A Novel Deep Learning Technique for Brain Tumor Detection and Classification Using Parallel CNN with Support Vector Machine †
Abstract
1. Introduction
2. Methods and Materials
2.1. Dataset Description
2.2. Pre-Processing
2.2.1. Image Reshaping
2.2.2. Image Augmentation
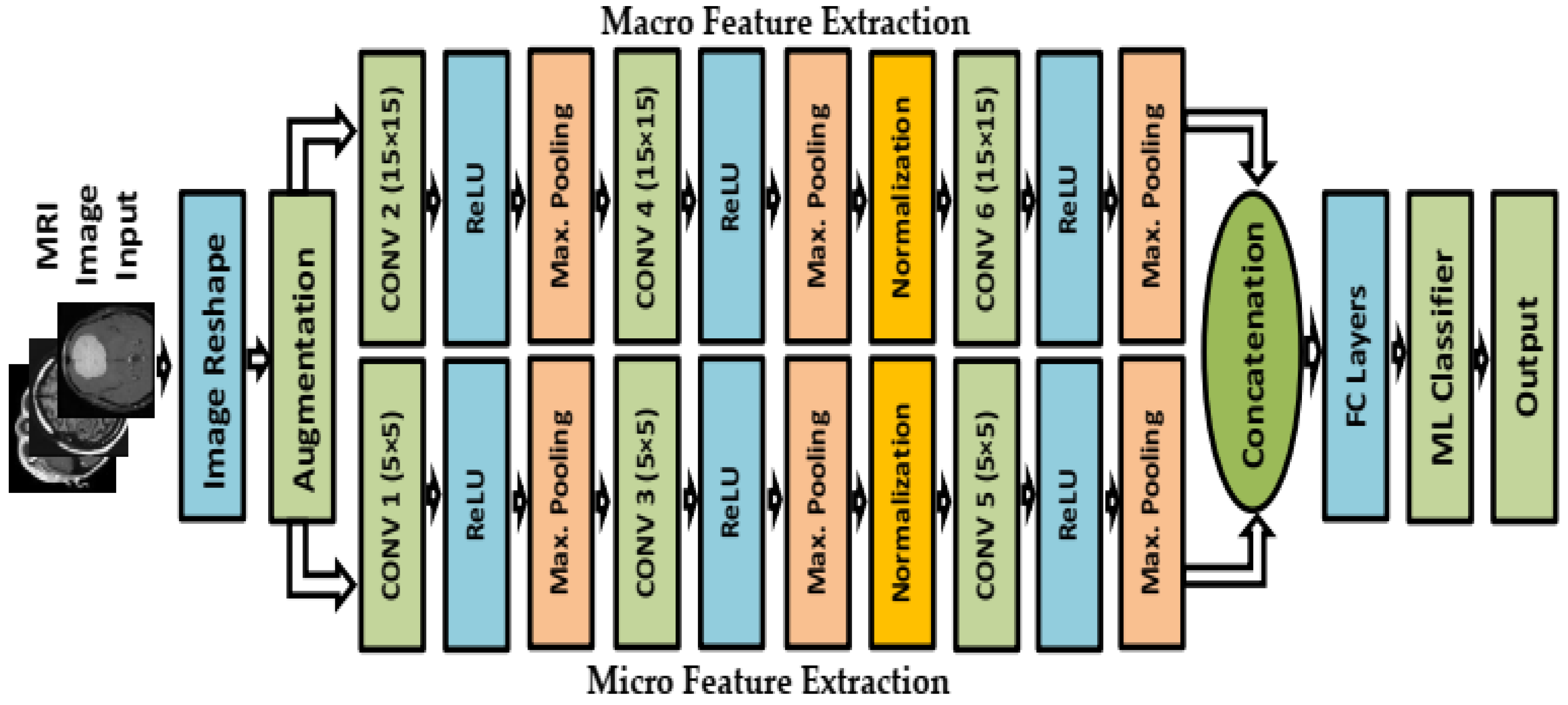
2.3. Feature Extraction Step
2.4. Classifier
3. Experimental RESULT and Analysis
3.1. Confusion Matrix
3.2. Performance Metrics
3.3. Pre-Trained Models
3.4. Comparison with Existing Research
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shanjida, S.; Islam, S.; Mohiuddin, M. Hybrid model-based Brain Tumor detection and classification using Deep CNN-SVM. In Proceedings of the 2024 6th International Conference on Electrical Engineering and Information & Communication Technology (ICEEICT), Dhaka, Bangladesh, 2–4 May 2024. [Google Scholar]
- Khazaei, Z.; Goodarzi, E.; Borhaninejad, V.; Iranmanesh, F.; Mirshekarpour, H.; Mirzaei, B.; Naemi, H.; Bechashk, S.M.; Darvishi, I.; Sarabi, R.E.; et al. The association between incidence and mortality of brain cancer and human development index (HDI). BMC Public Health 2020, 20, 1696. [Google Scholar] [CrossRef] [PubMed]
- Kalpana, R.; Chandrasekar, P. An optimized technique for brain tumor classification and detection with radiation dosage calculation in MR image. Microprocess. Microsyst. 2019, 72, 102903. [Google Scholar] [CrossRef]
- Ullah, U.; Ahmad, M.; Anwar, S.; Khattak, M.I. An Intelligent Hybrid Approach for brain tumor detection. Pak. J. Eng. Technol. 2023, 6, 42–50. [Google Scholar] [CrossRef]
- Isin, A.; Direko, C.; Sah, M. Review of MRI-Based Brain Tumor Image Segmentation Using Deep Learning Methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef]
- Rogers, L.; Barani, I.; Chamberlain, M.; Kaley, T.J.; McDermott, M.; Raizer, J.; Schiff, D.; Weber, D.C.; Wen, P.Y.; Vogelbaum, M.A. Meningiomas: Knowledge base, treatment outcomes, and uncertainties. A RANO review. J. Neurosurg. 2015, 122, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, M.; Sharifi, A.; Fard, M.J.; Soleimani, N. Detection of brain lesion location in MRI images using convolutional neural network and robust PCA. Int. J. Neurosci. 2021, 133, 55–66. [Google Scholar] [CrossRef]
- Mohsen, H.; El-Dahshan, E.S.A.; El-Horbaty, E.S.M.; Salem, A.B.M. Classification using deep learning neural networks for brain tumors. Future Comput. Inform. J. 2018, 3, 68–71. [Google Scholar] [CrossRef]
- Reza, A.W.; Hossain, M.S.; Wardiful, M.A.; Farzana, M.; Ahmad, S.; Alam, F.; Nandi, R.N.; Siddique, N. A CNN-Based strategy to classify MRI—Based Brain Tumors Using Deep Convolutional Network. Appl. Sci. 2023, 13, 312. [Google Scholar] [CrossRef]
- Garg, G.; Garg, R. Brain tumor detection and classification based on hybrid ensemble classifier. arXiv 2021, arXiv:2101.00216. [Google Scholar]
- Kabir, M.H.; Islam, M.S.; Mohiuddin, M.; Mortuza, M.G. Fuzzy-Based Smart Path Guidance System for Visually Impaired and Deaf Person. In Proceedings of the 2021 IEEE 4th International Conference on Computing, Power and Communication Technologies (GUCON), Kuala Lumpur, Malaysia, 24–26 September 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Seetha, J.; Raja, S.S. Brain Tumor Classification Using Convolutional Neural Networks. Biomed. Pharmacol. J. 2018, 11, 1457–1461. [Google Scholar] [CrossRef]
- Mohiuddin, M.; Islam, M.S. Rolling Element Bearing Faults Detection and Classification Technique Using Vibration Signals. Eng. Proc. 2022, 27, 53. [Google Scholar] [CrossRef]
- Saleh, A.; Sukaik, R.; Abu-Naser, S.S. Brain Tumor Classification Using Deep Learning. In Proceedings of the 2020 International Conference on Assistive and Rehabilitation Technologies (iCareTech), Gaza, Palestine, 28–29 August 2020. [Google Scholar]
- Mohiuddin, M.; Islam, S.; Uddin, J. Feature Optimization for Machine Learning Based Bearing Fault Classification. Indones. J. Electr. Eng. Inform. (IJEEI) 2024, 12, 610–624. [Google Scholar] [CrossRef]
- Kim, Y. Convolutional neural networks for sentence classification. In Proceedings of the 2014 Conference on Empirical Methods in Natural, Language Processing, EMNLP 2014. Doha, Qatar, 25–29 October 2014; pp. 1746–1755. [Google Scholar]
- Available online: https://www.kaggle.com/sartajbhuvaji/brain-tumor-classification-mri?select=Training (accessed on 28 October 2021).
- Wasule, V.; Sonar, P. Classifiacation of Brain MRI Using SVM and KNN Classifier. In Proceedings of the IEEE 3rd International Conference on Sensing, Signal Processing and Security (ICSSS), Chennai, India, 4–5 May 2017. [Google Scholar]
- Mohiuddin, M.; Islam, M.S.; Islam, S.; Miah, M.S.; Niu, M.-B. Intelligent Fault Diagnosis of Rolling Element Bearings Based on Modified AlexNet. Sensors 2023, 23, 7764. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1368–1372. [Google Scholar]
- Thejaswini, P.; Bhat, M.B.; Prakash, M.K. Detection and classification of tumor in brain MRI. Int. J. Eng. Manuf. (IJEM) 2019, 9, 11–20. [Google Scholar]
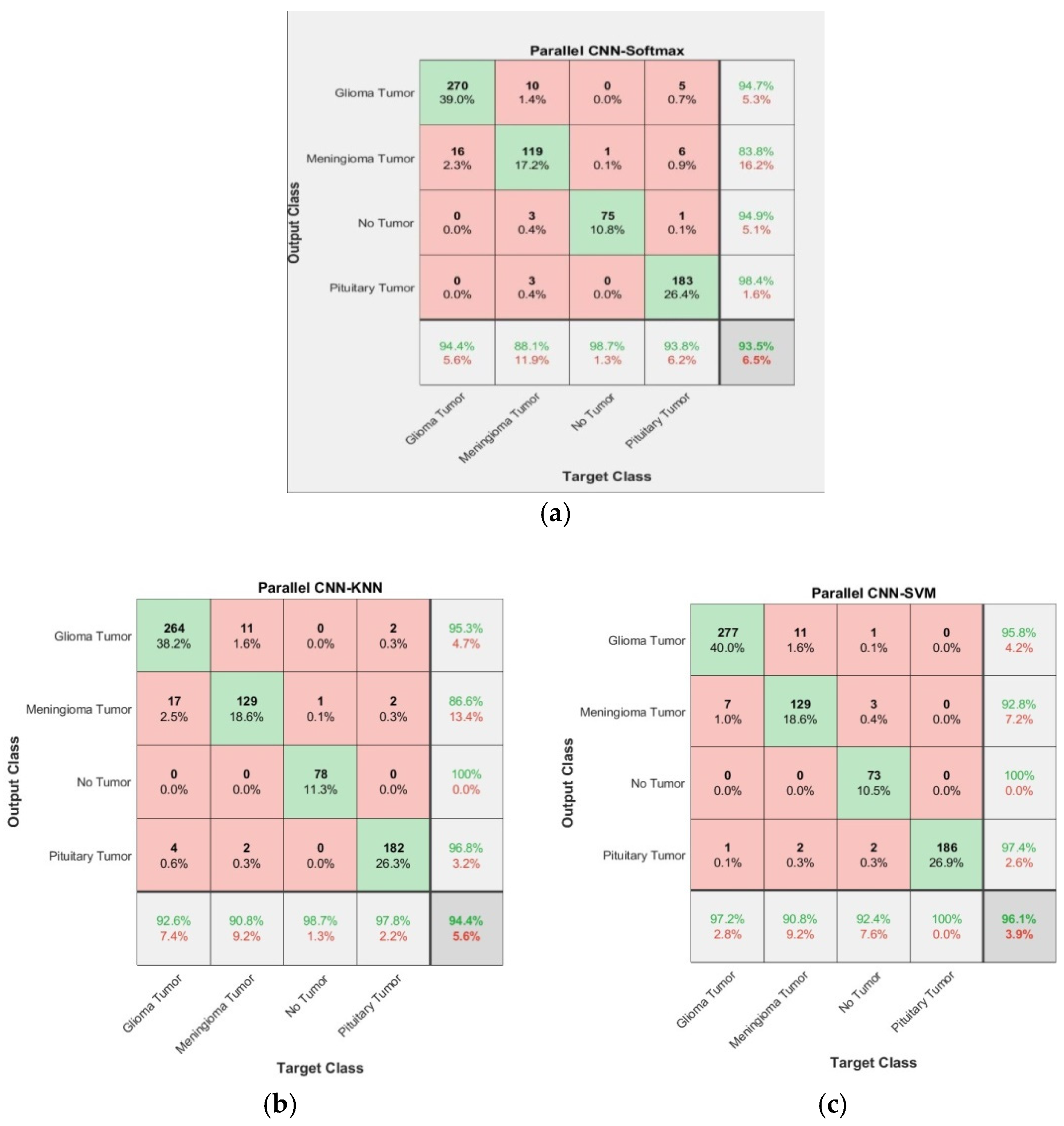
| Model | Accuary | Specificity | Sensitivity | Precision | F1-Score |
|---|---|---|---|---|---|
| PCNN-Softmax | 96.67% | 97.57% | 92.96% | 93.80% | 87.80% |
| PCNN-KNN | 97.26% | 97.86% | 94.93% | 95.11% | 90.44% |
| PCNN_SVM | 98.10% | 98.20% | 96.51% | 95.38% | 91.97% |
| Model | Google Net | VGG16 | VGG19 | ResNet18 | ResNet50 | PCNN-SVM |
|---|---|---|---|---|---|---|
| Accuracy | 93.90% | 94.80% | 95.30%, | 95.30% | 95.80% | 96.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanjida, S.; Mohiuddin, M.; Islam, M.S. A Novel Deep Learning Technique for Brain Tumor Detection and Classification Using Parallel CNN with Support Vector Machine. Eng. Proc. 2024, 82, 101. https://doi.org/10.3390/ecsa-11-20505
Shanjida S, Mohiuddin M, Islam MS. A Novel Deep Learning Technique for Brain Tumor Detection and Classification Using Parallel CNN with Support Vector Machine. Engineering Proceedings. 2024; 82(1):101. https://doi.org/10.3390/ecsa-11-20505
Chicago/Turabian StyleShanjida, Shaila, Mohammad Mohiuddin, and Md. Saiful Islam. 2024. "A Novel Deep Learning Technique for Brain Tumor Detection and Classification Using Parallel CNN with Support Vector Machine" Engineering Proceedings 82, no. 1: 101. https://doi.org/10.3390/ecsa-11-20505
APA StyleShanjida, S., Mohiuddin, M., & Islam, M. S. (2024). A Novel Deep Learning Technique for Brain Tumor Detection and Classification Using Parallel CNN with Support Vector Machine. Engineering Proceedings, 82(1), 101. https://doi.org/10.3390/ecsa-11-20505






