Abstract
This article presents an integrated system for automated diagnosis, combining convolutional neural networks (CNNs) for dermatological image analysis with a patient survey for clinical data collection. The goal is to evaluate the effectiveness of this hybrid approach compared to traditional diagnostic methods. The system was tested on a curated dataset composed of images collected from DermNet and publicly available dermatological image databases. The results demonstrate high diagnostic accuracy for inflammatory skin diseases, with the combined approach outperforming standalone methods. These findings highlight the potential of integrating machine learning with patient-reported data to enhance dermatological diagnostics. The proposed system can be implemented in clinical practice and integrated into existing medical platforms, aiding dermatologists in decision-making and improving patient care. Future research will focus on expanding the system to diagnose a broader range of skin conditions and incorporating additional clinical data to enhance its performance.
1. Introduction
Skin health is an essential element of a person’s overall well-being and has a significant impact on quality of life, emotional well-being, and social relationships. Skin condition often serves as a mirror of internal health, highlighting the importance of caring for and understanding skin problems. Skin conditions are often accompanied by physical symptoms such as itching, pain, and redness that interfere with daily activities [1].
In recent years, dermatology has entered a period of technological transformation, where traditional diagnostic approaches are increasingly supported by artificial intelligence (AI). Deep neural networks (DNNs), particularly convolutional neural networks (CNNs), have demonstrated impressive capabilities in medical image analysis. Esteva et al. (2017) showed that CNNs can classify skin lesions at a level comparable to experienced dermatologists [2]. AI applications in healthcare span both structured and unstructured data, utilizing classical machine learning algorithms as well as deep learning and natural language processing techniques.
Despite the progress in AI-based dermatological diagnostics, most existing research has concentrated on the detection of skin cancer [3,4]. In contrast, inflammatory skin diseases—although common and impactful—remain underrepresented in AI research. These conditions, while not typically life-threatening, can cause considerable psychological distress and social stigma, significantly affecting patients’ quality of life [5,6]. Their diagnosis is complicated by overlapping visual symptoms, variable clinical presentations, and reliance on subjective evaluation [7]. Purely image-based AI models may struggle with such complexity, particularly when lacking clinical context such as patient history or symptom perception.
To bridge this gap, this study proposes a hybrid diagnostic system that combines automated image analysis using CNNs with structured patient-reported information. The system collects data on symptom duration, lifestyle, medical history, and self-assessed severity to provide a richer diagnostic context [8,9,10]. This multimodal approach aims to improve diagnostic precision and personalization, aligning with the goals of precision medicine [11]. Beyond improving accuracy, the system may support practical applications in telemedicine and mobile health—especially in underserved areas with limited access to dermatological expertise [12,13].
This paper [14] presents the architecture of the proposed hybrid system, details its methodological framework, and shares preliminary results that demonstrate its potential to enhance AI-driven dermatological care.
2. Main Functionalities of the System
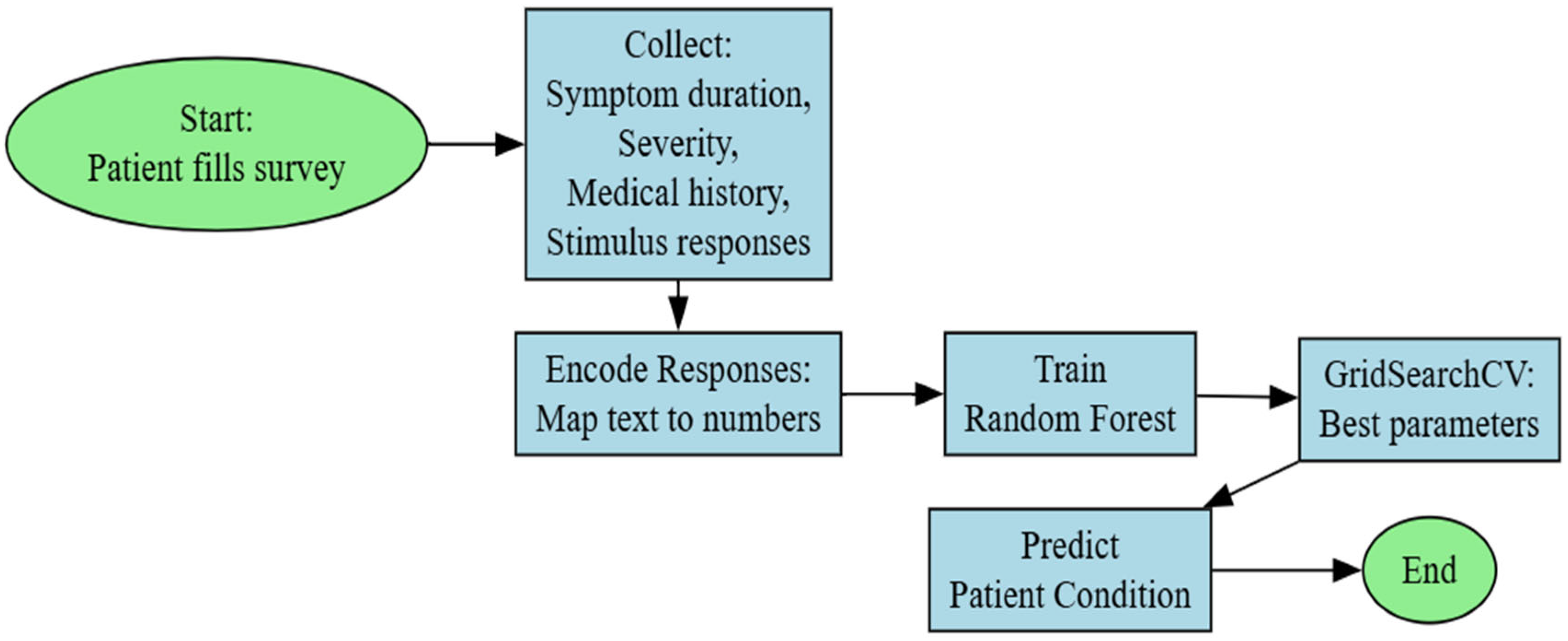
The main task of the system is to build a structure for collecting and processing user data, using machine learning, for diagnosing various skin problems. The system collects user responses from a survey, analyzes the user’s skin condition, and provides predictions for various skin problems, as well as possible recommendations (Figure 1).
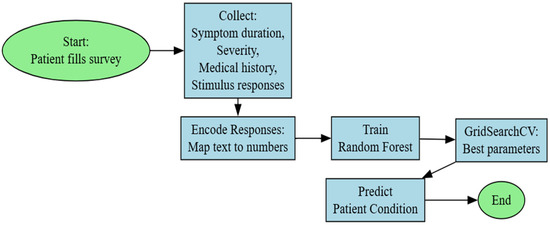
Figure 1.
Random forest model block diagram.
A survey is created that contains 26 questions about the user’s skin condition. The questions are grouped thematically into categories such as sensitivity, acne, dryness, and redness, as shown in Table 1. The resulting data is processed. For processing, a train_test is used to divide the data into training and test sets. Thus, the system is trained with one part of the data and is tested with another part of the data [15,16]. Using a machine learning model, the user’s skin problems are predicted. The models are used to assess the accuracy and present predictions for skin problems. Probabilities are calculated for each category of skin problems. A threshold (0.5) is used to determine whether a problem is present or not.

Table 1.
Thematic grouping of the patient-reported questionnaire used in the study.
3. Materials and Methods
This study proposes a multimodal approach for the automated diagnosis of inflammatory skin diseases by integrating two parallel components: deep neural networks (CNNs) for medical image analysis and a Random Forest-based assessment of subjective data collected via a patient survey.
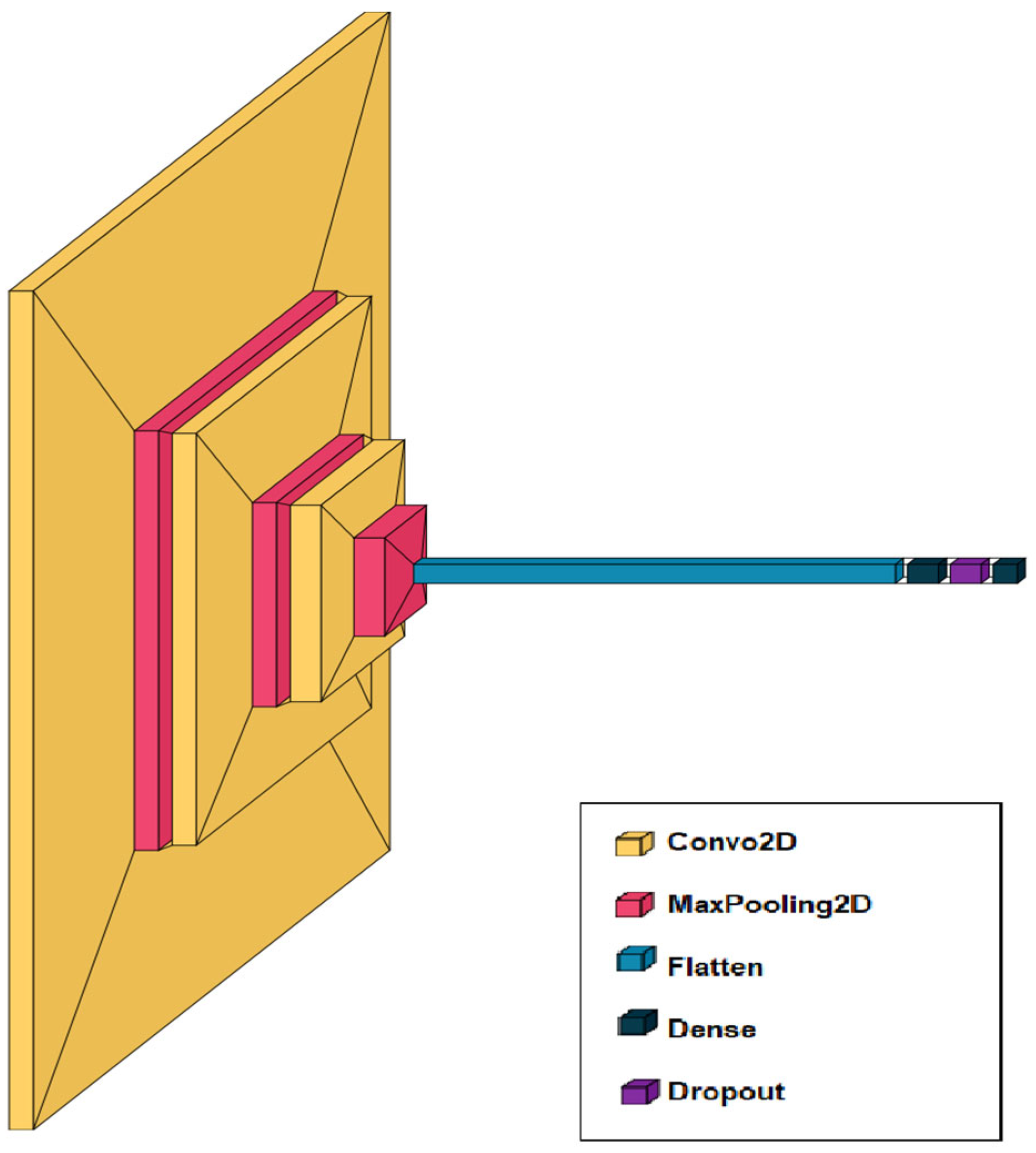
The network includes successive convolutional and pooling layers for feature extraction, followed by fully connected layers and dropout for classification and overfitting prevention (Figure 2).
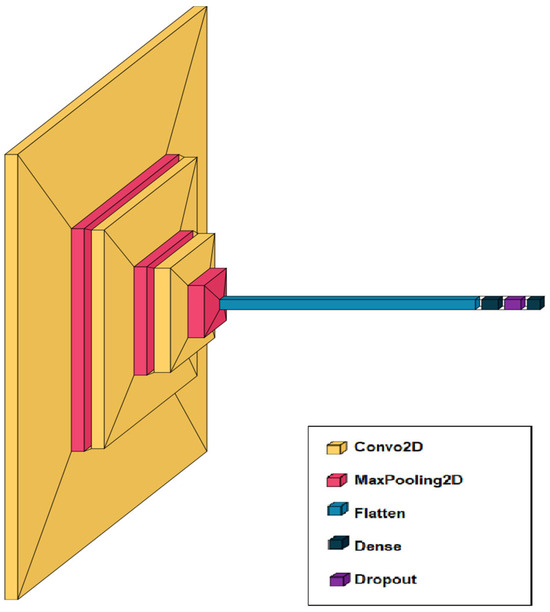
Figure 2.
Architecture of a convolutional neural network for recognizing skin diseases from images.
For the analysis of dermatological images, a convolutional neural network (CNN) was trained to automatically extract visual features from skin disease images. Preprocessing steps included pixel normalization and data augmentation techniques, such as rotation, horizontal and vertical shifting, scaling, and mirroring. These methods improve the model’s adaptability to variations in input data, enhancing its ability to generalize and classify different skin conditions.
To complement the image-based analysis, a structured patient survey was designed to collect subjective data, including symptom severity, medical history, and other relevant clinical indicators. The survey responses were encoded numerically using a predefined mapping system to convert categorical data into a usable format for analysis. A Random Forest algorithm was employed to process this multidimensional dataset, leveraging its ability to handle complex feature spaces. Hyperparameter optimization, including Grid Search, was used to identify the optimal configuration for maximizing diagnostic accuracy.
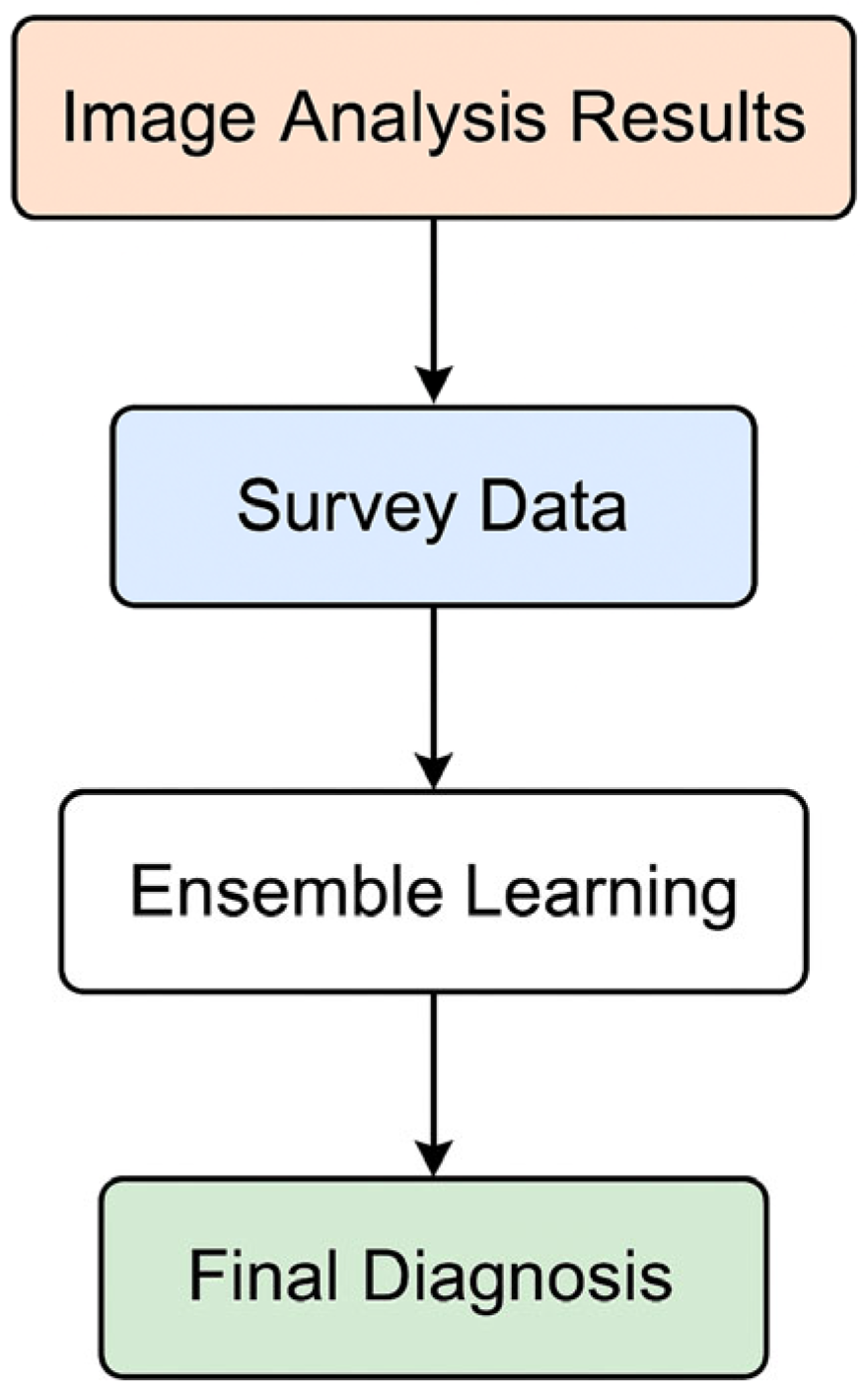
The final diagnosis was made by combining the results from both the image analysis and the patient survey evaluation. This multimodal approach enables the integration of objective visual data with subjective clinical and symptomatic information, resulting in a more accurate and personalized skin condition assessment. Ensemble learning techniques and data fusion methods were applied to mitigate the limitations of individual models, ultimately creating a more robust and reliable diagnostic system.
4. System Architecture
4.1. Module Diagram
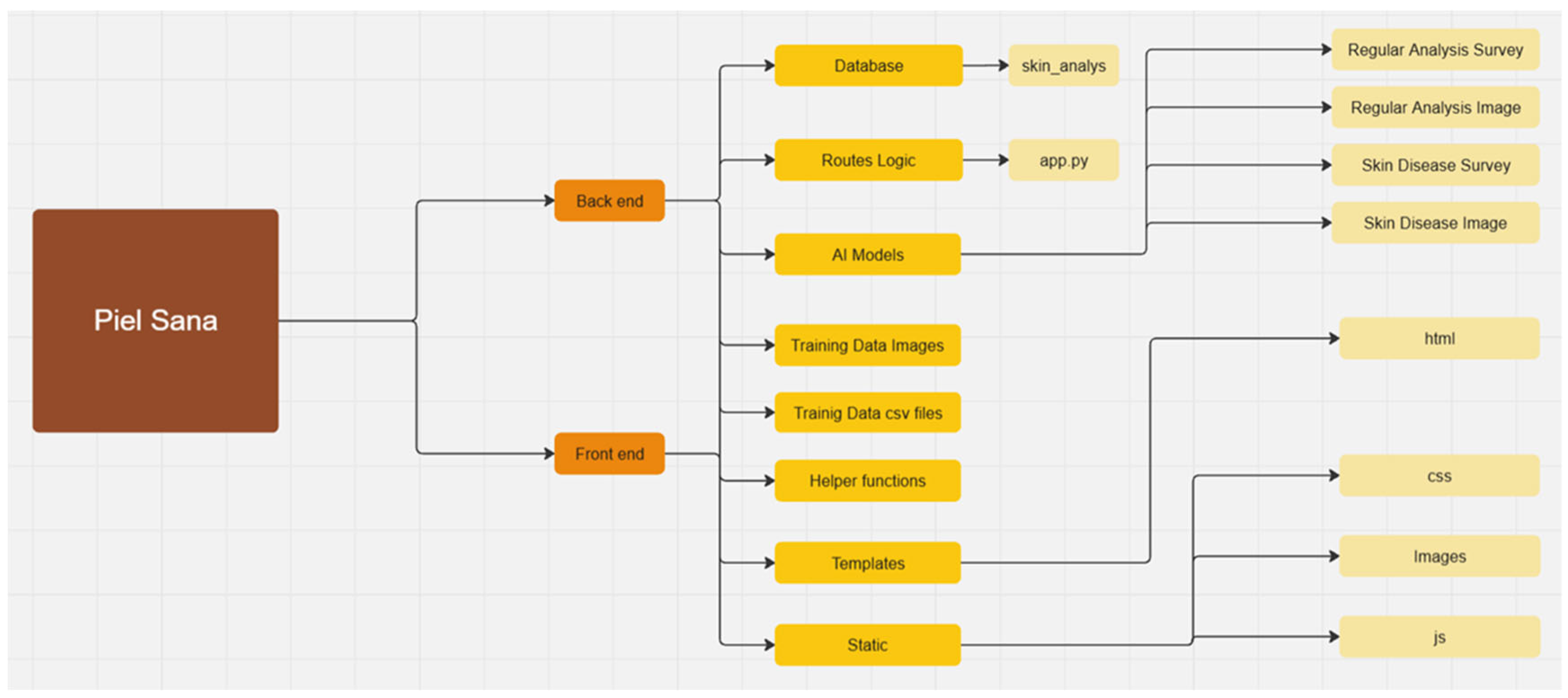
The diagram presents the modular structure of the “Piel Sana” platform (Figure 3), which consists of two main parts: backend (server part) and frontend user interfaces. This architecture provides a clear separation of functionalities, scalability and facilitates software maintenance and development.
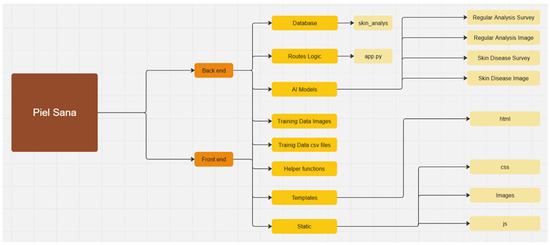
Figure 3.
Module diagram.
The backend is responsible for data processing, business logic, database interaction, and calculations related to AI models.
The frontend is responsible for visualization and user interaction.
4.2. Image Recognition Model (CNN)
The medical image analysis system is based on a convolutional neural network (CNN) designed to extract and classify visual features from images of skin diseases. The architecture begins with an input layer that accepts images of size 224 × 224 pixels with three color channels. It is followed by several sequential blocks of convolutional layers, each using 3 × 3 kernels and an increasing number of filters (64, 128, 256). After each convolutional block, a MaxPooling layer is applied to reduce the dimensionality of the feature maps, which helps decrease computational complexity and mitigate overfitting. A Flatten layer follows these blocks to transform the multidimensional outputs into a one-dimensional vector, which is then processed by one or more fully connected (Dense) layers. Dropout is applied to these layers to further reduce overfitting. The final layer uses softmax activation to generate a probability distribution over the classes corresponding to different skin conditions. This architecture is inspired by proven models in medical diagnostics (Figure 4) and allows for effective recognition of various inflammatory skin diseases.
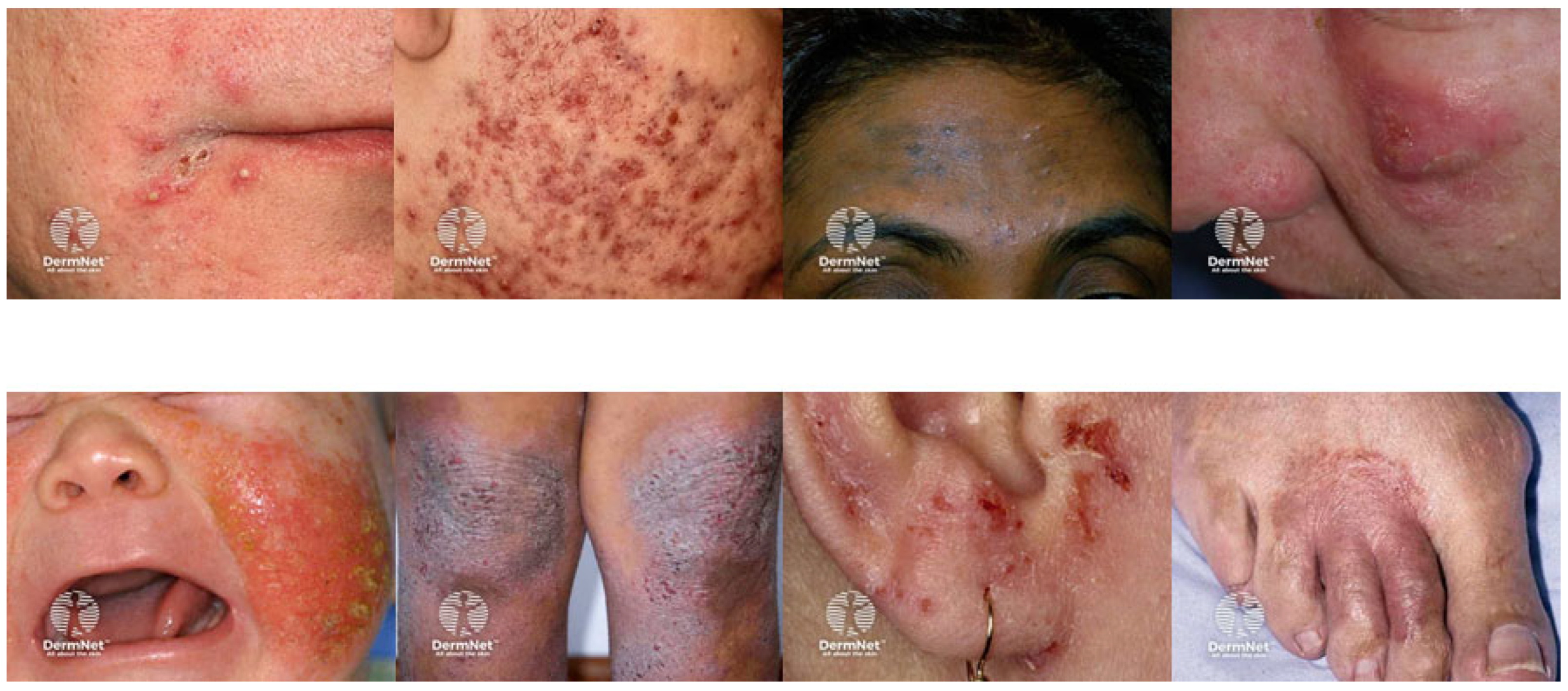
Figure 4.
Illustrative images of skin diseases [16,17,18,19,20,21].
4.3. Survey Development for Data Collection
To complement the objective analysis of images, the system incorporates a structured survey aimed at collecting subjective data from patients. The survey includes questions related to symptom duration, severity, medical history, and responses to specific stimuli (such as reactions to certain foods or medications). Each categorical response is encoded using a predefined response map, which transforms textual answers into numerical values that are suitable for machine learning analysis. A Random Forest algorithm is employed to analyze this data, chosen for its ability to handle heterogeneous, high-dimensional data and its robustness against noise and overfitting. Hyperparameter optimization, using techniques like GridSearchCV, is applied to identify the optimal model configuration, thereby improving the accuracy and reliability of the patient condition predictions.
4.4. Combining Results to Generate the Final Diagnosis
The final diagnosis is determined through a multimodal approach that integrates both image analysis results and survey data. The CNN model outputs probabilistic estimates for each skin disease class, while the Random Forest model, trained on survey data, generates assessments based on clinical and subjective indicators. Ensemble learning techniques, such as weighted averaging or stacking, are used to combine these two sets of results, creating a synergistic effect that incorporates both objective visual signs and subjective patient information. This integrated approach enables a more accurate and personalized diagnosis, considering the unique characteristics of each patient and enhancing the overall efficiency of the diagnostic system.
5. Results and Analysis
This study proposes a multimodal diagnostic framework for inflammatory skin diseases, integrating convolutional neural networks (CNNs) for image-based analysis and a Random Forest classifier for subjective clinical data (Figure 5). The performance of each component was evaluated individually, followed by an assessment of the fused system, to determine overall diagnostic accuracy, robustness, and clinical utility.
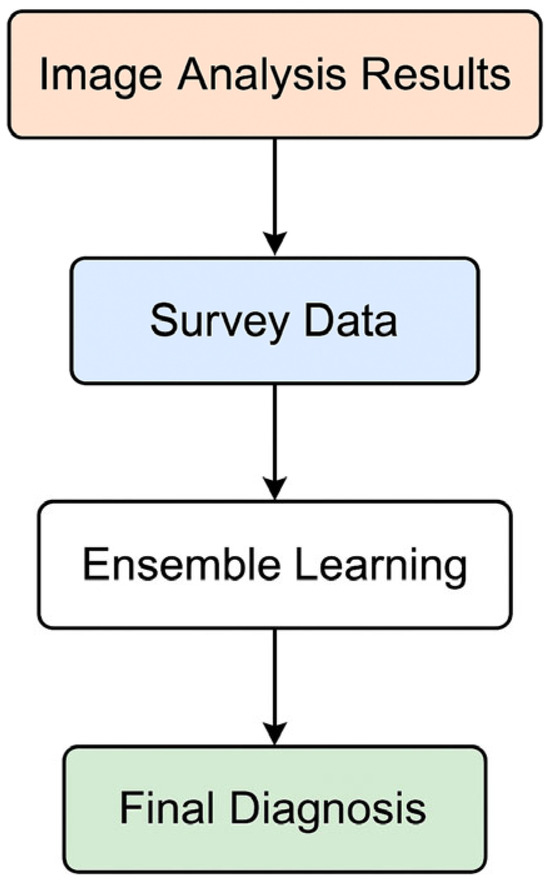
Figure 5.
AI classification workflow for disease diagnosis.
5.1. CNN-Based Dermatological Image Classification
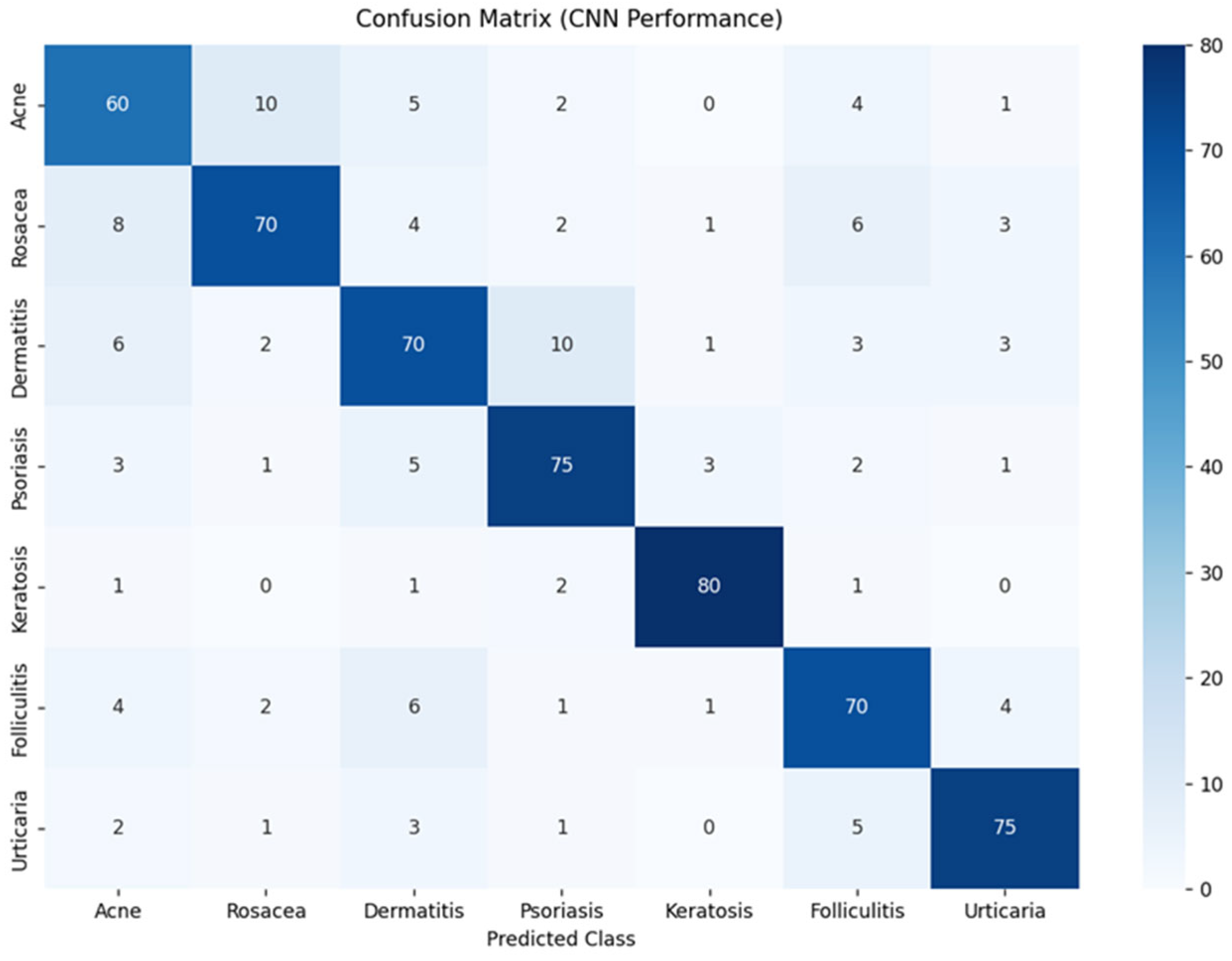
The CNN was developed to automatically classify dermatological conditions from clinical images. A robust preprocessing pipeline—including pixel normalization and data augmentation—was implemented to enhance the model’s generalizability across diverse imaging conditions, such as variations in illumination, scale, and orientation (Figure 6). The model achieved high classification accuracy, with overall performance ranging between 75% and 80% across diagnostic categories. Confusion matrix analysis revealed strong per-class accuracy for keratosis (80%) and psoriasis (75%), while higher rates of misclassification were observed among acne, rosacea, and dermatitis, likely due to overlapping visual characteristics among these conditions.
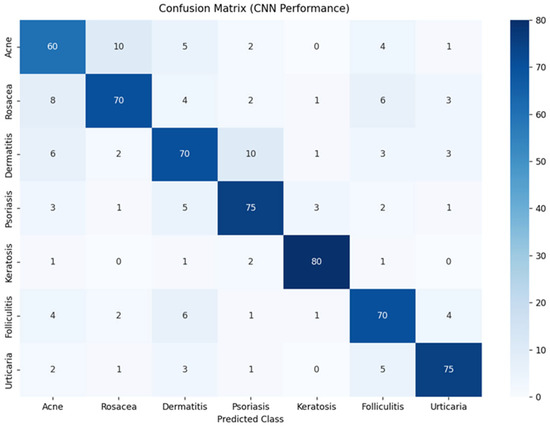
Figure 6.
Confusion matrix of neural network classification results.
Precision and recall scores ranged from 70% to 75%, indicating consistent ability to both detect true positives and minimize false positives. Despite some inter-class ambiguity, the CNN was able to distinguish clinically relevant features across most conditions.
F1-scores—balancing precision and recall—fell within the 0.72 to 0.75 range, reflecting the model’s overall balanced and reliable performance. The architecture, which includes convolutional layers, MaxPooling, and Dropout, in conjunction with data augmentation, contributed to the model’s ability to generalize effectively to unseen images while minimizing overfitting.
5.2. Subjective Data Analysis Using Random Forest
To complement the CNN-based image analysis, a structured patient questionnaire was employed to capture subjective information, including symptom severity, reactions to environmental triggers, and relevant medical history. The encoded survey responses were analyzed using a Random Forest classifier, which demonstrated near-perfect performance for common conditions such as psoriasis, dermatitis, and rosacea, achieving precision, recall, F1-scores, and overall accuracy exceeding 90% (Table 2). Notably, the classifier achieved perfect scores (1.0) across all metrics for rosacea, urticaria, and folliculitis, indicating clear separability of these conditions based on patient-reported data. Psoriasis, eczema, and dermatitis also yielded high performance, with F1-scores above 0.90, suggesting that the survey features successfully captured discriminative patterns relevant for classification.

Table 2.
Classification report for skin disease detection using random forest.
However, the performance for acne was notably lower in terms of precision (0.75), despite a high recall (0.92), resulting in an F1-score of 0.83. This indicates that while the model successfully identified most acne cases, it also generated more false positives, possibly due to overlapping features with other conditions such as folliculitis or eczema. This discrepancy highlights the challenge of distinguishing between visually and symptomatically similar diseases based solely on subjective input.
Beyond classification accuracy, the Random Forest model offered valuable interpretability, elucidating relationships between specific symptom patterns and diagnostic categories. Nevertheless, this advantage was tempered by the model’s sensitivity to data distribution. Reduced performance in underrepresented classes reinforces the importance of class balance and representative sampling in training datasets, especially when deploying such systems in real-world clinical or telemedicine contexts.
5.3. Integrated Diagnostic Framework via Data Fusion
The final diagnostic outcome was derived by integrating the outputs of the CNN and Random Forest models using ensemble techniques such as weighted averaging and model stacking. The integrated model demonstrated superior performance compared to individual methods, with results falling between the performance of CNN and Random Forest. Specifically, it achieved slightly higher accuracy than Random Forest and marginally lower error rates than CNN, leading to improved overall diagnostic reliability. This multimodal approach effectively balanced the strengths of both methods—leveraging CNN’s robust image analysis with Random Forest’s capacity to handle complex, patient-reported data. As a result, the integrated model provided a more stable and personalized diagnostic output, with reduced variance across different conditions.
5.4. Comparative Evaluation Against Traditional Methods
The CNN model demonstrated performance comparable to experienced dermatologists, particularly in terms of speed and consistency in identifying common conditions.
While the Random Forest algorithm effectively processed subjective data for common cases, it faced limitations with rare conditions, emphasizing the continued importance of clinician-led interpretation.
The fusion of CNN and Random Forest models provided a significant performance boost, offering a reliable, scalable diagnostic tool suitable for teledermatology and resource-limited settings.
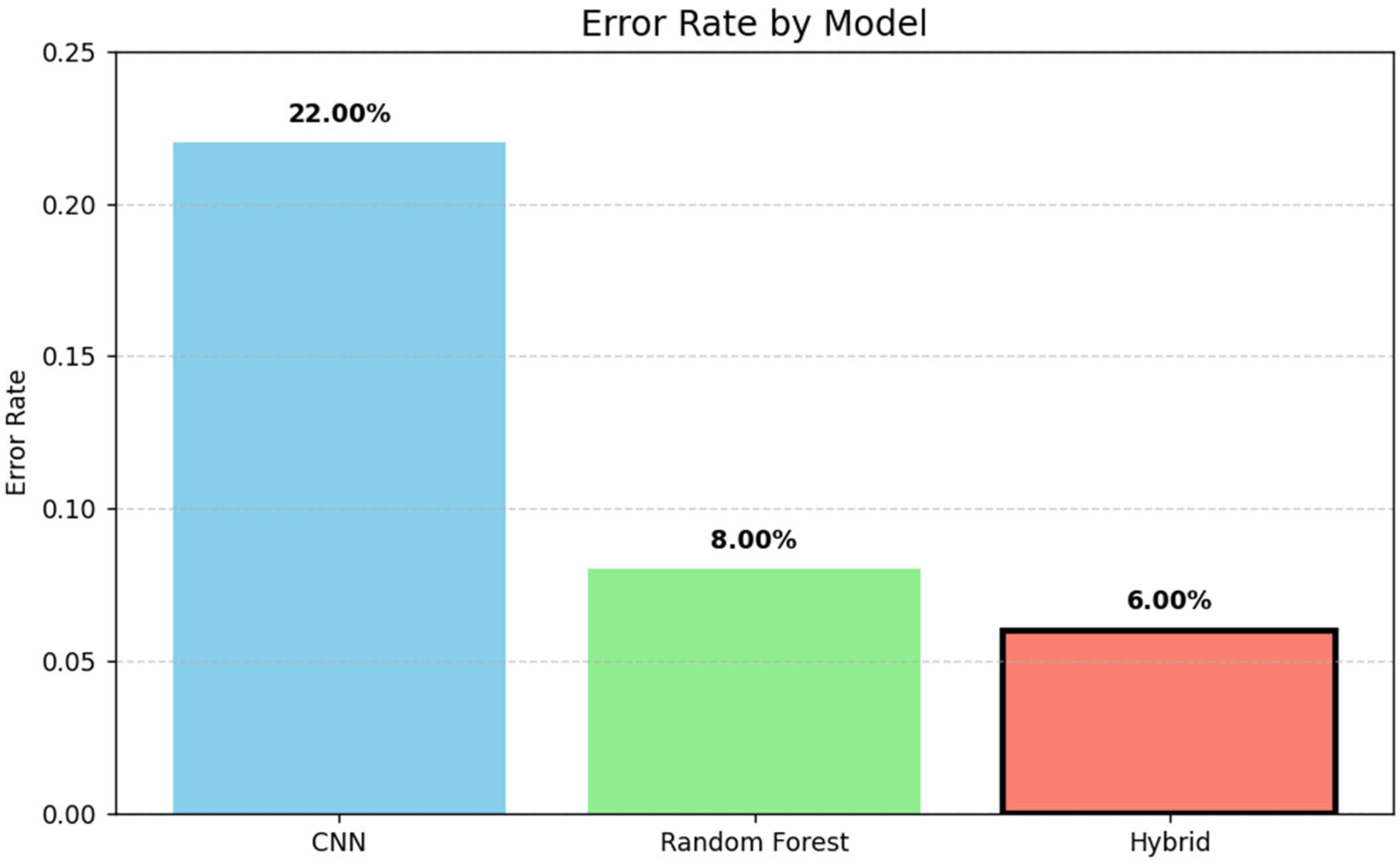
The bar chart in Figure 7 illustrates the error rates of three models: CNN, Random Forest, and a hybrid approach. The hybrid model achieved the lowest error rate (6%), followed by Random Forest (8%) and CNN (22%), demonstrating the effectiveness of combining techniques for improved performance.
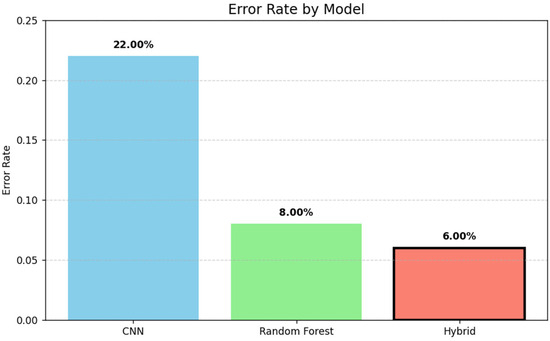
Figure 7.
Forest comparison of error rates across models.
6. Applications and Future Research
The proposed system has a wide range of potential applications in clinical practice. It can be integrated into existing medical platforms to assist dermatologists in the rapid and accurate diagnosis of inflammatory skin diseases. By combining automated medical image analysis using CNNs with clinical data collected through surveys, a more personalized assessment of a patient’s condition can be achieved. This approach can improve the diagnostic process and optimize therapeutic strategies, particularly in cases where traditional methods prove insufficient.
Future developments may explore expanding the system to diagnose additional diseases and incorporating supplementary patient data, such as biomarkers, genetic information, or wearable device data. This would enhance the scope and accuracy of diagnostic results. Furthermore, continuous model training and adaptation to new data will allow the system to update in real time, keeping pace with emerging clinical discoveries and changes in epidemiological patterns.
Additional research could focus on optimizing data fusion algorithms by employing ensemble learning techniques and hybrid models that integrate results from multiple sources. This multimodal approach not only improves diagnostic accuracy but also reduces the risk of errors associated with the limitations of individual diagnostic methods. Developing prototypes and conducting clinical pilot studies will allow for the evaluation of the system’s practical applicability and effectiveness in real-world medical settings.
By integrating this system into medical information platforms, a significant improvement in diagnosis and patient monitoring is expected, reducing diagnosis time and increasing access to high-quality medical services. This represents an important step toward more personalized and efficient healthcare, addressing the growing demand for rapid and accurate diagnostic solutions.
7. Conclusions
The proposed diagnostic system for inflammatory skin diseases demonstrates significant potential for improving the diagnostic process by integrating machine learning with patient data. The combination of convolutional neural networks (CNNs) for medical image analysis with algorithms such as Random Forest for processing survey data enables more accurate and personalized diagnostics. This approach not only enhances diagnostic precision but also provides a more comprehensive view of the patient’s condition by integrating both objective and subjective data.
The integration of machine learning with patient information in medical diagnostics represents a major advancement in modernizing healthcare. This approach enables the processing of large datasets and the extraction of meaningful patterns that can support clinicians in decision-making. By leveraging such systems, subjectivity in the diagnostic process is reduced, and the reliability of results is increased. In the future, as more data is accumulated and algorithms are refined, these systems are expected to become an integral part of clinical practice, leading to improved health outcomes and greater patient satisfaction.
Author Contributions
Conceptualization, A.S., M.K. and G.S.; methodology, A.S.; software, A.S.; validation, A.S., G.S. and M.K.; formal analysis, A.S.; investigation, G.S.; resources, M.K., G.S. and A.S.; data curation, M.K.; writing—original draft preparation, A.S.; writing—review and editing, G.S. and M.K.; visualization, A.S.; supervision, M.K.; project administration, M.K.; funding acquisition, A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study did not require ethical approval.
Informed Consent Statement
Informed oral consent was obtained from all subjects involved in the study during study research.
Data Availability Statement
Data sharing is not applicable due to privacy.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Smith, K.E.; Ahmed, A. Quality of Life. In Psychodermatology in Clinical Practice; Bewley, A., Lepping, P., Taylor, R., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Kim, M.; Lim, W.; Park, G.; Park, I.; Chang, S. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human–computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Dalgard, F.; Gieler, U.; Holm, J.Ø.; Bjertness, E.; Hauser, S. Self-esteem and body satisfaction among late adolescents with acne: Results from a population survey. J. Am. Acad. Dermatol. 2008, 59, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Anand, V.; Gupta, S.; Koundal, D.; Mahajan, S.; Pandit, A.; Zaguia, A. Deep Learning Based Automated Diagnosis of Skin Diseases Using Dermoscopy. Comput. Mater. Contin. 2021, 71, 3145–3160. [Google Scholar] [CrossRef]
- Choudhury, A.; Shahsavar, Y.; Shamszare, H. User Intent to Use DeepSeek for Health Care Purposes and Their Trust in the Large Language Model: Multinational Survey Study. JMIR Hum. Factors 2025, 12, e72867. [Google Scholar] [CrossRef] [PubMed]
- Escalé-Besa, A.; Vidal-Alaball, J.; Miró Catalina, Q.; Gracia, V.H.G.; Marin-Gomez, F.X.; Fuster-Casanovas, A. The Use of Artificial Intelligence for Skin Disease Diagnosis in Primary Care Settings: A Systematic Review. Healthcare 2024, 12, 1192. [Google Scholar] [CrossRef] [PubMed]
- Seth, D.; Cheldize, K.; Brown, D.; Freeman, E.F. Global Burden of Skin Disease: Inequities and Innovations. Curr. Dermatol. Rep. 2017, 6, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Asan, O. Role of Artificial Intelligence in Patient Safety Outcomes: Systematic Literature Review. JMIR Med. Inform. 2020, 8, e18599. [Google Scholar] [CrossRef] [PubMed]
- Pearce, F.J.; Rivera, S.C.; Liu, X.; Manna, E.; Denniston, A.K.; Calvert, M.J. The role of patient-reported outcome measures in trials of artificial intelligence health technologies: A systematic evaluation of ClinicalTrials.gov records (1997–2022). Lancet Digit. Health 2023, 5, e160–e167. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Tommasino, N.; Megna, M.; Cacciapuoti, S.; Villani, A.; Martora, F.; Ruggiero, A.; Genco, L.; Potestio, L. The Past, the Present and the Future of Teledermatology: A Narrative Review. Clin. Cosmet. Investig. Dermatol. 2024, 17, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, Z.; Dimitrova, V.; Warrington, L.; Velikova, G.; Absolom, K. Using artificial intelligence to predict patient outcomes from patient-reported outcome measures: A scoping review. Health Qual. Life Outcomes 2025, 23, 37. [Google Scholar] [CrossRef] [PubMed]
- Folliculitis. Available online: https://dermnetnz.org/imagedetail/11474-folliculitis (accessed on 25 August 2025).
- acne-face_79. Available online: https://dermnetnz.org/imagedetail/14025-acne-face-79 (accessed on 25 August 2025).
- acne-face_1_42. Available online: https://dermnetnz.org/imagedetail/11240-acne-face-1-42 (accessed on 25 August 2025).
- Atopic Dermatitis. Available online: https://dermnetnz.org/imagedetail/3231-atopic-dermatitis (accessed on 25 August 2025).
- Atopic Dermatitis, Ear. Available online: https://dermnetnz.org/imagedetail/13250-atopic-dermatitis (accessed on 25 August 2025).
- Atopic Eczema on the Dorsal Foot. Available online: https://dermnetnz.org/imagedetail/3237-atopic-dermatitis (accessed on 25 August 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).