Computational Approaches for Structure-Based Functional Annotation of an Uncharacterized Conserved Protein of Acinetobacter baumannii †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Selection and Sequence Retrieval
2.2. Characterization of the Selected Protein’s Physicochemical Properties
2.3. Identification of the Subcellular Location
2.4. The Selected Protein’s Functional Annotation
2.5. Protein-Protein Interaction
2.6. Identification and Verification of the Chosen Protein’s Predicted Secondary Structure
2.7. Prediction of the Three-Dimensional Structure and Validation of the Chosen Protein
2.8. Active Site Determination
3. Results and Discussion
3.1. Protein Sequence Retrieval
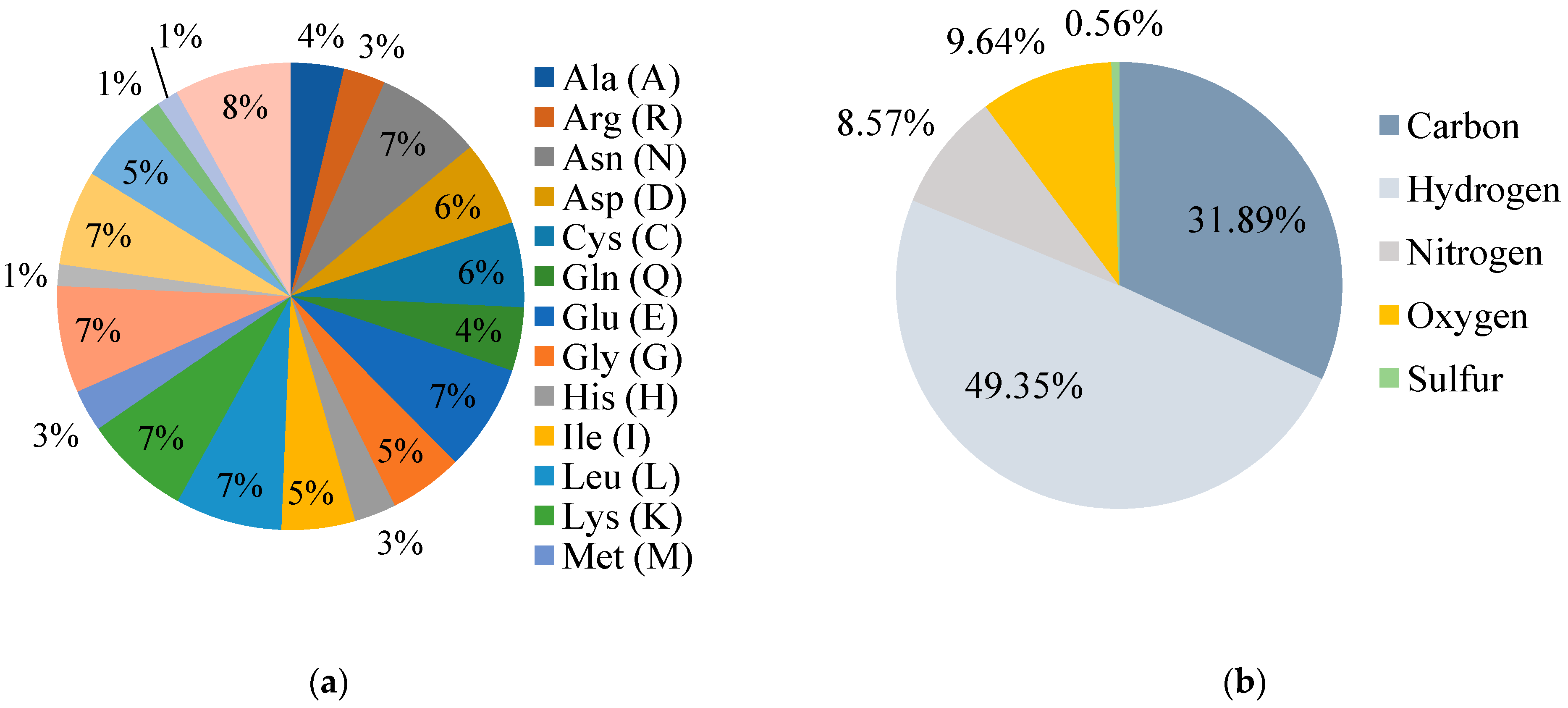
3.2. Physicochemical Properties
3.3. Subcellular Location Determination
3.4. Functional Annotation of the Selected Protein
3.5. Protein-Protein Interaction
3.6. Identification and Validation of the Predicted Secondary Structure of the Selected Protein
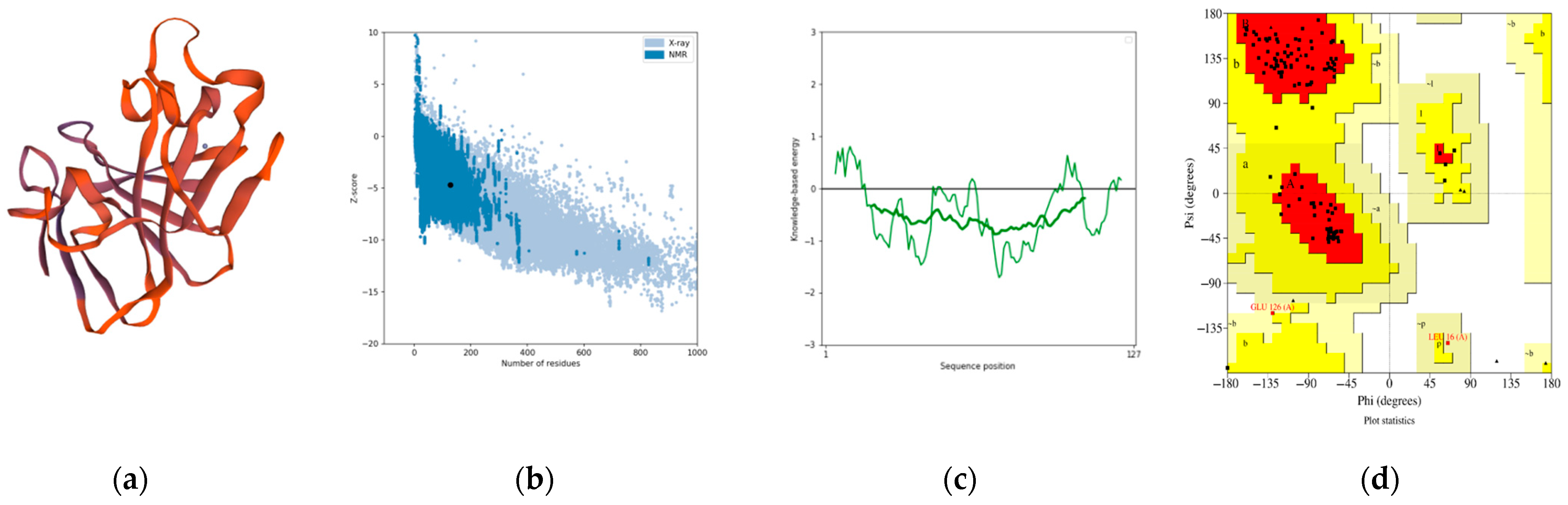
3.7. The Three-Dimensional Protein Structure Anticipation and Assessment
3.8. Active Site Determination
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Paul, A.K.; Dey, D.; Das, R.C.; Das, M.C. In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus. Chem. Proc. 2022, 12, 21. [Google Scholar] [CrossRef]
- Abhari, S.S.; Badmasti, F.; Modiri, L.; Aslani, M.M.; Asmar, M. Circulation of imipenem-resistant Acinetobacter baumannii ST10, ST2 and ST3 in a university teaching hospital from Tehran, Iran. J. Med. Microbiol. 2019, 68, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Quintanilla, M.; Pulido, M.R.; McConnell, M.J. First steps towards a vaccine against Acinetobacter baumannii. Curr. Pharm. Biotechnol. 2014, 14, 897–902. [Google Scholar] [CrossRef]
- Khan, R.A.; Hossain, R.; Siyadatpanah, A.; Al-Khafaji, K.; Khalipha, A.B.R.; Dey, D.; Asha, U.H.; Biswas, P.; Saikat, A.S.M.; Chenari, H.A.; et al. Diterpenes/Diterpenoids and Their Derivatives as Potential Bioactive Leads against Dengue Virus: A Computational and Network Pharmacology Study. Molecules 2021, 26, 6821. [Google Scholar] [CrossRef]
- Li, H.; Tan, H.; Hu, Y.; Pan, P.; Su, X.; Hu, C. Small protein A and phospholipase D immunization serves a protective role in a mouse pneumonia model of Acinetobacter baumannii infection. Mol. Med. Rep. 2017, 16, 1071–1078. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Ravella, D.; Kumar, M.U.; Sherlin, D.; Shankar, M.; Vaishnavi, M.K.; Sekar, K. SMS 2.0: An Updated Database to Study the Structural Plasticity of Short Peptide Fragments in Non-redundant Proteins. Genom. Proteom. Bioinform. 2012, 10, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. Gneg-mPLoc: A top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins. J. Theor. Biol. 2010, 264, 326–333. [Google Scholar] [CrossRef]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2019, 69, e90. [Google Scholar] [CrossRef] [PubMed]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Uddin, E.; Ahmad, T.; Mahmud, S.; Imran, A.S.; Ahmed, S.; Alyami, S.A.; Moni, M.A. Structural and Functional Elucidation of IF-3 Protein of Chloroflexus aurantiacus Involved in Protein Biosynthesis: An In Silico Approach. BioMed Res. Int. 2021, 2021, 9050026. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Islam, R.; Mahmud, S.; Imran, A.S.; Alam, M.S.; Masud, M.H.; Uddin, E. Structural and Functional Annotation of Uncharacterized Protein NCGM946K2_146 of Mycobacterium Tuberculosis: An In-Silico Approach. Proceedings 2020, 66, 13. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; E Bolton, E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 49, D10–D17. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Khalipha, A.B.R. Structure Prediction, Characterization, and Functional Annotation of Uncharacterized Protein BCRIVMBC126_02492 of Bacillus cereus: An In Silico Approach. Am. J. Pure Appl. Biosci. 2020, 2, 104–111. [Google Scholar] [CrossRef]
- Peng, X.; Wang, J.; Peng, W.; Wu, F.-X.; Pan, Y. Protein–protein interactions: Detection, reliability assessment and applications. Briefings Bioinform. 2017, 18, 798–819. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M.; Das, R.C.; Das, M.C. Computational Approaches for Structure-Based Molecular Characterization and Functional Annotation of the Fusion Protein of Nipah henipavirus. Chem. Proc. 2022, 12, 32. [Google Scholar] [CrossRef]
- Saikat, A.S.M. An In Silico Approach for Potential Natural Compounds as Inhibitors of Protein CDK1/Cks2. Chem. Proc. 2021, 8, 5. [Google Scholar] [CrossRef]
| Analysis Tool/Server | Location of Protein |
|---|---|
| PSORTb (v.3.0.2) | Cytoplasm |
| BUSCA | Cytoplasm |
| Gneg-mPLoc | Cell inner membrane |
| HMMTOP (v.2.0) | No transmembrane helices present |
| TMHMM (v.2.0) | No transmembrane helices present |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Asad, M.; Shorna, S.A.; Saikat, A.S.M.; Uddin, M.E. Computational Approaches for Structure-Based Functional Annotation of an Uncharacterized Conserved Protein of Acinetobacter baumannii. Eng. Proc. 2023, 37, 25. https://doi.org/10.3390/ECP2023-14679
Al Asad M, Shorna SA, Saikat ASM, Uddin ME. Computational Approaches for Structure-Based Functional Annotation of an Uncharacterized Conserved Protein of Acinetobacter baumannii. Engineering Proceedings. 2023; 37(1):25. https://doi.org/10.3390/ECP2023-14679
Chicago/Turabian StyleAl Asad, Mamun, Surya Afrin Shorna, Abu Saim Mohammad Saikat, and Md Ekhlas Uddin. 2023. "Computational Approaches for Structure-Based Functional Annotation of an Uncharacterized Conserved Protein of Acinetobacter baumannii" Engineering Proceedings 37, no. 1: 25. https://doi.org/10.3390/ECP2023-14679
APA StyleAl Asad, M., Shorna, S. A., Saikat, A. S. M., & Uddin, M. E. (2023). Computational Approaches for Structure-Based Functional Annotation of an Uncharacterized Conserved Protein of Acinetobacter baumannii. Engineering Proceedings, 37(1), 25. https://doi.org/10.3390/ECP2023-14679








