Resilience of UNet-Based Models Under Adversarial Conditions in Medical Image Segmentation †
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Zhang, L.; Yang, J.; Teng, F. Role of artificial intelligence in medical image analysis: A review of current trends and future directions. J. Med. Biol. Eng. 2024, 44, 231–243. [Google Scholar] [CrossRef]
- Xu, Y.; Quan, R.; Xu, W.; Huang, Y.; Chen, X.; Liu, F. Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering 2024, 11, 1034. [Google Scholar] [CrossRef]
- Archana, R.; Jeevaraj, P.E. Deep learning models for digital image processing: A review. Artif. Intell. Rev. 2024, 57, 11. [Google Scholar] [CrossRef]
- Gabdullin, M.T.; Mukasheva, A.; Koishiyeva, D.; Umarov, T.; Bissembayev, A.; Kim, K.S.; Kang, J.W. Automatic cancer nuclei segmentation on histological images: Comparison study of deep learning methods. Biotechnol. Bioprocess Eng. 2024, 29, 1034–1047. [Google Scholar] [CrossRef]
- Jiang, H.; Diao, Z.; Shi, T.; Zhou, Y.; Wang, F.; Hu, W.; Yao, Y.D. A review of deep learning-based multiple-lesion recognition from medical images: Classification, detection and segmentation. Comput. Biol. Med. 2023, 157, 106726. [Google Scholar] [CrossRef]
- Trigka, M.; Dritsas, E. A Comprehensive Survey of Deep Learning Approaches in Image Processing. Sensors 2025, 25, 531. [Google Scholar] [CrossRef] [PubMed]
- Kshatri, S.S.; Singh, D. Convolutional neural network in medical image analysis: A review. Arch. Comput. Methods Eng. 2023, 30, 2793–2810. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Merhof, D. Medical image segmentation review: The success of u-net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef] [PubMed]
- Mukasheva, A.; Koishiyeva, D.; Sergazin, G.; Sydybayeva, M.; Mukhammejanova, D.; Seidazimov, S. Modification of U-Net with Pre-Trained ResNet-50 and Atrous Block for Polyp Segmentation: Model TASPP-UNet. Eng. Proc. 2024, 70, 16. [Google Scholar] [CrossRef]
- Zoetmulder, R.; Gavves, E.; Caan, M.; Marquering, H. Domain-and task-specific transfer learning for medical segmentation tasks. Comput. Methods Programs Biomed. 2022, 214, 106539. [Google Scholar] [CrossRef]
- Ma, X.; Niu, Y.; Gu, L.; Wang, Y.; Zhao, Y.; Bailey, J.; Lu, F. Understanding adversarial attacks on deep learning based medical image analysis systems. Pattern Recognit. 2021, 110, 107332. [Google Scholar] [CrossRef]
- Lee, W.; Ju, M.; Sim, Y.; Jung, Y.K.; Kim, T.H.; Kim, Y. Adversarial Attacks on Medical Segmentation Model via Transformation of Feature Statistics. Appl. Sci. 2024, 14, 2576. [Google Scholar] [CrossRef]
- Maliamanis, T.V.; Apostolidis, K.D.; Papakostas, G.A. How resilient are deep learning models in medical image analysis? The case of the moment-based adversarial attack (Mb-AdA). Biomedicines 2022, 10, 2545. [Google Scholar] [CrossRef]
- Pal, S.; Rahman, S.; Beheshti, M.; Habib, A.; Jadidi, Z.; Karmakar, C. The Impact of Simultaneous Adversarial Attacks on Robustness of Medical Image Analysis. IEEE Access 2024, 12, 66478–66494. [Google Scholar] [CrossRef]
- Mahbod, A.; Polak, C.; Feldmann, K.; Khan, R.; Gelles, K.; Dorffner, G.; Ellinger, I. Nuinsseg: A fully annotated dataset for nuclei instance segmentation in H&E-stained histological images. Sci. Data 2024, 11, 295. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 9351. [Google Scholar]
- Li, Z.; Zhang, H.; Li, Z.; Ren, Z. Residual-Attention UNet++: A Nested Residual-Attention U-Net for Medical Image Segmentation. Appl. Sci. 2022, 12, 7149. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Rueckert, D. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing properties of neural networks. arXiv 2013, arXiv:1312.6199. [Google Scholar] [CrossRef]
- Kurakin, A.; Goodfellow, I.; Bengio, S. Adversarial machine learning at scale. arXiv 2016, arXiv:1611.01236. [Google Scholar] [CrossRef]
- Madry, A.; Makelov, A.; Schmidt, L.; Tsipras, D.; Vladu, A. Towards deep learning models resistant to adversarial attacks. arXiv 2017, arXiv:1706.06083. [Google Scholar] [CrossRef]
- Dong, Y.; Liao, F.; Pang, T.; Su, H.; Zhu, J.; Hu, X.; Li, J. Boosting adversarial attacks with momentum. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 9185–9193. [Google Scholar] [CrossRef]
- Xie, C.; Zhang, Z.; Zhou, Y.; Bai, S.; Wang, J.; Ren, Z.; Yuille, A.L. Improving transferability of adversarial examples with input diversity. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 2725–2734. [Google Scholar] [CrossRef]
- Lin, Z.; Peng, A.; Wei, R.; Yu, W.; Zeng, H. An enhanced transferable adversarial attack of scale-invariant methods. In Proceedings of the 2022 IEEE International Conference on Image Processing (ICIP), Bordeaux, France, 16–19 October 2022; pp. 3788–3792. [Google Scholar] [CrossRef]
- Lian, Y.; Tang, Y.; Zhou, S. Research on three-step accelerated gradient algorithm in deep learning. Stat. Theory Relat. Fields 2022, 6, 40–57. [Google Scholar] [CrossRef]
- Koishiyeva, D.; Bissembayev, A.; Iliev, T.; Kang, J.W.; Mukasheva, A. Classification of Skin Lesions using PyQt5 and Deep Learning Methods. In Proceedings of the 2024 5th International Conference on Communications, Information, Electronic and Energy Systems (CIEES), Veliko Tarnovo, Bulgaria, 20–22 November 2024; pp. 1–7. [Google Scholar] [CrossRef]
- Ming, Q.; Xiao, X. Towards accurate medical image segmentation with gradient-optimized dice loss. IEEE Signal Process. Lett. 2023, 31, 191–195. [Google Scholar] [CrossRef]
- Dietrich, N.; Gong, B.; Patlas, M.N. Adversarial artificial intelligence in radiology: Attacks, defenses, and future considerations. Diagn. Interv. Imaging, 2025; in press. [Google Scholar] [CrossRef]
- Tolkynbekova, A.; Koishiyeva, D.; Bissembayev, A.; Mukhammejanova, D.; Mukasheva, A.; Kang, J.W. Comparative Analysis of the Predictive Risk Assessment Modeling Technique Using Artificial Intelligence. J. Electr. Eng. Technol. 2025; in press. [Google Scholar] [CrossRef]
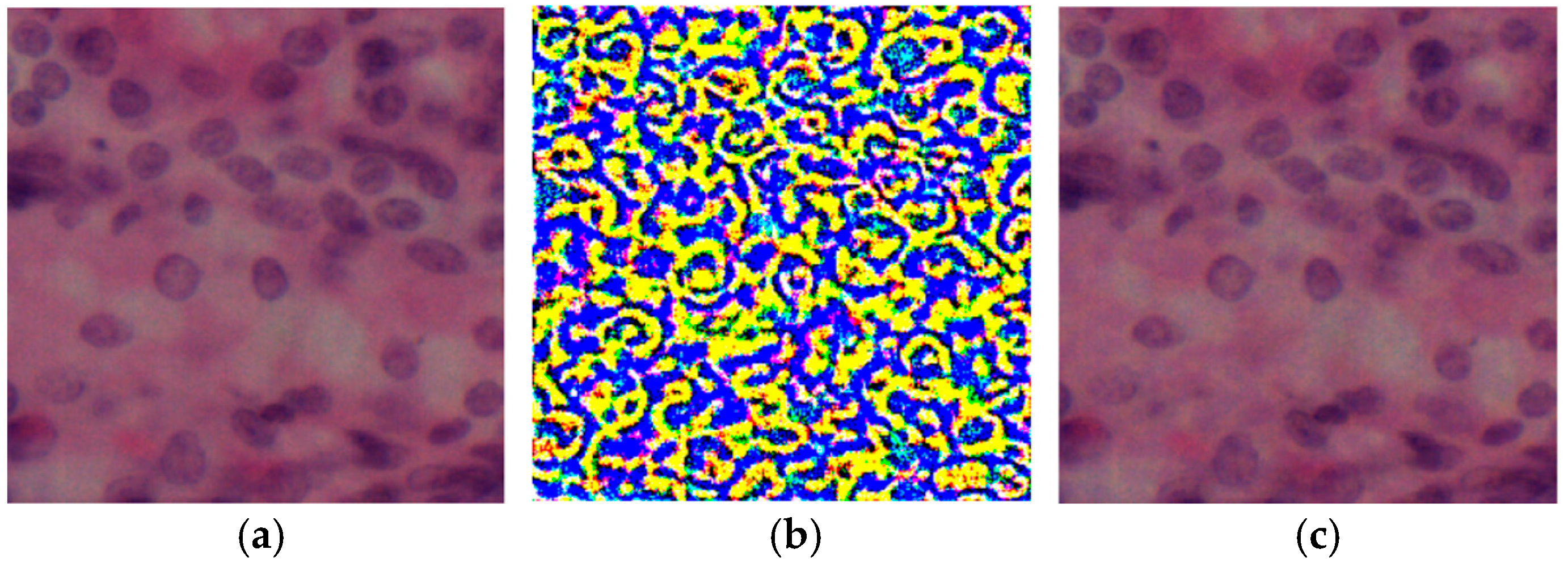



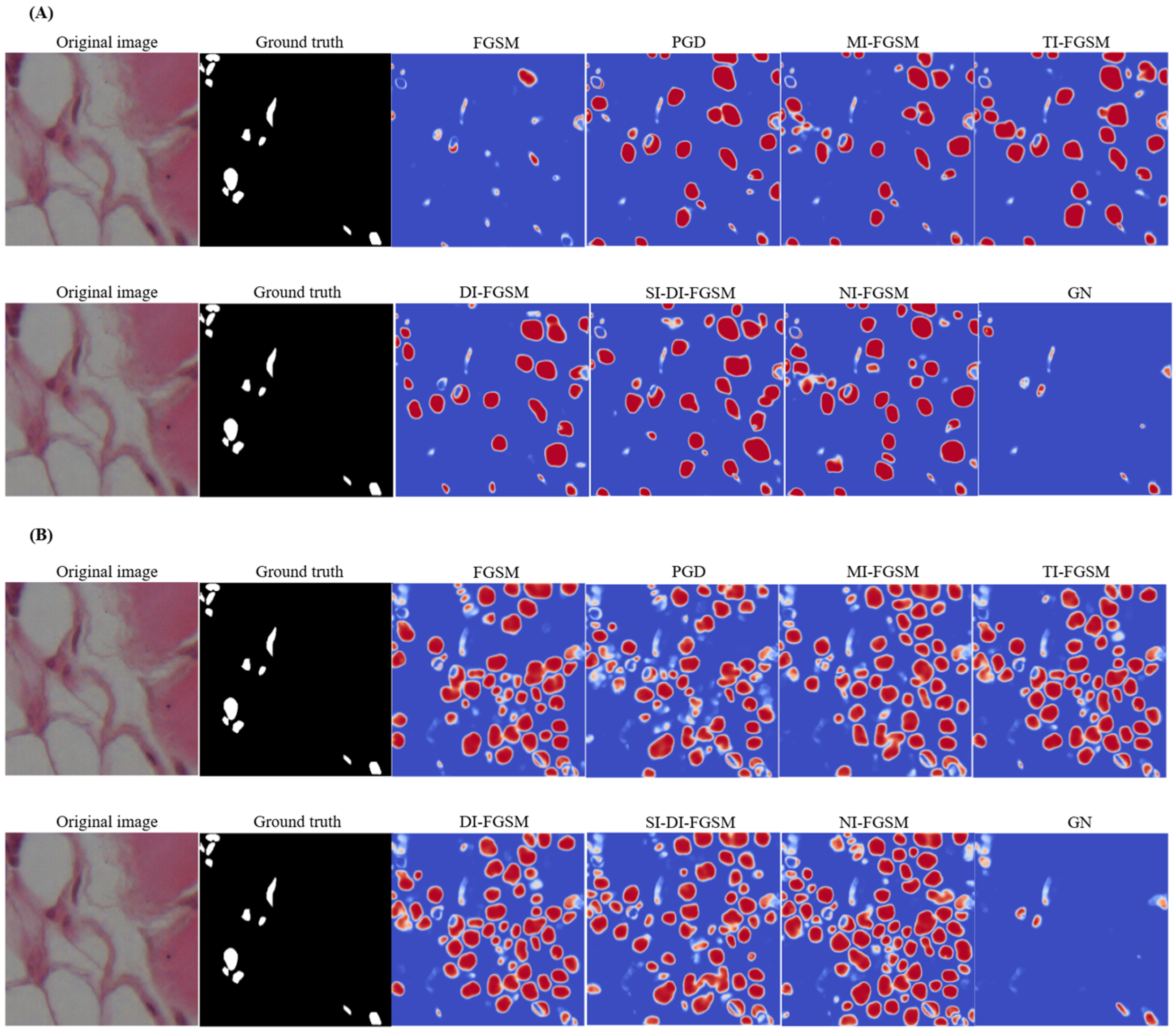
| Metrics | UNet | Att-Unet++ |
|---|---|---|
| Dice | 0.6424 | 0.7160 |
| Mean IOU | 0.4732 | 0.6190 |
| Accuracy | 0.9009 | 0.9292 |
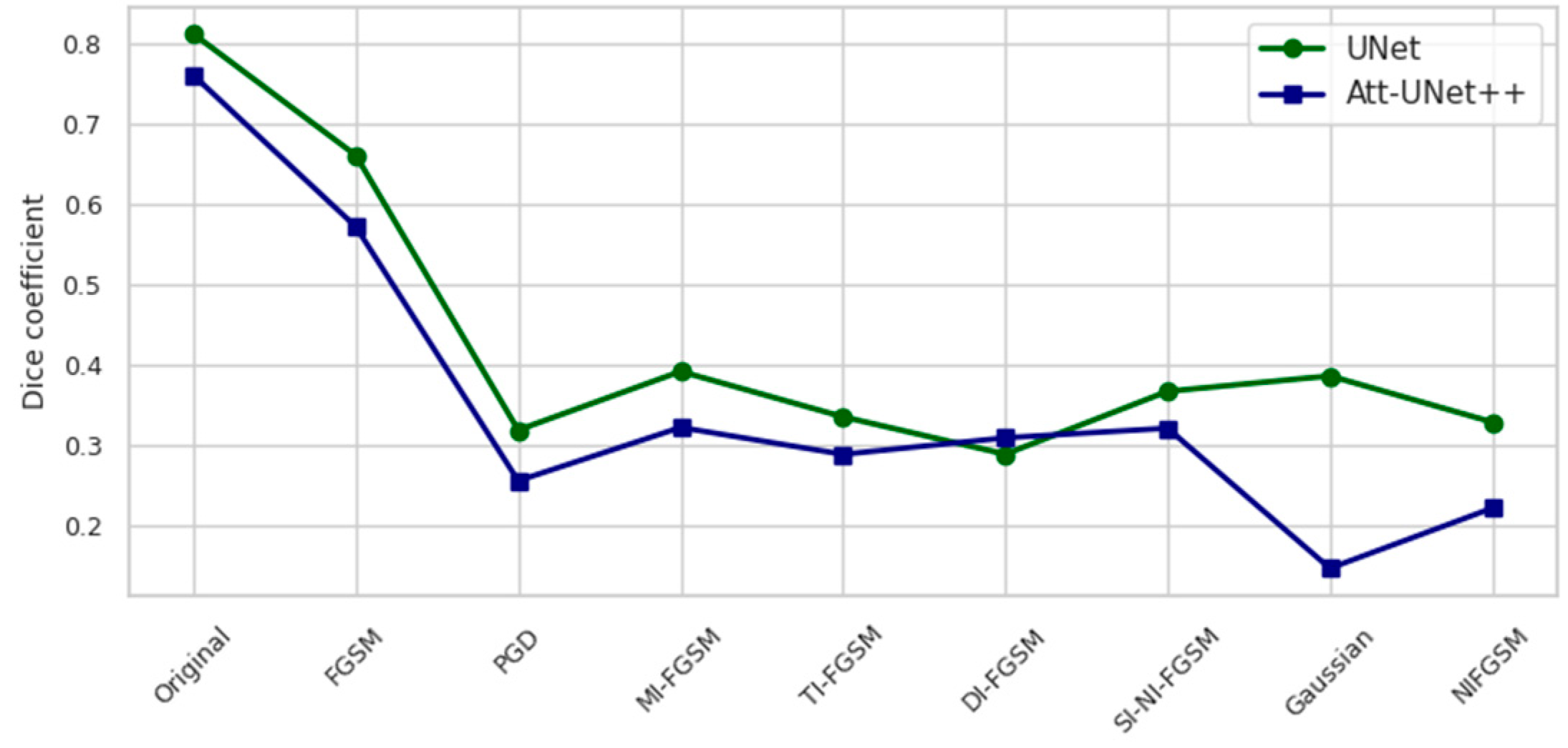
| Attack | UNet | Att-Unet++ |
|---|---|---|
| Original image | 0.8118 | 0.7603 |
| FGSM | 0.6607 | 0.5717 |
| PGD | 0.3185 | 0.2553 |
| MI-FGSM | 0.3185 | 0.3217 |
| TI-FGSM | 0.3350 | 0.2882 |
| DI-FGSM | 0.2884 | 0.3088 |
| SI-DI-FGSM | 0.3667 | 0.3208 |
| NI-FGSM | 0.3859 | 0.2212 |
| GN | 0.3284 | 0.1463 |
| Attack | UNet | Att-Unet++ |
|---|---|---|
| Original | 0.6845 | 0.6550 |
| FGSM | 0.4899 | 0.4153 |
| PGD | 0.1893 | 0.1428 |
| MI-FGSM | 0.2441 | 0.1896 |
| TI-FGSM | 0.2003 | 0.1690 |
| DI-FGSM | 0.1674 | 0.1788 |
| SI-DI-FGSM | 0.2246 | 0.1899 |
| NI-FGSM | 0.1968 | 0.1215 |
| GN | 0.2329 | 0.0658 |
| Attack | UNet | Att-Unet++ |
|---|---|---|
| Original | 0.3408 | 0.2127 |
| FGSM | 1.0201 | 0.6375 |
| PGD | 3.7275 | 2.3940 |
| MI-FGSM | 2.5136 | 1.4659 |
| TI-FGSM | 3.4177 | 2.0134 |
| DI-FGSM | 3.4365 | 1.9829 |
| SI-DI-FGSM | 3.1738 | 1.9152 |
| NI-FGSM | 4.0243 | 2.6552 |
| GN | 2.0101 | 1.5234 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koishiyeva, D.; Kang, J.W.; Iliev, T.; Bissembayev, A.; Mukasheva, A. Resilience of UNet-Based Models Under Adversarial Conditions in Medical Image Segmentation. Eng. Proc. 2025, 104, 3. https://doi.org/10.3390/engproc2025104003
Koishiyeva D, Kang JW, Iliev T, Bissembayev A, Mukasheva A. Resilience of UNet-Based Models Under Adversarial Conditions in Medical Image Segmentation. Engineering Proceedings. 2025; 104(1):3. https://doi.org/10.3390/engproc2025104003
Chicago/Turabian StyleKoishiyeva, Dina, Jeong Won Kang, Teodor Iliev, Alibek Bissembayev, and Assel Mukasheva. 2025. "Resilience of UNet-Based Models Under Adversarial Conditions in Medical Image Segmentation" Engineering Proceedings 104, no. 1: 3. https://doi.org/10.3390/engproc2025104003
APA StyleKoishiyeva, D., Kang, J. W., Iliev, T., Bissembayev, A., & Mukasheva, A. (2025). Resilience of UNet-Based Models Under Adversarial Conditions in Medical Image Segmentation. Engineering Proceedings, 104(1), 3. https://doi.org/10.3390/engproc2025104003







