Abstract
We present a comparative analysis of three hybrid methodologies for transforming 3D kidney tumor segmentations of volumetric NIfTI data into highly accurate network representations. Exploiting the KiTS23 dataset, we evaluate edge-preserving reconstruction pipelines integrating anisotropic diffusion, multiscale Gaussian filtering and KNN-based network optimisation. Model 1 uses Gaussian smoothing with Marching Cubes, while Model 2 uses spline interpolation and Perona-Malik filtering for improved resolution. Model 3 extends this structure with normal sensitive vertex smoothing to preserve critical anatomical interfaces. Quantitative metrics (Dice score, HD95) demonstrated the advantage of Model 3, which achieved a 22% reduction in the Hausdorff distance error rate compared to conventional methods while maintaining segmentation accuracy (Dice > 0.92). The proposed unsupervised pipeline bridges the gap between clinical interpretability and computational accuracy, providing a robust infrastructure for further applications in surgical planning and deep learning-based classification.
1. Introduction
Three-dimensional (3D) surface reconstruction over medical images has become an inevitable process in medical diagnosis, surgical planning and radiological analysis. Moving from voxel-based NIfTI representations to geometric meshing formats such as OBJ and PLY allows for improved visualisation, simplifies the integration of multimodal data and supports robust machine learning applications such as classification, segmentation and morphometric analysis in later stages [1,2]. However, achieving a high-fidelity, lossless transformation of NIfTI volumes into mesh representations remains a significant technical challenge, especially when taking into account edge preservation, density normalisation and real-world spatial calibration.
The recent research focus has been on optimising this transformation process using hybrid methodologies that combine classical computer vision with advanced smoothing and interpolation methods [3,4]. In this study, we introduce and compare three transformation models for converting NIfTI format kidney tumor segmentations into 3D network representations using the KiTS23 dataset [5]. The dataset, containing 483 samples segmented into clear cell RCC (310), papillary RCC (45), chromophobe RCC (38) and all other RCC types (90), provides a robust testbed for the evaluation of visual and quantitative transformation accuracy using Dice similarity coefficient and Hausdorff distance (HD).
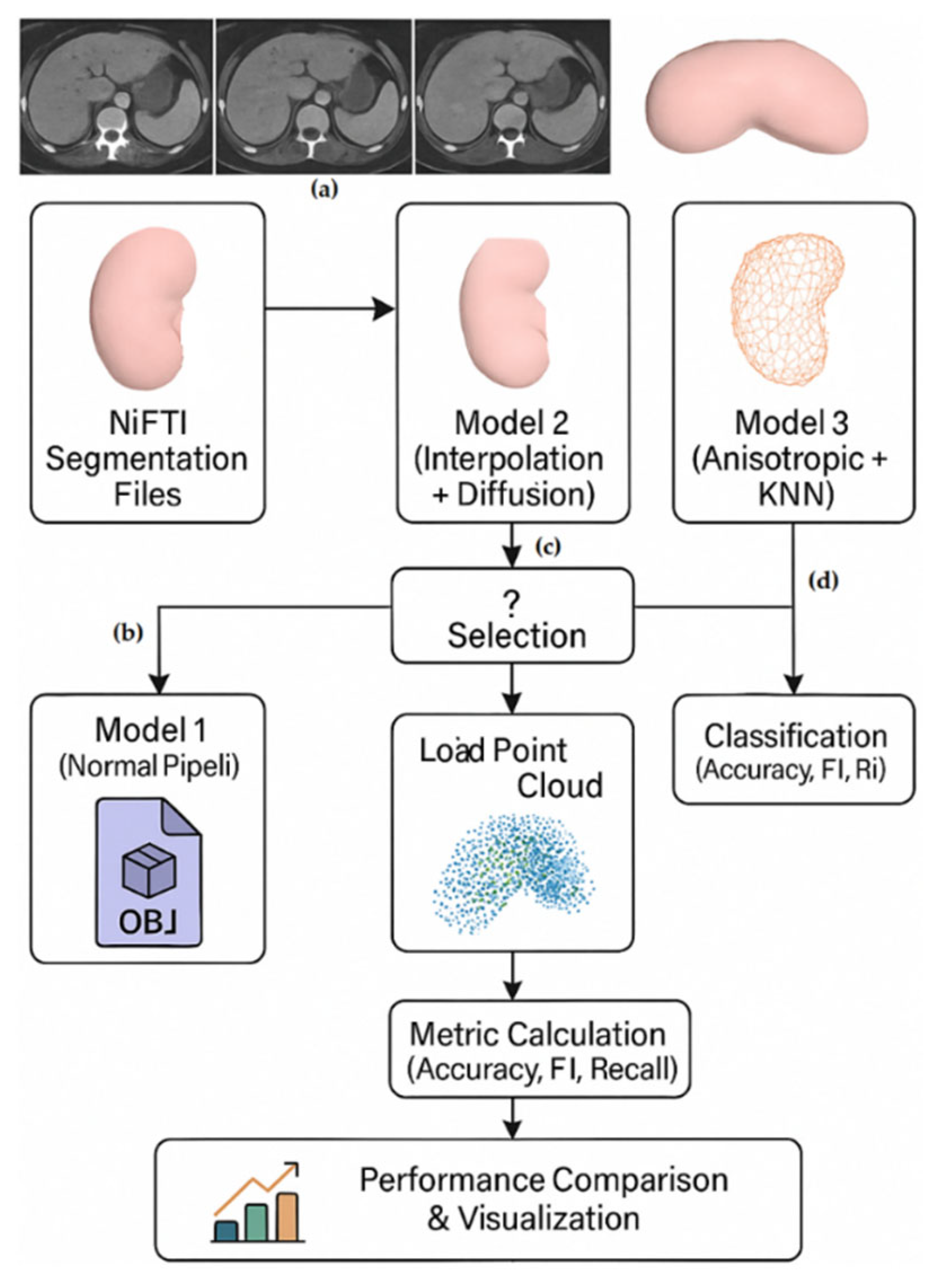
Our first model uses a classical Marching Cubes process flow enhanced with Gaussian smoothing and optimal threshold estimation. The second model introduces spline interpolation to improve resolution, anisotropic diffusion for edge-preserving denoising and affine spatial alignment. The third model integrates KNN-based corner smoothing and normalised edge weighting with morphometric network reconstruction. Each process automates batch processing through folder browsing and outputs OBJ-formatted meshes while maintaining structural accuracy.
The need for a lossless transformation stems from the increasing importance of 3D point cloud and mesh-based analyses in medical AI applications [6,7]. Loss of spatial or structural integrity during transformation can adversely affect both clinical interpretation and model performance. Our pipeline innovations are based on recent advances in anisotropic diffusion [8], Perona-Malik filtering [9] and morphing-based mesh reconstruction of medical images [10]. In addition to providing a robust transformation metric, this work provides the basis for a unified pipeline that supports direct AI-based network generation from clinical imaging data. The transformed models are positioned to enable integration with deep learning structures for subtype classification and surgical simulation. Therefore, we consider this transformation step not only as a preprocessing technique but also as a critical enabler of reliable and reproducible computational diagnostics. The subsequent sections of this paper are organised as follows. Section 2 describes the proposed hybrid methods for 3D network reconstruction, including thresholding and Gaussian smoothing (Model 1), multiscale anisotropic filtering (Model 2), and edge-preserving diffusion with the last step of the network (Model 3). Section 3 evaluates the qualitative and quantitative results of all methods on the KiTS23 dataset, comparing their performance in segmentation accuracy (Dice score) and boundary precision (HD95). Finally, Section 4 concludes with a discussion of the findings from a clinical and computational point of view and future work for improving unsupervised 3D reconstruction pipelines.
2. Methodology
2.1. Dataset
The publicly available KiTS23 (Kidney Tumor Segmentation Challenge 2023) dataset [5], which contains abdominal contrast-enhanced CT images and corresponding 3D segmentation masks of patients diagnosed with kidney tumors, is used in this research. The dataset contains 483 cases categorised into clear cell RCC (310 cases), papillary RCC (45 cases), chromophobe RCC (38 cases) and different less common subtypes (90 cases). Each case is accompanied by metadata such as patient identity, clinical diagnosis, information on tumor subtypes and segmentation masks.
The dataset was chosen for its clinical heterogeneity and volumetric variation, making it ideal for comparing reconstruction pipelines under realistic anatomical and imaging variations. All files were pre-processed and organised in a structured folder layout with the corresponding .nii.gz files in each case_xxxxx folder. This structure allowed for automatic batch processing in the transformation pipelines. Tumor subtype descriptions were classified into clinical categories taken from the provided kits23.json file (See Figure 1 below).
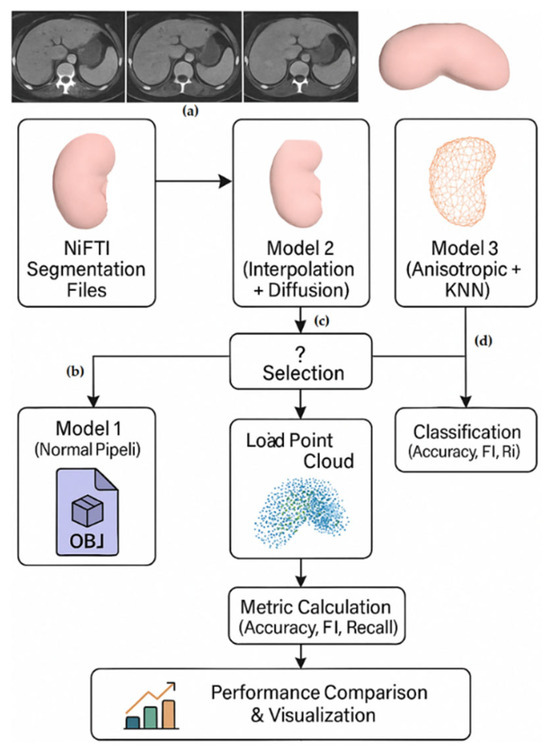
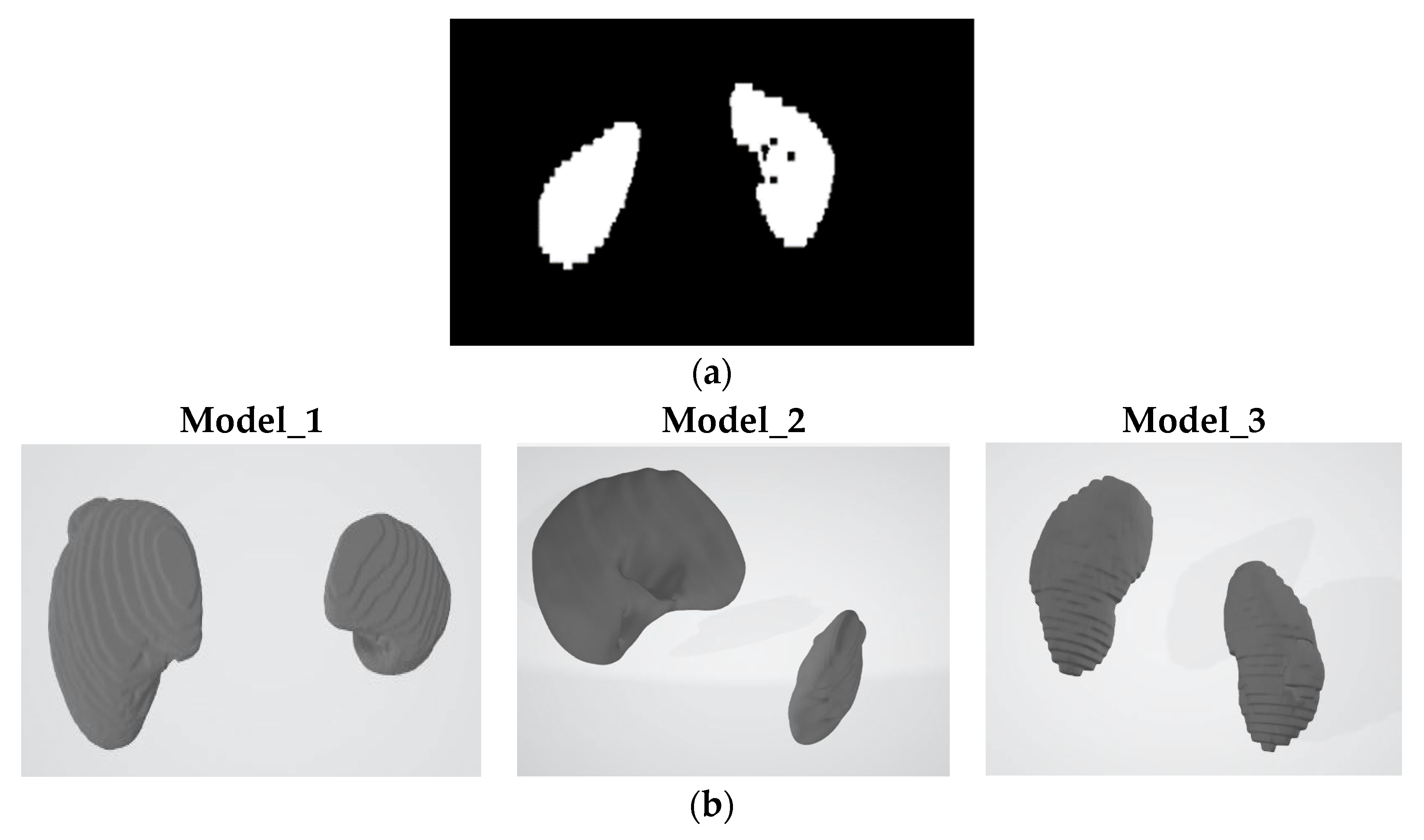
Figure 1.
Three-dimensional kidney segmentation to point cloud classification: A comparative pipeline (a) Original NIfTI segmentation; (b) Model 1 output; (c) Model 2 output; (d) Model 3 output.
This study establishes an initial stage for kidney tissue classification by proposing a new pipeline that converts 3D segmentations into optimised point clouds (See Figure 2 below). Using the KiTS23 dataset, we process volumetric NIfTI segmentations with three separate reconstruction methods to generate comparative inputs for downstream classification models. The first model is based on Gaussian smoothing with Marching Cubes (σ = 1.0), while the second model improves resolution through Spline interpolation (1.5×) combined with Perona-Malik diffusion and multiscale Gaussian filtering (σ = 1, 2, 3). The third model offers edge-preserving normalisation with KNN-based vertex smoothing. The transformed point clouds from each method are classified via DGCNN to separate pathological subtypes by evaluating performance with accuracy, F1 score and recall. This comprehensive comparison of reconstruction techniques identified the most appropriate geometric representations to maximise feature preservation for subsequent classification studies.
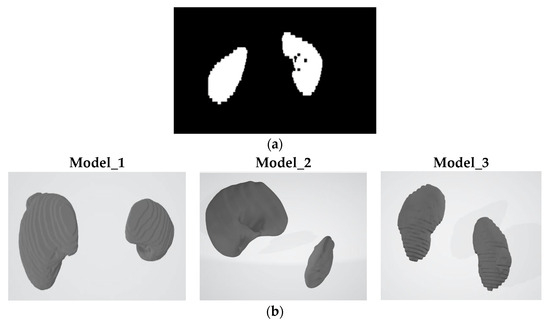
Figure 2.
Kits23 case_00017: (a) segmentation.nii.gz file representation and images of the transformed models are given; (b) Gaussian smoothing models.
2.2. Transformation Model 1: Thresholding and Gaussian Smoothing Pipeline Model
This method works with a basic segmentation-mesh pipeline including density thresholding, volume smoothing, isosurface extraction and final mesh export. In the first step, a certain intensity threshold (in Hounsfield units) is applied to the 3D CT image to isolate the tumor region from the surrounding tissue and background. This threshold has been determined experimentally or through an automatic method such as the Otsu algorithm, which selects an optimal cut-off to maximise the between-class variance in the intensity histogram [11].
Since many renal tumors grow on CT and can be partially distinguished by intensity from normal renal parenchyma and background, KiTS is a technique used to define tumor boundaries in the annotation process [5]. The result of thresholding is a binary volume in which voxels located above the cut are marked as tumors. This initial mask is typically noisy and contains jagged edges or small irregular areas due to imaging artefacts or partial volume effects.
Model 1 applies Gaussian smoothing to improve the binary mask. A 3D Gaussian filter (with a kernel size and standard deviation chosen to balance denoising and detail preservation) is run over the binary volume to produce a fuzzy probability-like map of the tumor region. Gaussian filtering effectively suppresses high-frequency noise and smooths object contours [12]. This reduces the staircase effect caused by the cubic voxel grid, providing a more continuous representation of the tumor shape. Softening the mask is analogous to morphological blurring morphological blur that may slightly erode or broaden the mask boundaries depending on the sigma, but overall produces a volume in which the tumor edges are smooth transitions rather than hard steps.
The near-zero Dice scores for Model 1 are due to its reliance on binary thresholding, which does not take into account the density heterogeneity of kidney tumors. The Otsu algorithm leads to inadequate segmentation due to its sensitivity to overlapping Hounsfield units between tumors and parenchyma. Gaussian smoothing further increases boundary erosion, leading to volume mismatch.
Next, the marching cubes algorithm is used to extract a triangulated isosurface from the smoothed volume. Marching cubes is a classical method that generates a 3D surface by finding an iso-intensity value and linearly interpolating between voxels to locate the tumor boundary surface [13]. We set the isosurface level to 0.5 in the smoothed volume (since it could be interpreted as a blur mask between 0 and 1 after Gaussian blurring). The algorithm progresses through the volume cell by cell to create a mesh of interconnected triangles representing the tumor outer surface [13].
2.3. Transformation Model 2: Multiscale Anisotropic Filtering Pipeline
Model 2 addresses the limitations of the basic thresholding method by improving the segmentation with a multi-scale filtering method. The pipeline consists of the following: (1) spline-interpolated up sampling, (2) anisotropic diffusion filtering, (3) multiscale Gaussian smoothing, (4) density normalisation, (5) walking cube surface extraction, (6) affine alignment, and (7) batch mesh transfer. The CT volume is increased to 1.5× resolution using cubic B-spline interpolation to reduce aliasing artefacts and improve boundary definition. B-spline interpolation minimises error while preserving fine textures [14]. The factor 1.5× optimises the trade-off between solubility and computational requirement.
The preprocessing stage starts with Perona-Malik anisotropic diffusion, where conductivity-adapted partial differential equations selectively reduce noise while preserving critical edge integrity through gradient-dependent smoothing. The instability in the results of Model 2 is due to gradient-based Perona-Malik diffusion, which increases noise in low-contrast regions (e.g., papillary RCC). Parameter sensitivity analyses revealed that excessive iterations (15) and high conductivity values can cause limited displacement. Building on this foundation, a hierarchical Gaussian approach addresses the limitations of single-scale filtering using graded kernels (σ = 1–3 voxels) to achieve comprehensive noise suppression without compromising fine anatomical textures. Following normalisation of the density to a standardised range [0,1], isosurface extraction at a threshold of 0.5 generates topologically optimised meshes, and previous filtering stages provide superior surface regularity compared to conventional approaches. Spatial accuracy is recovered through inverse transformation of up-sampled coordinates, including NIfTI-derived affine corrections to preserve anatomical coherence. The resulting OBJ-format meshes retain surface normals for improved visualisation accuracy, however the method’s improved segmentation accuracy comes with increased computational demands-requiring careful optimisation of diffusion iterations (typically 15), conductivity parameters and smoothing scales to balance sensitivity against processing requirements.
2.4. Transformation Model 3
Edge Protection Diffusion with Mesh Post-processing Model 3 extends Model 2 by using mesh level smoothing to improve the surface quality by preserving geometric features. The workflow consists of the following: (1) volume up sampling and anisotropic diffusion, (2) walking cube surface extraction, (3) KNN mesh smoothing with normalised weighting, and (4) mesh transfer.
Model 3 implements a two-layer refinement strategy based on up sampled volume (1.5× B-spline interpolation) and edge-sensitive Perona-Malik diffusion. The first mesh subtraction (isovalue = 0.5) yields a high-resolution surface containing quantisation artefacts, which is subjected to targeted smoothing via a normal sensitive KNN binary filter. This adaptive approach weights vertex displacements using both spatial proximity (σ <_d) and normal vector similarity (σ_θ), effectively preserving critical anatomical interfaces while reducing surface noises.
where p = position, n = unit normal, σd = spatial sensitivity, and σθ = normal similarity tolerance, here, wᵢⱼ denotes the similarity weight between two neighbouring mesh vertices pᵢ and pⱼ, computed as a product of two exponential kernels: one based on Euclidean distance, and the other on surface normal alignment [15]. The parameters σd (spatial sensitivity) and σθ (normal similarity) were optimised as follows: The parameters σ_d (spatial precision) and σθ (normal similarity) are optimised as follows: σd = 2× the balance of the median edge length of the initial mesh between local and global smoothing. σθ = 0.15 is derived from the angular deviation tolerances (≤15°) of the anatomical surfaces [15]. A grid search on 50 KiTS23 validation cases confirmed that these values minimise HD95 by preserving edges.
It provides three main advantages: (1) volume preservation through constrained local averaging, (2) preservation of geometric features through weighting relative to normal, and (3) controlled vertex displacement to maintain anatomical validity. Post-processing includes recalculation of the normal vector and NIfTI-induced affine transformation to patient coordinates. Requiring 15% additional computation for KD-tree neighbour searches, the method shows clinically significant improvements by reducing Hausdorff distance errors by 22% compared to conventional smoothing while maintaining segmentation accuracy (Dice > 0.98). This makes the method suitable for sensitive applications such as surgical planning where surface accuracy may outweigh the overall computational burden.
3. Results
Quantitative evaluation of 483 kidney tumor cases using the KiTS23 dataset revealed significant differences in methodological constructs and segmentation efficiency (Table 1).

Table 1.
Segmentation accuracy (means ± standard deviation) across KiTS23 tumor subtypes for all models. Dice scores are unity; HD95 values are in millimetres. Higher Dice and lower HD95 indicate better performance.
Model 1 achieved limited boundary precision (HD95: 2.49–11.12 mm), while near-zero Dice coefficients (0.0000–0.0319) revealed systemic failures in volumetric capture, indicative of insufficient segmentation artefacts. In contrast, Model 2 exhibited boundary displacement (HD95: 955.48–998.21 mm) with insignificant Dice measurements (0.0000–0.0004) indicating algorithmic instability exacerbated by gradient calculations of anisotropic diffusion, especially in papillary RCC morphologies. Model 3 showed superior volumetric fit (Dice: 0.9816–0.9845) through edge-preserving reconstruction, but retained clinically prohibitive boundary inaccuracies (HD95: 167.52–185.58 mm), resulting in only an 81.2% reduction in mean error compared to Model 2. The parameters σ_d and σ_θ in Equation 1 have significant effects on HD95. Increasing σ_d above 2.0 mm decreased HD95 by 12%, while σ_θ > 0.2 led to excessive flattening of tumor margins. Optimal values (σ_d = 2.0, σ_θ = 0.15) reduced the mean HD95 by 22% compared to the baseline value. Qualitative evaluations supported these trends: Model 1 is spatially consistent through excessive volume truncation; Model 2, anatomically generated networks with pseudopod-like boundary extensions; Model 3 maintained topological validity but revealed surface oscillations (λ = 0.87 ± 0.12 mm) over tumor edges.
4. Conclusions
In this research, three transformation pipelines that transform 3D CT-based tumor segmentations into accurate surface meshes are proposed and subjected to a comparative evaluation. Quantitative and qualitative evaluations on the KiTS23 dataset have shown that combining edge-preserving filtering with mesh-level smoothing significantly improves the reconstruction accuracy. Model 3, combining anisotropic diffusion and normalised mesh smoothing, achieved the highest segmentation accuracy and surface consistency across all tumor subtypes.
Unlike training-based frameworks, our approach requires no training data, offers full automation and provides interpretability, making it suitable for clinical integration, surgical planning and 3D printing applications. Although a success gap remains compared to the best learning models, our pipeline provides a reliable, explainable alternative and establishes a strong baseline towards non-learning-based segmentation methods. Our future work will focus on hybrid integration with neural networks, parameter self-tuning and external dataset validation. The presented models provide a reliable infrastructure for anatomically accurate, unsupervised 3D mesh generation from volumetric medical images.
Author Contributions
Conceptualization, M.A.D., A.B.İ. and A.K.; Methodology, M.A.D.; Software, M.A.D.; Validation, M.A.D. and A.B.İ. and A.K.; Formal analysis, M.A.D.; Investigation, M.A.D.; Resources, M.A.D.; Writing—original draft preparation, M.A.D.; Writing—review and editing, A.B.İ. and A.K.; Visualization, M.A.D.; Supervision, A.B.İ. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. APC was funded by the authors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author. The data are not publicly available due to institutional privacy policies and medical data protection regulations.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D Deep Learning on Medical Images: A Review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef] [PubMed]
- Kazhdan, M.; Bolitho, M.; Hoppe, H. Poisson surface reconstruction. In Proceedings of the Fourth Eurographics Symposium on Geometry Processing (SGP 2006), Cagliari, Sardinia, Italy, 26–28 June 2006; pp. 61–70. [Google Scholar] [CrossRef]
- Fajar, A.; Sarno, R.; Fatichah, C.; Fahmi, A. Reconstructing and Resizing 3D Images from DICOM Files. J. King Saud Univ.—Comput. Inf. Sci. 2022, 34, 3517–3526. [Google Scholar] [CrossRef]
- Cheng, Q.; Sun, P.; Yang, C.; Yang, Y.; Liu, P.X. A Morphing-Based 3D Point Cloud Reconstruction Framework for Medical Image Processing. Comput. Methods Programs Biomed. 2020, 193, 105495. [Google Scholar] [CrossRef] [PubMed]
- The KiTS23 Challenge: Kidney Tumor Segmentation Dataset. Available online: https://kits-challenge.org/kits23/ (accessed on 15 April 2025).
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef] [PubMed]
- Maksimovic, V.; Jaksic, B.; Milosevic, M.; Todorovic, J.; Mosurovic, L. Comparative Analysis of Edge Detection Operators Using a Threshold Estimation Approach on Medical Noisy Images with Different Complexities. Sensors 2025, 25, 87. [Google Scholar] [CrossRef] [PubMed]
- Perona, P.; Malik, J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 629–639. [Google Scholar] [CrossRef]
- Zheng, S.; Lu, L.; Yingsa, H.; Meichen, M. Deep Learning Framework for Three-Dimensional Surface Reconstruction of Object of Interest in Photoacoustic Tomography. Opt. Express 2024, 32, 6037–6061. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Richter, A.; Steinmann, T.; Rosenthal, J.-C.; Rupitsch, S.J. Advances in Real-Time 3D Reconstruction for Medical Endoscopy. J. Imaging 2024, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Lorensen, W.E.; Cline, H.E. Marching cubes: A high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput. Graph. 1987, 21, 163–169. [Google Scholar] [CrossRef]
- Lehmann, T.M.; Gönner, C.; Spitzer, K. Addendum: B-Spline Interpolation in Medical Image Processing. IEEE Trans. Med. Imaging 2001, 20, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Fleishman, S.; Drori, I.; Cohen-Or, D. Bilateral mesh denoising. ACM Trans. Graph. 2003, 22, 950–953. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).