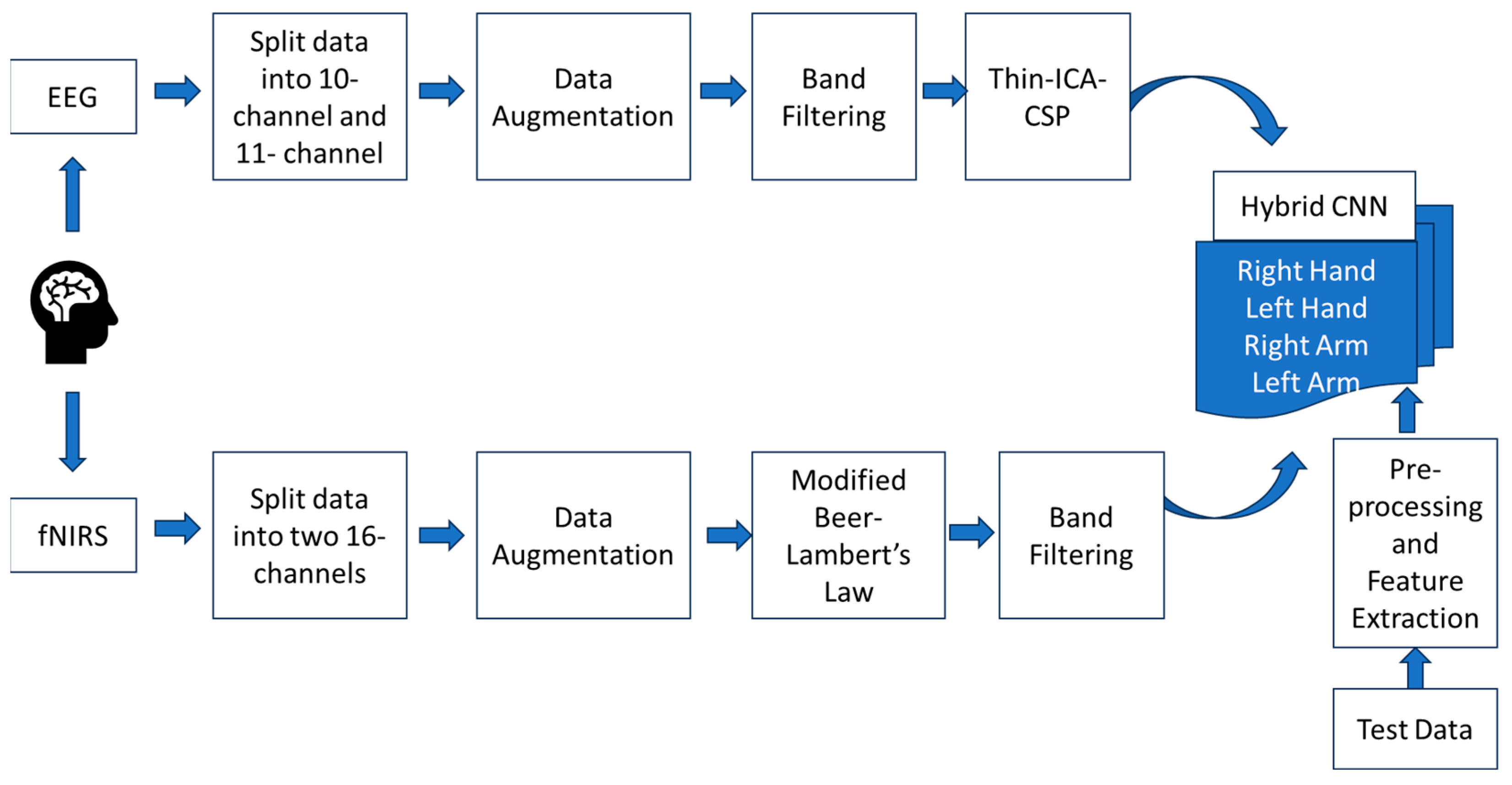
2. Methodology
The dataset was obtained from CORE datasets, consisting of EEG and fNIRS data for the right/left arm and right/left hand from 15 healthy male subjects within the age group of 23–50 years. fNIRS was obtained at two wavelengths, W1 = 760 nm (red) and W2 = 850 nm (infrared). The EEG consists of 21 electrodes of the 10–20 electrode system, namely, F3, Fz, F4, Fc5, Fc1, Fc2, Fc6, T3, C3, C1, Cz, C2, C4, T4, Cp5, Cp1, Cp2, Cp6, P3, Pz, and P4. The electrode position shows that the electrodes are distributed in the frontal (F) and parietal (P) parallel such that the signal acquisition is restricted to the motor cortex. The fNIRS was paired so that the same source could be used for multiple detectors. The list of 34 fNIRS channels includes Fc3A, Fc1A, Fc3, Fc3M, Fc1M, C3A, C1A, C3L, C3M, C1M, Cp3A, Cp1A, Cp3L, Cp3M, Cp1M, Cp3P, Cp1P, Fc2A, Fc4A, Fc2M, Fc4M, Fc4L, C2A, C4A, C2M, C4M, C4L, Cp2A, Cp4A, Cp2M, Cp4M, Cp4L, Cp2P, and Cp4P (A = Anterior, P = Posterior, M = Medial, L = Lateral).
The data were randomly split into two sets, maintaining a similar count on each hemisphere.
Table 1 shows the list of channels for the set 1 and set 2 data. The pre-processing and augmentation of the data were performed separately and simultaneously for both sets. The overall methodology is shown in
Figure 1.
The dataset needed to be augmented since deep learning models require a large amount of data. The dataset consisted of 6 s rest and 6 s task for every trial, and 25 such trials were performed for each class. The 6 s window of task performance was augmented for a 3 s time window with an overlap of 1 s.
The EEG signals of both sets were band-filtered using an Infinite Impulse Response (IIR) filter with a frequency band of 8–30 Hz and were normalized. This frequency contains both motor imagery (8–13 Hz) and motor execution (13–30 Hz) frequencies. This also omits power line interference. The normalization was performed by subtracting the data from its mean and dividing by the standard deviation. The fNIRS signals were also augmented similarly to the EEG signals. The wavelength information was converted to changes in hemoglobin concentration, that is, oxygenated and deoxygenated hemoglobin (HbO and HbR). This was performed by using the Modified Beer–Lamberts Law (MBLL). However, since these were slow varying signals, they were band-filtered using an IIR filter, between 0.01 and 0.1 Hz, because this band is in-phase. Both EEG and fNIRS use a 5th-order filter since it has a constant group delay.
The HbO and HbR data were considered as features for the fNIRS signal [
8]. However, the features for EEG data were obtained by combining Independent Component Analysis (ICA) and Common Spatial Pattern (CSP) using the Thin-ICA CSP method [
4]. CSP is known to give good results on two-class problems [
9]. Integrating this with ICA can improve spatial features that are better applicable for use in CNNs. Since CSP is better on 2-class problems, the multi-class problem was initially considered a binary problem as right/left and arm/hand. The obtained filters were used as an initialization matrix for ICA. The term Thin-ICA denotes that only second- and higher-order statistics were considered. Hence, from Thin-ICA, two features for each class were extracted.
These features were combined and fed to the hybrid CNN model. In this study, there were two ways in which the data were presented to the model. The first method was with a redundancy in the EEG data alone, and the second method was without the redundancy. This was performed to check if the amount of data was sufficient for producing good classification accuracy. The combined features were first given to a three-layer CNN with 256, 128, and 64 filters. This was then presented to three layers of Bi-LSTM with two 128 and one 64 filters. Bi-LSTM was particularly chosen in this hybrid model due to its proven performance in EEG classification [
10]. This was further given to 4 dense layers with 128 and 32 filters. Max pooling and elu activation were followed throughout the layers. Softmax activation was applied in the last dense layer. The Adam optimizer was used along with 5-fold cross-validation.
3. Results and Discussion
The dataset for this work was taken from
https://figshare.com/search?q=EEG-fNIRS+hybrid+SMR+BCI+data (accessed on 1 October 2023). The data were split into two groups, as shown in
Table 1. EEG signals were band-filtered, and features were extracted using the Thin-ICA algorithm. fNIRS, on the other hand, was also band-filtered after converting the signals to optical densities. However, no other feature extraction methods were performed on the HbO/HbR data, as they were considered as features themselves. These were then combined using zero padding since the number of channels (column data) is less in EEG than in fNIRS, i.e., 10 and 17 channels, respectively. The EEG data were zero-padded and masked before giving them to the model so that the zeros would not affect the classification accuracy. The amount of training and validation was split at 60% and 40% initially, which gave an accuracy of 73%. Hence, 80% of the data was split for training, and 20% of the data was set for validation. A 5-fold cross-validation was performed to ensure good classification.
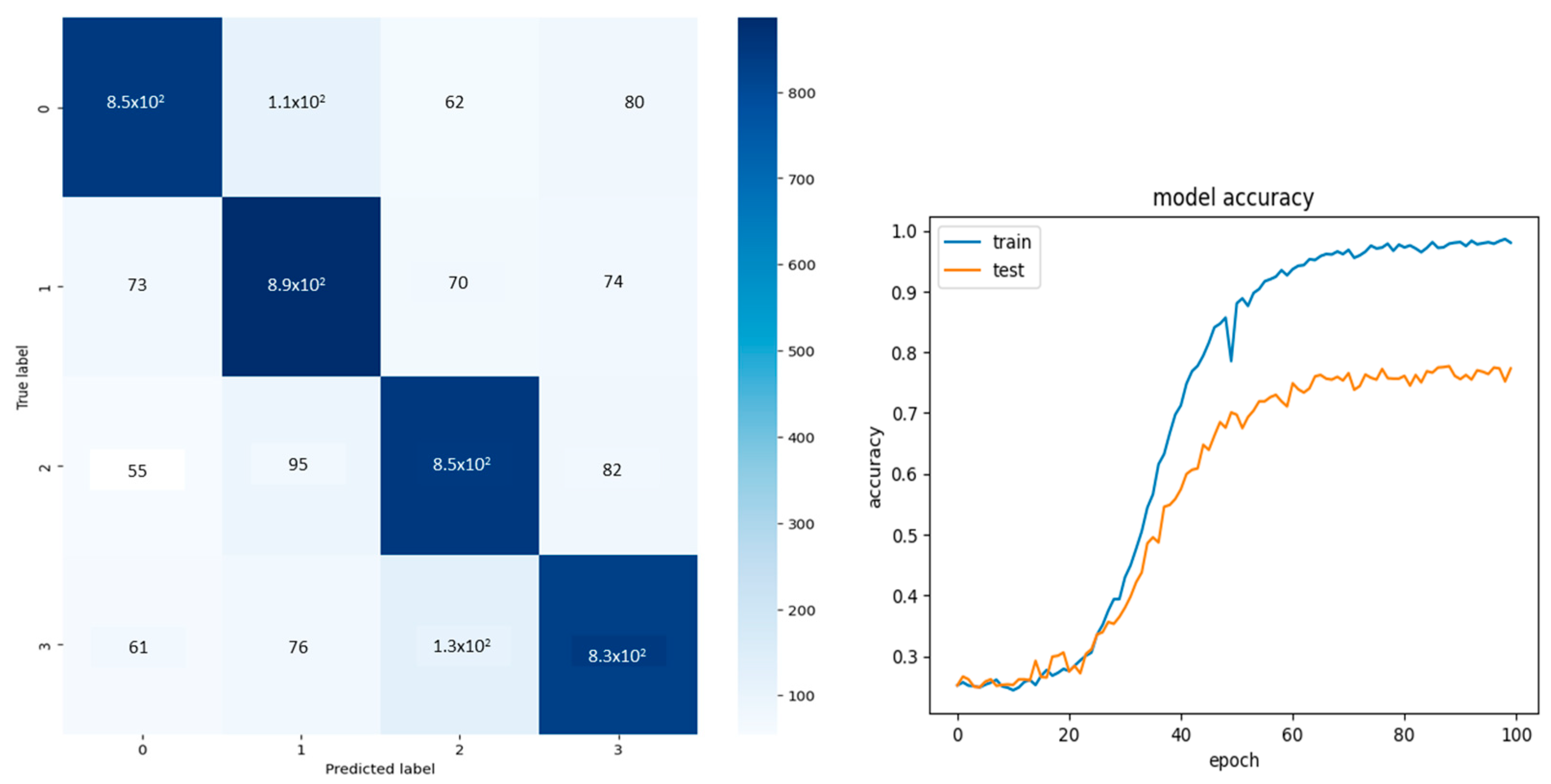
Two sets of input were passed through the model separately, and the results are shown in
Figure 2, which shows an accuracy of 78%. The confusion matrix labels 0, 1, 2, and 3 denote the four classes: right hand, left hand, right arm, and left arm.
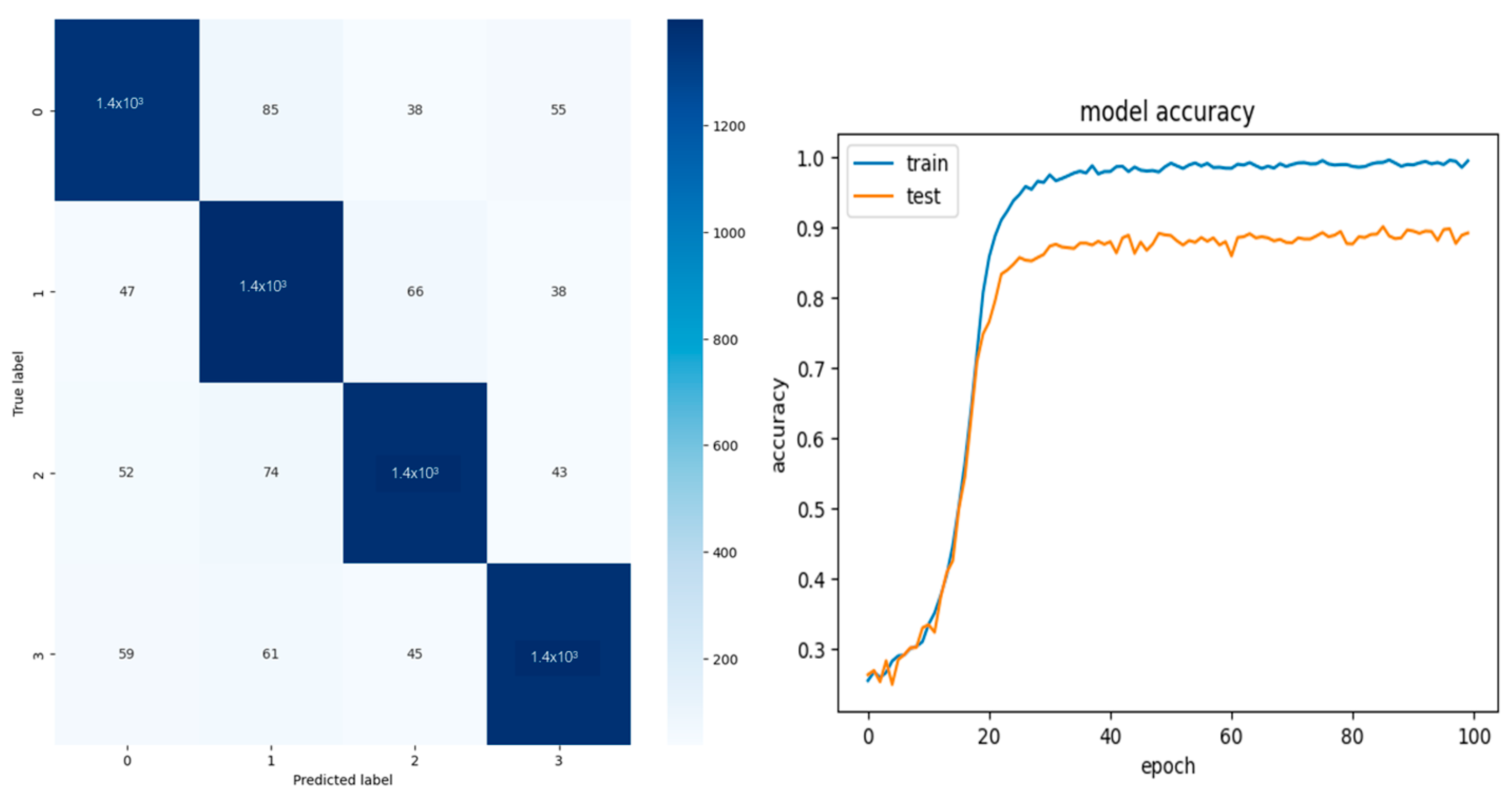
To improve accuracy, a redundancy was induced only in the EEG data, assuming data insufficiency. The classification results and their confusion matrix of the redundant data of sets 1 and 2 are shown in
Figure 3 and
Figure 4, respectively. The performance metrics for both the redundant and non-redundant data are shown in
Table 2.
Table 2 shows that a 10% increase in performance was noted when redundancy was introduced. The previous studies that used the entire channel gave an accuracy of 99% [
8]. However, the main aim of this study was to identify if every channel contributes to the spatial features by halving the data to consist of an equal number of channels on the left and right hemispheres.
Figure 4 show that the model is underfitting, which may be due to limited input data since, in the Thin-ICA CSP algorithm, the total number of independent components extracted was only two instead of five (due to the reduced number of channels), so singular value decomposition can be performed. The performance metrics are low compared to the whole dataset; however, they show equal performance. This shows that each channel has contributed to the features; hence, the acquisition system can be used in switched modes, if needed, with an equal distribution of channels on the left and right hemispheres.