1. Introduction
Retinal disorders, such as diabetic retinopathy, cataract, and glaucoma, are among the leading causes of vision loss and blindness worldwide [
1,
2]. These conditions have a significant impact on quality of life and place a burden on the healthcare system [
3]. In addition to disease, the use of normal data in diagnostic studies provides a basis for distinguishing between pathological and healthy conditions [
4]. Complete and accurate diagnosis of these conditions is essential for effective treatment and prevention of recurrence [
5]. However, physical retinal examination requires a lot of time, effort, and sensitivity to individual differences, necessitating the use of automated diagnostic tools [
6].
Deep learning, particularly convolutional neural networks (CNNs), has helped the field of medical image analysis by providing powerful solutions for image classification and feature extraction [
7]. Among the available CNN architectures, the VGG19 model stands out due to its robustness and effectiveness in capturing complex features in data [
8]. The use of transfer learning, which uses a previously trained model to adapt to new tasks, is increasing model efficiency, particularly when dealing with large amounts of data [
9]. The goal of this study is to improve the accuracy and efficiency of diagnosing retinal diseases by implementing transfer learning with the VGG19 model [
10,
11]
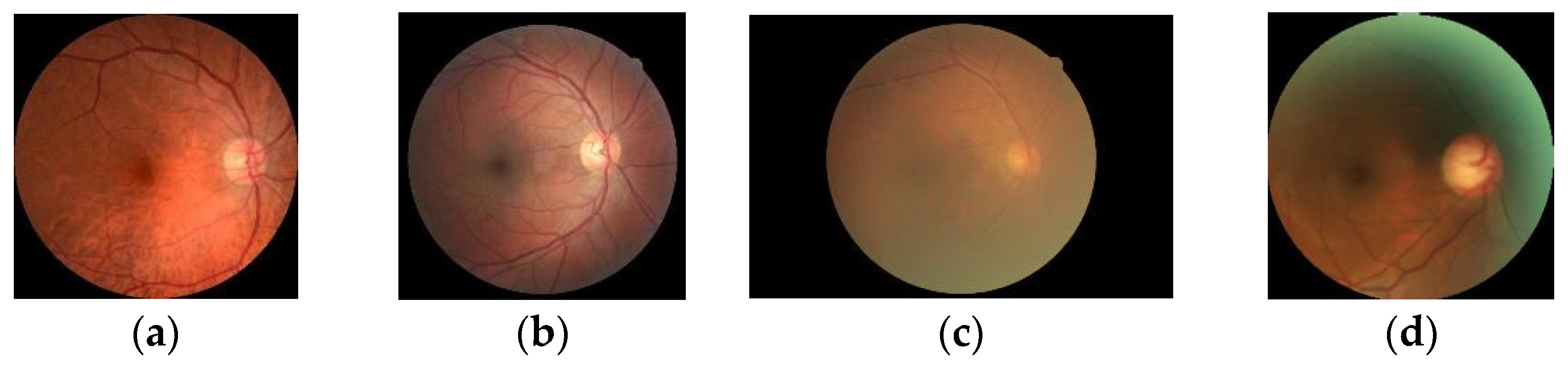
This study focuses on the identification of retinal diseases in four categories: normal, diabetic, cataract, and glaucoma [
12]. Including normal data allows for clearer distinctions between healthy and unhealthy conditions, which improves the diagnostic accuracy [
13]. The collection of retinal data available to the public is used to evaluate the performance of the VGG19 architecture in classifying this condition [
14]. This study also aims to investigate the model’s ability to generalize across multiple disease categories and its potential application in clinical practice [
15].
With this goal in mind, our research aims to contribute to the rapidly growing field of healthcare solutions provided by AI [
16]. The goal of this project is to foster the development of efficient, accurate and reliable diagnostic tools that can reduce the amount of global waste generated and improve the quality of environmental protection [
17].
1.1. Previous Related Research
Previous research [
18] focused on one type of diabetic retinopathy (DR), which is caused by diabetes and leads to vision loss, using transfer learning and a previously trained VGGNet network to analyze retinal fundus image features, as shown in
Figure 1 below.
Likewise, the present research will focus on three types of retinal diseases using the VGG19 model and transfer learning from a dataset containing normal eyes, diabetic retinopathy, cataracts, and glaucoma. The purpose of this study is to improve the classification accuracy by comparing the models for three types of eye conditions.
1.2. VGG19 Architecture
In simple terms, the VGG19 model works with a three-part architecture: backbone, pooling, and head, which is used to classify retinal diseases such as normal, diabetic retinopathy, cataract, and glaucoma using ImageNet transfer learning [
20]. The backbone uses a 3 × 3 matrix with filters that gradually increase from 64 to 512, as shown in the
Figure 2 below.
Pooling reduces the spatial dimensions and uses maximum pooling to ensure computer efficiency while retaining important information. Finally, the head converts the features into one dimension and optimizes them using a spreadsheet, followed by a softmax spreadsheet to determine the probability of reaching the target class level.
This architecture allows for efficient feature extraction and accurate classification for multi-layer systems.
2. Materials and Methods
This research was carried out in several stages, as shown in
Figure 3 below.
In
Figure 3, there are learning tasks that are completed sequentially from beginning to end, interconnected, and structured to ensure that all the learning is logical.
2.1. Dataset Retrieval
The data used in this study was obtained from Kaggle, a website that provides high-quality data and information for machine learning. The dataset consists of retinal images divided into four categories: 1038 cataract, 1098 diabetic retinopathy, 1007 glaucoma, and 1074 normal vision images, totaling 4217 images [
19]. But to maximize the training time and due to hardware limitations, not all the data is included in the VGG19 model training process. The data used consists of 400 images for 4 classes, with each class consisting of 100 images. This labeled table is essential for model training and evaluation to ensure accurate retinal disease diagnosis, as shown in
Figure 4 below.
Figure 4 shows that the dataset consists of four types of retinal diseases, with a total of 400 images used to develop the retinal disease diagnosis model.
2.2. Training Dataset
In this study, the dataset was divided into two parts: 80% for training and 20% for validation, which will be used also as testing images. The total number of images was 400 images, which were classified into four categories: normal, cataract, diabetic retinopathy, and glaucoma. Each class level has approximately 80 images for training and 20 images for validation, as illustrated in
Figure 5.
To evaluate the model performance, key metrics such as the accuracy (A), recall (R), F1 score (F1), and precision (P), which are based on standard formulas, are used.
3. Results
The model was previously trained in the Jupyter Notebook version 7.3.2 environment via the Google Colab training process. The number of images was 400, divided into four categories: normal, diabetic retinopathy, cataract, and glaucoma, with 320 images used for training and 80 images for validation, which were also used as test images. The modeling was carried out with the VGG19 transfer learning model, with parameters used to optimize the model performance. The evaluation was carried out using the accuracy, loss, precision, recall and F1 score metrics to assess the effectiveness of the model in diagnosing retinal diseases based on medical images. The model performance against the test data is shown in
Table 1 below.
The results of training the VGG19 model with transfer learning show that the model has the best performance in detecting the normal class, with precision of 1.00 and an F1 score of 0.95, while the worst performance is in the glaucoma class, with precision of 0.74 and an F1 score of 0.79. The highest F1 score values are found in the diabetic retinopathy (0.95) and normal classes, which shows that the model is more effective in detecting diabetic retinopathy than other diseases. Therefore, improving the model architecture or using data augmentation techniques may be a solution to improve the accuracy of retinal disease diagnosis.
We can visualize the model performance for the training and validation data using a curve diagram, and for the test data using an ROC curve, as shown in
Figure 6 below.
From
Figure 6 above, looking at the ROC curve results, VGG19 modeling is able to predict the classes according to the actual class values quite well, as seen from the high true positive rate in each class. In the cataract class, it is 1.00; in diabetic retinopathy, it is 0.99; in glaucoma, it is 0.94; and in normal, it is 0.98.
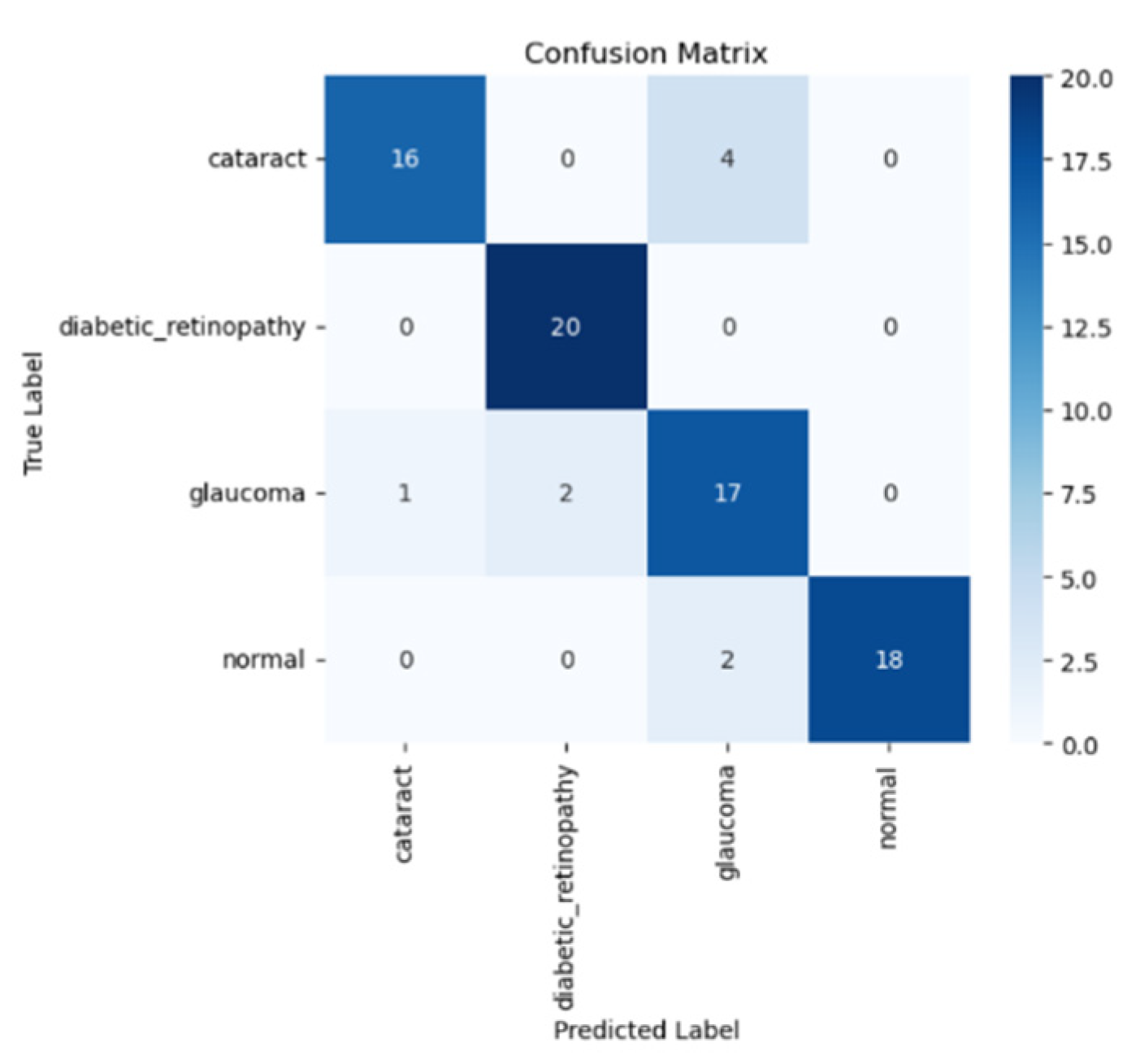
Next, we will visualize the performance model using the test data using the confusion matrix, as shown in
Figure 7 below.
The confusion matrix of the VGG19 model with transfer learning is displayed in the figure above, showing how the model classifies four types of eye disease: normal, diabetic retinopathy, cataract, and glaucoma. Based on the confusion matrix, the model has the best performance in detecting diabetic retinopathy, with a total of 20 predictions, in addition to normal 18 predictions, glaucoma 17 predictions and cataract 16 predictions.
Despite the fact that this model has limitations in differentiating various types of eye diseases, especially cataract, it excels in classifying diabetic retinopathy and normal eyes, as characterized by the high prediction rate. This shows that the model is quite effective in determining the typical patterns of these two diseases. The classification that occurs can be performed based on similar visual features between categories or by synchronizing the amount of data in each class. Therefore, improving the quality of datasets and using data augmentation techniques can help improve the model accuracy, especially in terms of the class categorization.
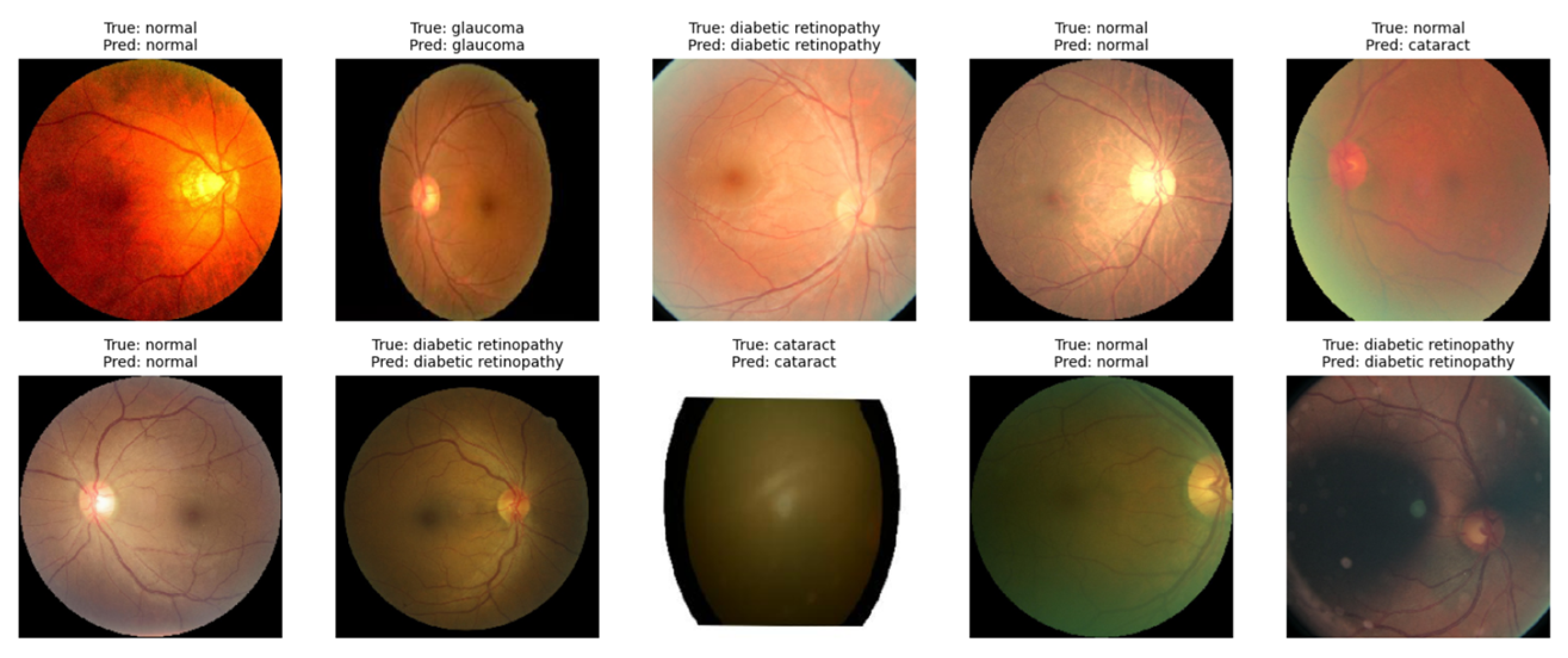
At the end of testing, the VGG19 model with transfer learning was tested to classify retinal diseases. In the image below, the model successfully identifies images with some predictions based on their category, as stated in
Figure 8 below.
The results show that the model can classify all 10 images precisely, indicating its suitability with all the true labels of the data. More extensive evaluation using larger datasets and model refinement techniques could improve the accuracy in terms of detecting different types of retinal disease.
4. Conclusions
The use of the VGG19 model and the transfer learning method for retinal disease classification has been completed. The study concluded that the VGG19 model had the best performance in detecting normal eyes, with a precision of 1.00 and a recall of 0.90, while the worst performance was found in detecting glaucoma, with a precision of 0.74 and a recall of 0.79. The highest F1 score was found in the diabetic retinopathy and normal classes (0.95), which shows that the model is more effective in classifying these types of disease compared to other types of disease.
Evaluation using the confusion matrix showed that the model achieved accuracy results of 0.91 for 320 training data and 0.88 for 80 validation data. The loss value was 0.18 for the training data and 0.31 for the validation data. Although the results are impressive, this model still has challenges in categorizing several types of retinal diseases, including glaucoma. The high probability of classification occurring is caused by visual similarities between categories or inconsistent data distribution in a dataset. Therefore, if this method is to be used consistently in medical practice, it is necessary to consider various other factors, such as clinical need, data availability, and priority of use based on the available resources.
Overall, this research found that the VGG19 model with transfer learning has the potential to be used as a diagnostic tool for retinal diseases based on medical images. However, to improve the model performance, further research can be conducted by expanding the dataset, using data augmentation techniques, and exploring more complex network architectures. With further development, this model will be able to make a significant contribution to the field of ophthalmology and enable more accurate and efficient detection of retinal diseases.
Author Contributions
Conceptualization, S., R., R.A.P. and I.L.K.; Methodology, I.L.K., S. and K.; Software, S., R.A.P., R. and I.L.K.; Validation, I.L.K. and K.; Formal Analysis, S.; Resources, S., R. and R.A.P.; Writing—Original Draft Preparation, S., R., I.L.K. and R.A.P.; Writing—Review and Editing, S., I.L.K. and K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kuper, H.; Foster, A. Impact of VISION 2020 on Global Blindness. Can. J. Ophthalmol. 2006, 41, 669–671. [Google Scholar] [CrossRef] [PubMed]
- Vardhan, K.B.; Nidhish, M.; Kiran, C.S.; Shameem, D.N.; Charan, V.S.; Bhavadharini, R.M. Eye Disease Detection Using Deep Learning Models with Transfer Learning Techniques. EAI Endorsed Trans. Scalable Inf. Syst. 2024, 11. [Google Scholar] [CrossRef]
- Assi, L.; Chamseddine, F.; Ibrahim, P.; Sabbagh, H.; Rosman, L.; Congdon, N.; Evans, J.; Ramke, J.; Kuper, H.; Burton, M.J.; et al. A Global Assessment of Eye Health and Quality of Life: A Systematic Review of Systematic Reviews. JAMA Ophthalmol. 2021. [Google Scholar] [CrossRef]
- Aberegg, S.K.; Johnson, S.A. When Alternative Diagnoses Are More Likely Than Pulmonary Embolism: A Paradox. Ann. Am. Thorac. Soc. 2020, 17, 670–672. [Google Scholar] [CrossRef]
- de Carlo, T.E.; Romano, A.; Waheed, N.K.; Duker, J.S. A Review of Optical Coherence Tomography Angiography (OCTA). Eye Vis. 2015, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Marrugo, A.G.; Millán, M.S.; Cristóbal, G.; Gabarda, S.; Šorel, M.; Šroubek, F. Toward Computer-Assisted Diagnosis and Telemedicine in Ophthalmology. SPIE Newsroom 2012. [Google Scholar] [CrossRef]
- Greenspan, H.; Van Ginneken, B.; Summers, R.M. Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Sharma, A.; Mittal, S. Deep Learning-Based Ocular Disease Screening: Comparative Performance of CNN and VGG19. In Proceedings of the 2024 International Conference on Artificial Intelligence and Emerging Technology (Global AI Summit), Greater Noida, India, 5–7 September 2024; pp. 436–440. [Google Scholar]
- Rodman, E.; Williams, N.W. Transfer Learning: A New Paradigm for Big Data Research. Available online: https://ssrn.com/abstract=4555572 (accessed on 20 June 2025).
- Proceedings of the 4th International Conference on Communication and Electronics Systems (ICCES 2019), Coimbatore, India, 17–19 July 2019; IEEE: Piscataway, NJ, USA, 2019.
- Choudhary, A.; Ahlawat, S.; Urooj, S.; Pathak, N.; Lay-Ekuakille, A.; Sharma, N. A Deep Learning-Based Framework for Retinal Disease Classification. Healthcare 2023, 11, 212. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, P.; Sharma, P. Efficient Eye Disease Detection: A Deep Learning Approach with InceptionResNetV2 for Retinal Image Classification. In Proceedings of the 2024 International Conference on Advances in Computing, Communication and Materials (ICACCM), Dehradun, India, 13–14 September 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Pepe, M.S.; Thompson, M.L. Combining Diagnostic Test Results to Increase Accuracy. Biostatistics 2000, 1, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Singla, S.; Gupta, R. Diabetic Retinopathy Diagnosis Using VGG-19 Deep Learning Model. In Proceedings of the 2024 3rd International Conference for Advancement in Technology (ICONAT), Goa, India, 19–20 January 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Kalivaraprasad, B.; Prasad, M.V.D.; Bharathi, H. Enhancing Disease Diagnosis through Transfer Learning: Improving Accuracy and Generalization. J. Electr. Syst. 2024, 20, 1424–1433. [Google Scholar] [CrossRef]
- Rastogi, M. The Growth and Potential of AI Applications in Medicine and Healthcare. Indian J. Appl. Res. 2023, 13, 17–18. [Google Scholar] [CrossRef]
- Street, A.; Vernooij, E.; Rogers, M.H. Diagnostic Waste: Whose Responsibility? Glob. Health 2022, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Sudha, V.; Ganeshbabu, T.R. A Convolutional Neural Network Classifier VGG-19 Architecture for Lesion Detection and Grading in Diabetic Retinopathy Based on Deep Learning. Comput. Mater. Continua 2021, 66, 827–842. [Google Scholar] [CrossRef]
- Doddi, G.V. Eye Diseases Classification (Eye Disease Retinal Images). Available online: https://www.kaggle.com/datasets/gunavenkatdoddi/eye-diseases-classification (accessed on 20 June 2025).
- Ogden, J.; Lee, D.M.; Moradi, H. Transfer Learning for Classification of Retinal Disease Using Fundus Imaging. In Proceedings of the 2023 IEEE 11th International Conference on Healthcare Informatics (ICHI), Houston, TX, USA, 26–29 June 2023; pp. 510–512. [Google Scholar] [CrossRef]
- Computer Vision 101. Available online: https://media.geeksforgeeks.org/wp-content/uploads/20240712183746/VGG--19-Architecture-.webp (accessed on 20 June 2025).
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).