Molecular Docking Study of Flavonoids to Block the Aryl Hydrocarbon Receptor †
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
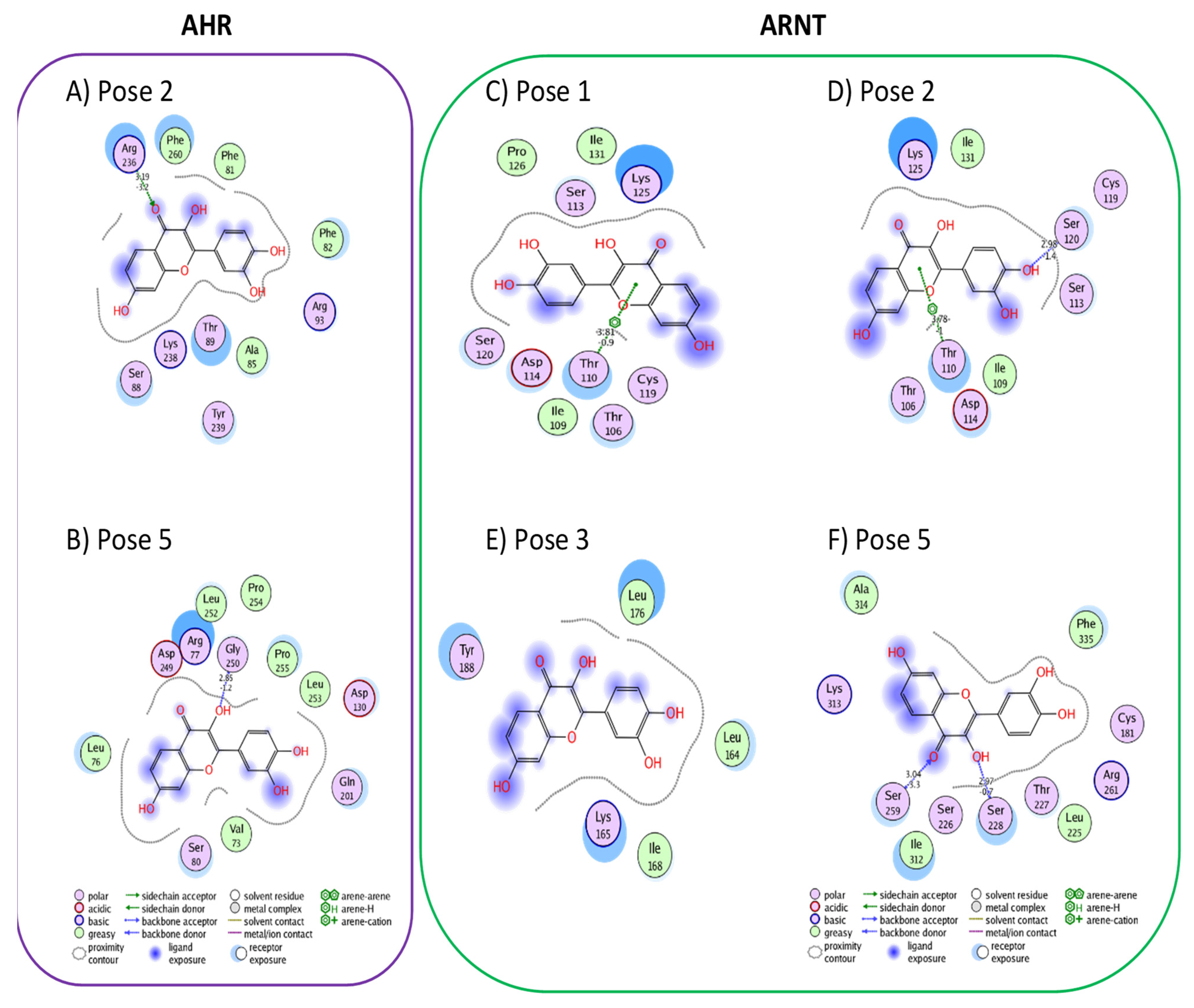
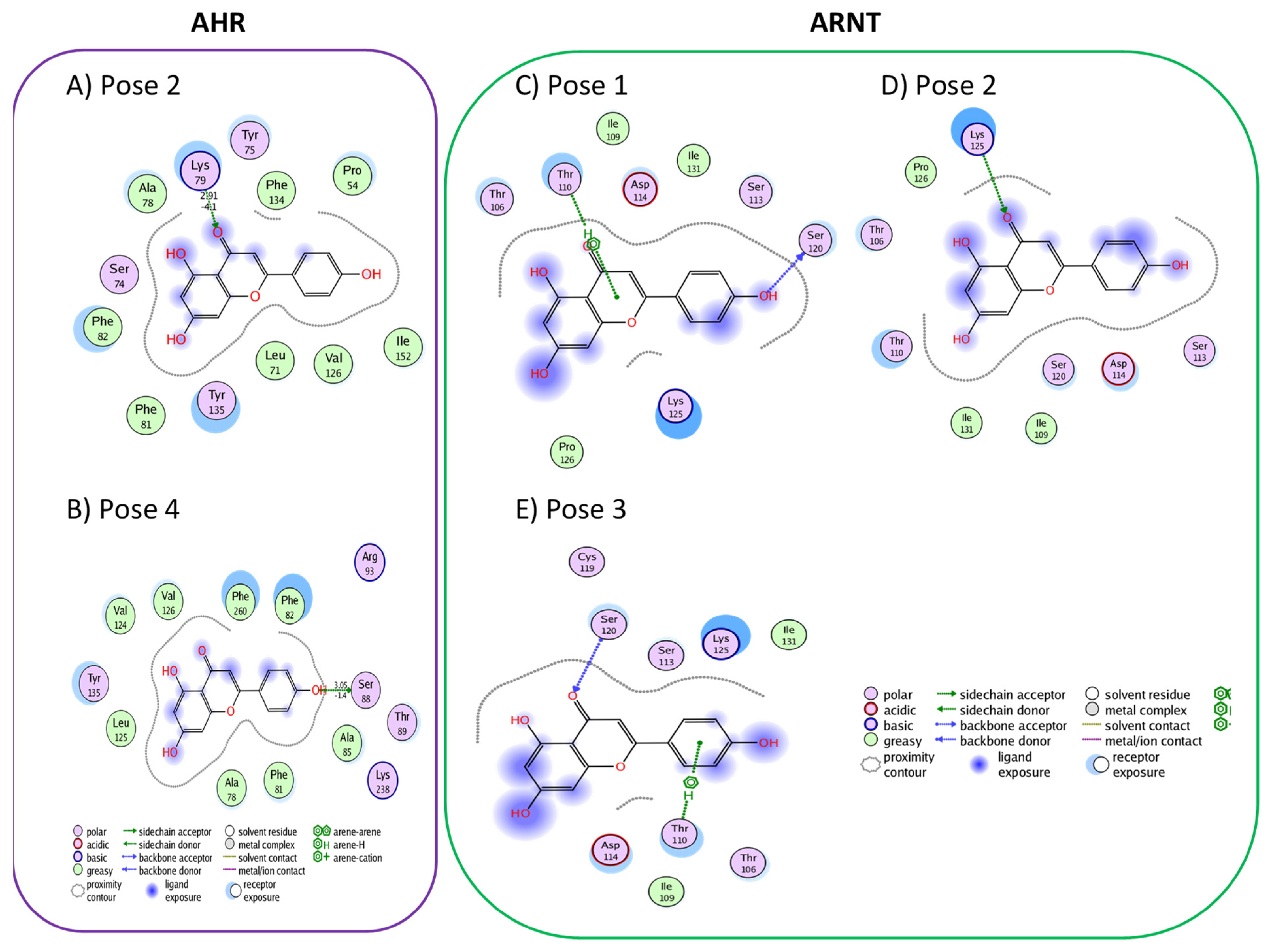
3.1. Active Flavonoids with a Probability of Blockage Higher Than 50%
3.2. Active Flavonones with a Probability of Blockage Higher Than 50%
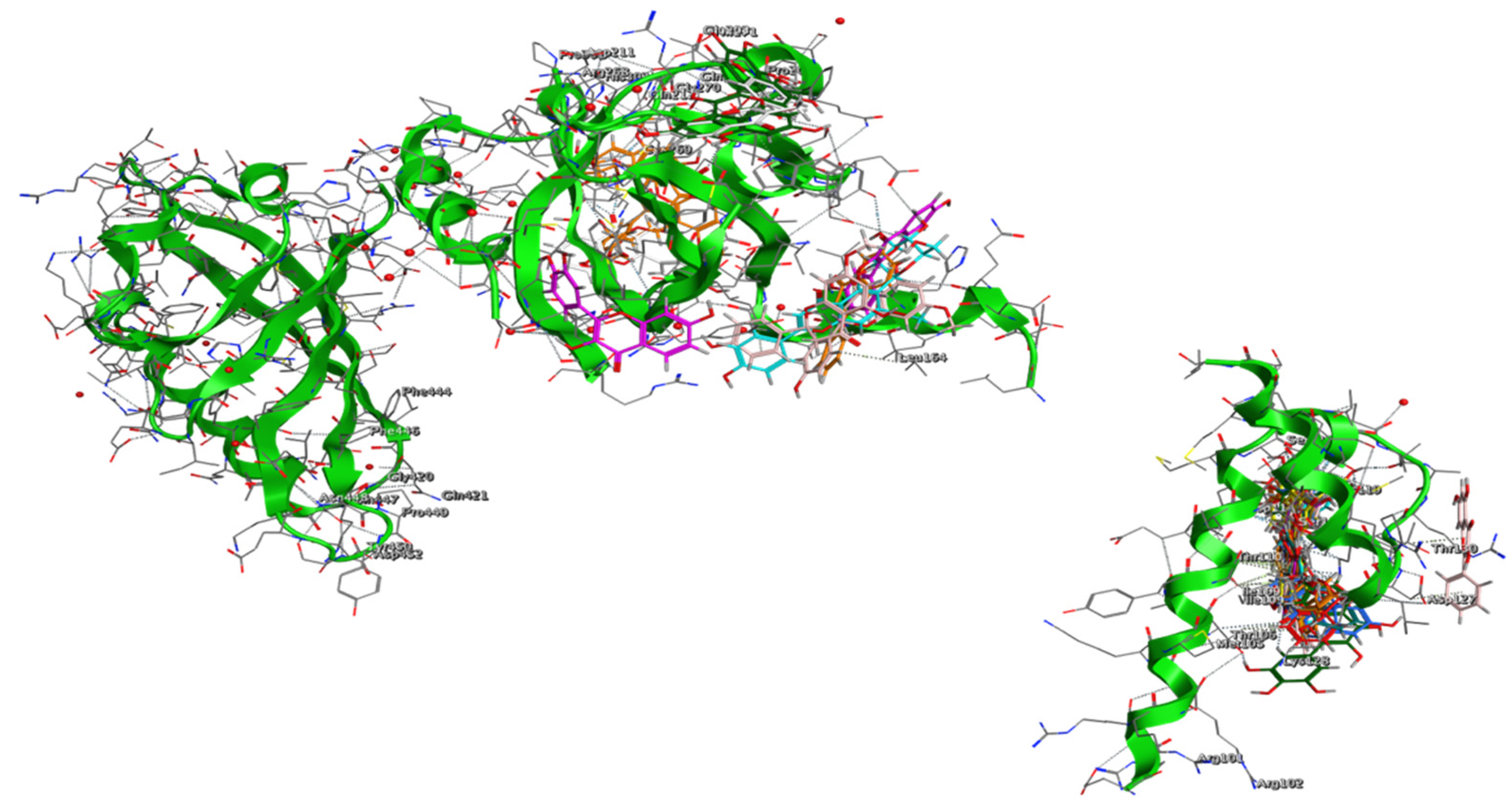
3.3. Proof of Concept
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Collado García, O.; Álvarez Gil, M.; Martínez Sáez, S. Aryl Hydrocarbon Receptor Agonists as Contaminants in the Feed for Production Animals. J. An. Prod. 2022, 34. Available online: https://revistas.reduc.edu.cu/index.php/rpa/article/view/e4044 (accessed on 14 October 2021).
- Salzano, M.; Marabotti, A.; Milanesi, L.; Facchiano, A. Human aryl-hydrocarbon receptor and its interaction with dioxin and physiological ligands investigated by molecular modelling and docking simulations. Biochem. Biophys. Res. Commun. 2011, 413, 176–181, Erratum in: Biochem. Biophys. Res. Commun. 2012, 418, 852. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.S.; Soshilov, A.A.; He, G.; DeGroot, D.E.; Zhao, B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011, 124, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Potluri, N.; Kim, Y.; Rastinejad, F. Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol. Cell. Biol. 2013, 33, 4346–4356. [Google Scholar] [CrossRef] [PubMed]
- Corrada, D.; Soshilov, A.A.; Denison, M.S.; Bonati, L. Deciphering Dimerization Modes of PAS Domains: Computational and Experimental Analyses of the AhR:ARNT Complex Reveal New Insights Into the Mechanisms of AhR Transformation. PLoS Comput. Biol. 2016, 12, e1004981. [Google Scholar] [CrossRef] [PubMed]
- Corrada, D.; Denison, M.S.; Bonati, L. Structural modeling of the AhR:ARNT complex in the bHLH-PASA-PASB region elucidates the key determinants of dimerization. Mol. Biosyst. 2017, 13, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Goya-Jorge, E.; Jorge Rodríguez, M.E.; Veitía, M.S.-I.; Giner, R.M. Plant Occurring Flavonoids as Modulators of the Aryl Hydrocarbon Receptor. Molecules 2021, 26, 2315. [Google Scholar] [CrossRef]
- Ray, S.K.; Mukherjee, S. Evolving Interplay Between Dietary Polyphenols and Gut Microbiota-An Emerging Importance in Healthcare. Front. Nutr. 2021, 8, 634944. [Google Scholar] [CrossRef]
- Guardado, E.; Molina, E.; João, M.M.; Uriarte, E. Antioxidant and Pro-Oxidant Effects of Polyphenolic Compounds and Structure-Activity Relationship Evidence. Nutr. Well-Being Health 2012, 2, 23–48. [Google Scholar] [CrossRef]
- Llauradó Maury, G.; Méndez Rodríguez, D.; Hendrix, S.; Escalona Arranz, J.C.; Fung Boix, Y.; Pacheco, A.O.; García Díaz, J.; Morris-Quevedo, H.J.; Ferrer Dubois, A.; Aleman, E.I.; et al. Antioxidants in Plants: A Valorization Potential Emphasizing the Need for the Conservation of Plant Biodiversity in Cuba. Antioxidants 2020, 9, 1048. [Google Scholar] [CrossRef]
- Vangone, A.; Spinelli, R.; Scarano, V.; Cavallo, L.; Oliva, R. COCOMAPS: A web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 2011, 27, 2915–2916. [Google Scholar] [CrossRef] [PubMed]
- Kortemme, T.; Baker, D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. USA 2002, 99, 14116–14121. [Google Scholar] [CrossRef] [PubMed]
- Kortemme, T.; Kim, D.E.; Baker, D. Computational alanine scanning of protein-protein interfaces. Sci STKE 2004, 2004, 12. [Google Scholar] [CrossRef] [PubMed]
- Szöllősi, D.; Erdei, Á.; Gyimesi, G.; Magyar, C.; Hegedűs, T. Access path to the ligand binding pocket may play a role in xenobiotics selection by AhR. PLoS ONE 2016, 11, e0146066. [Google Scholar] [CrossRef] [PubMed]
- Schulte, K.W.; Green, E.; Wilz, A.; Platten, M.; Daumke, O. Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure 2017, 25, 1025–1033. [Google Scholar] [CrossRef]
- Tuomisto, J.; Airaksinen, R.; Pekkanen, J.; Tukiainen, E.; Kiviranta, H.; Tuomisto, J.T. Comparison of questionnaire data and analyzed dioxin concentrations as a measure of exposure in soft-tissue sarcoma studies. Toxicol. Lett. 2017, 270, 8–11. [Google Scholar] [CrossRef][Green Version]
- Hattori, Y.; Takeda, T.; Nakamura, A.; Nishida, K.; Shioji, Y.; Fukumitsu, H.; Yamada, H.; Ishii, Y. The aryl hydrocarbon receptor is indispensable for dioxin-induced defects in sexually-dimorphic behaviors due to the reduction in fetal steroidogenesis of the pituitary-gonadal axis in rats. Biochem. Pharmacol. 2018, 154, 213–221. [Google Scholar] [CrossRef]
- Danjou, A.M.; Coudon, T.; Praud, D.; Lévêque, É.; Faure, É.; Salizzoni, P.; Le Romancer, M.; Severi, G.; Mancini, F.R.; Leffondré, K.; et al. Long-term airborne dioxin exposure and breast cancer risk in a case-control study nested within the French E3N prospective cohort. Environ. Int. 2019, 124, 236–248. [Google Scholar] [CrossRef]
- Mengoni, M.; Braun, A.D.; Gaffal, E.; Tüting, T. The aryl hydrocarbon receptor promotes inflammation-induced dedifferentiation and systemic metastatic spread of melanoma cells. Int. J. Cancer 2020, 147, 2902–2913. [Google Scholar] [CrossRef]
- Sadik, A.; Patterson, L.F.; Öztürk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfänder, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 is a metabolic immune checkpoint that activates the AHR and promotes tumor progression. Cell 2020, 182, 1252–1270. [Google Scholar] [CrossRef]
- Haidar, R.; Henkler, F.; Kugler, J.; Rosin, A.; Genkinger, D.; Laux, P.; Luch, A. The role of DNA-binding and ARNT dimerization on the nucleo-cytoplasmic translocation of the aryl hydrocarbon receptor. Sci. Rep. 2021, 11, 18194. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Amadou, A.; Mercier, C.; Praud, D.; Faure, E.; Iwaz, J.; Severi, G.; Mancini, F.R.; Coudon, T.; Fervers, B.; et al. The impact of left truncation of exposure in environmental case–control studies: Evidence from breast cancer risk associated with airborne dioxin. Eur. J. Epidemiol. 2022, 37, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Panda, R.; Cleave, A.S.S.; Suresh, P.K. In silico predictive studies of mAHR congener binding using homology modelling and molecular docking. Toxicol. Ind. Health 2014, 30, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Collado García, O.; Rodríguez Torrens, H.; Barreto Argilagos, G. The Role of Aryl Hydrocarbon Receptor-Microbiota Interactions in the Litter and Pre-Fattening Pigs’ Health. J. Anim. Prod. 2022, 34. Available online: https://revistas.reduc.edu.cu/index.php/rpa/article/view/e4156 (accessed on 14 October 2021).
- Valkov, E.; Sharpe, T.; Marsh, M.; Greive, S.; Hyvönen, M. Targeting Protein–Protein Interactions and Fragment-Based Drug Discovery. Top. Curr. Chem. 2012, 317, 145–179. [Google Scholar] [CrossRef]
- Corbi-Verge, C.; Kim, P.M. Motif mediated protein-protein interactions as drug targets. Cell. Commun. Signal. 2016, 14, 8. [Google Scholar] [CrossRef]
- van Dun, S.; Ottmann, C.; Milroy, L.G.; Brunsveld, L. Supramolecular Chemistry Targeting Proteins. J. Am. Chem. Soc. 2017, 139, 13960–13968. [Google Scholar] [CrossRef]
- Ashida, H.; Fukuda, I.; Yamashita, T.; Kanazawa, K. Flavones and flavonols at dietary levels inhibit a transformation of aryl hydrocarbon receptor induced by dioxin. FEBS Lett. 2000, 476, 213–217. [Google Scholar] [CrossRef]
- Zhang, S.; Qin, C.; Safe, S.H. Flavonoids as aryl hydrocarbon receptor agonists/antagonists: Effects of structure and cell context. Environ. Health Perspect 2003, 111, 1877–1882. [Google Scholar] [CrossRef]
- Jin, U.H.; Park, H.; Li, X.; Davidson, L.A.; Allred, C.; Patil, B.; Jayaprakasha, G.; Orr, A.A.; Mao, L.; Chapkin, R.S.; et al. Structure-Dependent Modulation of Aryl Hydrocarbon Receptor-Mediated Activities by Flavonoids. Toxicol. Sci. 2018, 164, 205–217. [Google Scholar] [CrossRef]
- Gasaly, N.; Riveros, K.; Gotteland, M. Fitoquímicos: Una nueva clase de prebióticos. Rev. Chil. Nutr. 2020, 47, 317–327. [Google Scholar] [CrossRef]
- Han, H.; Safe, S.; Jayaraman, A.; Chapkin, R.S. Diet-Host-Microbiota Interactions Shape Aryl Hydrocarbon Receptor Ligand Production to Modulate Intestinal Homeostasis. Annu. Rev. Nutr. 2021, 41, 455–478. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Edgell, C.J.; McDonald, C.C.; Graham, J.B. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 1983, 80, 3734–3737. [Google Scholar] [CrossRef]
- Luo, C.; Zou, P.; Ji, G.; Gu, A.; Zhao, P.; Zhao, C. The aryl hydrocarbon receptor (AhR) 1661G>A polymorphism in human cancer: A meta-analysis. Gene 2013, 513, 225–230. [Google Scholar] [CrossRef]
- Kolluri, S.K.; Jin, U.H.; Safe, S. Role of the aryl hydrocarbon receptor in carcinogenesis and potential as an anti-cancer drug target. Arch. Toxicol. 2017, 91, 2497–2513. [Google Scholar] [CrossRef]
- Wang, Z.; Snyder, M.; Kenison, J.E.; Yang, K.; Lara, B.; Lydell, E.; Bennani, K.; Novikov, O.; Federico, A.; Monti, S.; et al. How the AHR Became Important in Cancer: The Role of Chronically Active AHR in Cancer Aggression. Int. J. Mol. Sci. 2020, 22, 387. [Google Scholar] [CrossRef]
- Matos, M.J. Cumarinas: Versatilidad estructural y aplicaciones en Química Farmacéutica. Ph.D. Thesis, Universidad de Santiago de Compostela, Santiago, Spain, 2013; p. 342. [Google Scholar]
- Park, H.; Jin, U.H.; Orr, A.A.; Echegaray, S.P.; Davidson, L.A.; Allred, C.D.; Chapkin, R.S.; Jayaraman, A.; Lee, K.; Tamamis, P.; et al. Isoflavones as Ah Receptor Agonists in Colon-Derived Cell Lines: Structure-Activity Relationships. Chem. Res. Toxicol. 2019, 32, 2353–2364. [Google Scholar] [CrossRef]
- Bonati, L.; Corrada, D.; Giani Tagliabue, S.; Motta, S. Molecular modeling of the AhR structure and interactions can shed light on ligand-dependent activation and transformation mechanisms. Curr. Opin. Toxicol. 2017, 2, 42–49. [Google Scholar] [CrossRef]











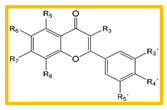 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Comp | R3 | R5 | R6 | R7 | R8 | R3’ | R4´ | R5´ | AHR:ARNT Prob (%) |
| 1 | H | H | H | OH | H | H | OH | H | 60 |
| 2 | OH | H | H | OH | H | OH | OH | H | 60 |
| 3 | CH3 | OH | H | OH | H | OH | OH | H | 60 |
| 4 | H | OH | H | OH | H | H | OH | H | 50 |
| 5 | H | H | H | H | H | H | H | H | 50 |
| 6 | OH | OH | H | OH | H | H | H | H | 50 |
| 7 | H | OH | H | OCH3 | Glu | OH | OH | H | 40 |
| 8 | OH | OH | H | OH | H | H | OH | H | 30 |
| 9 | H | OH | Glu | OCH3 | H | H | OH | H | 30 |
| 10 | H | H | H | OH | H | H | OCH3 | H | 20 |
| 11 | OH | OH | H | OH | H | OH | OH | OH | 20 |
| 12 | OH | OH | H | OH | H | OH | OH | H | 20 |
| 13 | H | OH | Glu | OH | H | H | OH | H | 20 |
| 14 | OH | H | H | OH | H | H | H | H | 10 |
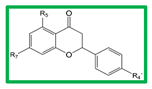 | ||||
|---|---|---|---|---|
| Comp | R5 | R7 | R4´ | AHR:ARNT Prob (%) |
| 15 | OH | OCH3 | OH | 60 |
| 16 | OCH3 | OH | H | 50 |
| 17 | OH | OCH3 | H | 30 |
| Concentration (µg/mL) | Determination 1 | Determination 2 | Mean | % CV |
|---|---|---|---|---|
| 8 | 14,455 | 19,804 | 17,129 | 2208 |
| 16 | 12,551 | 9956 | 11,254 | 1630 |
| 32 | 14,018 | 15,413 | 14,716 | 670 |
| 64 | 8487 | 9921 | 9204 | 1102 |
| 128 | 2486 | 2740 | 2613 | 686 |
| 256 | −1702 | −1501 | −1602 | −885 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, O.C.; De Winter, H.; Cos, P.; Matos, M.J.; Uriarte, E.; Maury, G.L.; De Waele, J.; Santiago, G.C.; Molina, E. Molecular Docking Study of Flavonoids to Block the Aryl Hydrocarbon Receptor. Chem. Proc. 2022, 8, 77. https://doi.org/10.3390/ecsoc-25-11766
García OC, De Winter H, Cos P, Matos MJ, Uriarte E, Maury GL, De Waele J, Santiago GC, Molina E. Molecular Docking Study of Flavonoids to Block the Aryl Hydrocarbon Receptor. Chemistry Proceedings. 2022; 8(1):77. https://doi.org/10.3390/ecsoc-25-11766
Chicago/Turabian StyleGarcía, Oscar Collado, Hans De Winter, Paul Cos, Maria João Matos, Eugenio Uriarte, Gabriel Llaurado Maury, Jorrit De Waele, Glay Chinea Santiago, and Enrique Molina. 2022. "Molecular Docking Study of Flavonoids to Block the Aryl Hydrocarbon Receptor" Chemistry Proceedings 8, no. 1: 77. https://doi.org/10.3390/ecsoc-25-11766
APA StyleGarcía, O. C., De Winter, H., Cos, P., Matos, M. J., Uriarte, E., Maury, G. L., De Waele, J., Santiago, G. C., & Molina, E. (2022). Molecular Docking Study of Flavonoids to Block the Aryl Hydrocarbon Receptor. Chemistry Proceedings, 8(1), 77. https://doi.org/10.3390/ecsoc-25-11766








