Abstract
Stokesia laevis (common name Stokes aster) ethanolic extract (Slae26) containing 5 mg GAE/mL extract was investigated to establish cytotoxicity and anti-proliferative effects. The assays were performed on normal murine fibroblast cell line L929 and malignant murine melanoma cell line B16, respectively; for the first time in literature data, potential cytotoxic and anti-proliferative effects of the ethanolic extract from S. laevis on both, normal murine fibroblast cell line L929, and murine melanoma cell line B16 have been proved. The study is supplemented by molecular docking simulations of the major components of Slae26 against human tyrosinase receptor, to evaluate possible melanogenesis inhibition.
1. Introduction
According to the specialized data, the cutaneous malignant melanoma is the most aggressive type of skin cancer [1]. Data also indicate that the skin melanoma is the most commonly occurring cancer worldwide; the most affected are Australian and New Zealand peoples, followed by Caucasian peoples [2]. Due to high aggressiveness and chemical therapy resistance [3], there are many attempts to find new therapy combinations and antitumor agents or synergistic compounds able to fight against skin melanoma cancer resistance. Among these, several plant species and specific plant compounds indicated promising results, inhibitory activity by in vitro testing of human and murine melanoma cell lines and murine melanoma models, respectively [4].
Aiming to extend the authors’ previously published work [5], the present paper aims to study the cytotoxic and anti-proliferative potential of the standardized ethanolic extract (Slae26) from Stokesia laevis (J. Hill, fam. Asteraceae), 5 mg GAE/mL extract, on normal murine fibroblast cell line L929 and malignant murine melanoma cell line B16, respectively; the predominant compounds in Slae26 (HPTLC analysis) are caffeic acid and luteolin derivates [5].
2. Materials and Methods
Briefly, the test vegetal extract was obtained from dried and powdered aerial part of S. laevis extracted with 70% (v/v) ethanol at the boiling temperature. The resulted ethanolic extract was analyzed concerning quantitative and qualitative aspects [5], then prepared as standardized 5 mg total phenols content expressed as gallic acid derivates [GAE] per 1 mL 40% ethanol solution, v/v, namely Slae26; the Slae26 dilution series (0.5, 1, 5, 10, 25, 50 and 100 μg GAE/mL, test vegetal samples) and corresponding 40% ethanol solvent dilution series (0.04, 0.08, 0.8, 2, 4 and 8% ethanol, v/v, control samples) were used for further in vitro cell studies.
The in vitro cytotoxicity and anti-proliferative assays were done according to the Technical Bulletin of Promega Corporation CellTiter 96 AQueous One solution Cell Proliferation Assay as described in previous study [5]. Briefly, after 20 h (cytotoxicity test) and, respectively, 20 and 44 h (antiproliferative test) of cell exposure, the absorbance of the test samples (Slae26 dilution series) face to control sample (40% ethanol dilution series) were measured at 490 nm (using Chameleon V Plate Reader, LKB Instruments, Mount Waverley, Australia). The recorded values were used for the estimation of the cell viability (see Equation (1)). Results are calculated as mean ± SD, n = 3.
The molecular docking study was realized using CLC Drug, Discovery Work Bench. Protein fragment, human tyrosinase related protein 1 [6] in complex with kojic acid (PDB ID 5M8M) [7] was imported from Protein Data Bank. Ligands’ structures, components of Slae26 (Caffeic acid, Chlorogenic acid, Luteolin, Luteolin-5-O-glucoside, Luteolin-7-O-glucoside, Luteolin-6-C-glucoside, Luteolin-8-C-glucoside, Luteolin-7,3′-di-O-glucoside and Luteolin 3,4′-di-O-glucoside) were imported from PubChem (https://pubchem.ncbi.nlm.nih.gov, accessed on 5 November 2020), and docked into 107.01 Å3 binding pocket.
3. Results
3.1. Cytotoxicity and Anti-Proliferative Assays
Figure 1 shows the results on the Slae26 dilution series tested on (a) murine fibroblast cell line L929 and (b) murine melanoma cell line B16, compared to the control negative cell series (40% ethanol solvent series). Therefore, the cytotoxicity test on the normal murine fibroblast cell line L929 indicated that Slae26 test sample concentrations less than 25 μg/mL induced moderate stimulating effects on the L929 cell line viability (up to 20% increase), after that there were noticed augmented inhibitory activity (up to 84% cell viability decrease at 100 μg/mL); the anti-proliferative test indicated that, less than 10 μg/mL extract at 24 h/h, and less than 5 μg/mL at 48 h, Slae26 induced stimulating effects (up to 25%, and up to 7% cell viability increase, respectively), after which the same decrease in cell viability was observed (inhibition of cell viability up to 65% and 81%, at 24 h and 48 h, respectively).
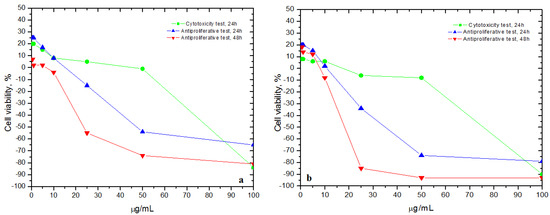
Figure 1.
Cytotoxic and antiproliferative effects (cell viability, %) of Slae26 dilution series tested on (a) murine fibroblast cell line L929 and (b) murine melanoma cell line B16, compared to control negative cell lines (40% ethanol solvent series); n = 3, ±SD (%).
In the case of malignant murine melanoma cell line B16, the cytotoxicity test shown the same stimulating effects on the B16 cell line viability (up to 8% stimulation of cell viability at the test sample concentrations less than 10 μg/mL), followed by a severe decrease in cell viability at higher concentrations (up to 90% cell viability decrease at 100 μg/mL); similarly, the anti-proliferative test indicated less than 10 μg/mL extract at 24 h, and less than 5 μg/mL at 48 h, Slae26 extract induced stimulating effects on B16 cell line viability (up to 20% and up to 18%, at 24 h and 48 h, respectively), followed by a sharp decrease in cell viability at 24 and 48 h (up to 79% and up to 93% inhibition of cell viability at 24 h and 48 h at 100 μg/mL, respectively).
3.2. Molecular Docking Results
All investigated structures reveal greater docking score than the co-crystallized ligand (kojic acid). The chart of obtained values for docking score is presented in Figure 2. The best result is given for L-7-O-glucoside (66.76). For all compounds, the interactions as hydrogen bond type and length formed with the amino acids residues form the active binding site of the tyrosinase, are listed in Table 1, along with results obtained for the natural ligand (KOJ A514 = Kojic acid). Two of hydrogen bonds interactions formed with the same amino acid residues, as kojic acid forms, SER394 and HIS215, respectively, occur in the complexes of investigated ligands with human tyrosinase related protein 1 fragment, excepting di-glucosides (L-3′,4′-di-O-glucoside and L-3′,4′-di-O-glucoside). Only HIS215 is present in their interacting amino acids group, but don’t establish interactions with the di-glucosides.

Figure 2.
Docking scores for Luteolin (L) derivatives, caffeic and chlorogenic acids, against human tyrosinase related protein 1 (PDB ID 5M8M).

Table 1.
Intermolecular interactions between ligands and 5M8M and docking results.
Figure 3 illustrates the arbitrary numbering scheme for constitutive atoms of investigated structures, as given for the most stable conformers of each structures, obtained after energy minimization using Spartan Software [8]. Figure 4 depicts the intermolecular interactions of investigated ligands with 5M8M, illustrating the hydrogen bonds formed by the hydroxyl (O sp3) or carboxyl group (O sp2) of investigated structures and amino acids residues from the binding pocket of the protein fragment, directly interacting.

Figure 3.
Atoms labels numbering for: (a) L; (b) L-7-O-glucoside; (c) L-5-O-glucoside; (d) L-6-C-glucoside; (e) L-8-C-glucoside; (f) L-3′,4′-di-O-glucoside; (g) L-7,3′-di-O-glucoside; (h) caffeic acid; (i) chlorogenic acid.


Figure 4.
Intermolecular interactions of investigated ligands with 5M8M, as hydrogen bonds formed by: (a) Luteolin (L); (b) L-7-O-glucoside; (c) L-5-O-glucoside; (d) L-6-C-glucoside; (e) L-8-C-glucoside; (f) L-3′,4′-di-O-glucoside; (g) L-7,3′-di-O-glucoside; (h) caffeic acid and (i) chlorogenic acid.
4. Discussion
Very extensive studies summing 582 extracted samples obtained from 370 plants [9] indicated several plants derived products and specific phytocompounds (there were investigated 118 separate compounds) as being able to act as inhibitors of B16F-10 melanoma metastatic cell line viability. Therefore, the most effective plant derived products were Mangifera indica-barks, leaves and seeds extracts, Annona cherimola, Annona muricata and Annona squamosa-bark, leaves, stems and twigs extracts, Anthriscus sylvestris-fruits, leaves and roots extracts, Osmorhiza aristata-aerial parts extracts, Araucaria heterophylla-leaves extracts, Tylophora ovata and Tylophora tanakae-fresh leaves, twigs and aerial parts extracts, Crepidiastrum lanceolatum-aerial parts extracts, Garcinia subelliptica-barks extracts, Luffa acutangula and Momordica cochinchinensis-seeds extracts, Juniperus rigida and Thuja occidentalis-leaves extracts, Persea americana-leaves extracts and Coptis japonica-rhizomes extracts. Plant derived products inhibitory activity (EC50) was mostly evaluated as less than 100 µg/mL (70%), 12% of them being evaluated as less than 12.5 µg/mL. Moreover, lignan compounds were proved as the most effective inhibitors of B16F-10 melanoma metastatic cell line viability; deoxypodophyllotoxin and morensin proved the most augmented antiproliferative effects being evaluated at EC50 = 0.21 and 0.23 µg/mL respectively. Other polyphenols compounds such as apigenin (EC50 = 25 µg/mL), luteolin (EC50 = 21 µg/mL), baicalein (EC50 = 11 µg/mL), gallic acid and derivates (EC50 = 2–9 µg/mL) and hydroxycinnamic acid derivates, also were proved to provide certain anti-proliferative effects against B16F-10 melanoma metastatic cell line viability [9].
The present work suggests certain cytotoxic and antiproliferative activity of 40% ethanolic extract (Slae26) from Stokesia laevis plant species (the aerial part), upon normal murine fibroblast cell line L929 and murine melanoma cell line B16; also, HPTLC analysis of Slae26 indicated the presence of two major polyphenols subclasses, caffeic acid and luteolin derivates, punctually the predominance of caffeic acid, chlorogenic acid, luteolin and luteolin-7-O-glucoside [5]. Also, the docking results indicated similar interactions for the co-crystallized kojic acid and the nine vegetal compounds tested (punctually, Luteolin (L), L-7-O-glucoside, L-5-O-glucoside, L-6-C-glucoside, L-8-C-glucoside, L-3′,4′-di-O-glucoside, L-7, 3′-di-O-glucoside, Caffeic acid and Chlorogenic acid); furthermore, there were noticed interactions with the same amino acid residues, by hydrogen bonds formed with O sp3 of SER394, and N sp2 of HIS215, respectively, except for di-glucosides. In addition, due to the numerous hydroxyl groups of our investigated structures, more interactions in the protein-complex occur and higher docking score are revealed. Therefore, docking results suggest the ability of luteolin and caffeic acid derivatives to act as potential skin melanoma cancer inhibitors.
5. Conclusions
For the first time in literature data, potential cytotoxic and anti-proliferative effects of the ethanolic extract from S. aster on both, normal murine fibroblast cell line L929, and murine melanoma cell line B16 have been proved. Molecular docking approach on the major components of Slae26 against human tyrosinase receptor has reveal possible melanogenesis inhibition.
Author Contributions
Conceptualization, L.C.P. and A.S.; methodology, A.S. and G.N.; software, L.P., A.S. and B.A.; validation, G.N. and L.P.; formal analysis, B.A.; investigation, A.S., G.N. and I.T.; resources, G.N.; data curation, L.C.P.; writing—original draft preparation, A.S.; writing—review and editing, L.C.P.; visualization, G.N.; supervision, L.C.P.; project administration, L.C.P.; funding acquisition, L.C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the ANCSI POC-A1-A1.2.3-G-2015 (Project ID P_40_406, SMIS 105542, Contract no. 60/05.09.2016).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Domingues, B.; Lopes, J.M.; Soares, P.; Populo, H. Melanoma treatment in review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Skin Cancer Statistics. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/skin-cancer-statistics (accessed on 24 October 2020).
- Kalal, B.S.; Upadhya, D.; Pai, V.R. Chemotherapy resistance mechanisms in advanced skin cancer. Oncol. Rev. 2017, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Chinembiri, N.T.; du Plessis, L.H.; Gerber, M.; Hamman, J.H.; du Plessis, J. Review of natural compounds for potential skin cancer treatment. Molecules 2014, 19, 11679–11721. [Google Scholar] [CrossRef] [PubMed]
- Pirvu, L.; Neagu, G.; Terchescu, I.; Albu, B.; Stefaniu, A. Comparative studies of two vegetal extracts from Stokesia laevis and Geranium pratense: Polyphenol profile, cytotoxic effect and antiproliferative activity. Open Chem. 2020, 18, 488–502. [Google Scholar] [CrossRef]
- Lai, X.; Wichers, H.J.; Soler-Lopez, M.; Dijkstra, B.W. Structure of human tyrosinase related protein 1 reveals a binuclear zinc active site important for melanogenesis. Angew. Chem. Int. Ed. 2017, 56, 9812–9815. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Soler-Lopez, M.; Wichers, H.J.; Dijkstra, B.W. Crystal Structure of Human Tyrosinase Related Protein 1 in Complex with Kojic Acid; PDB ID: 5M8M. Deposited on: 2016-10-29. Available online: https://www.rcsb.org/structure/5M8M (accessed on 5 November 2020).
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Gilbert, A.T.B.; Slipchenko, L.V.; Levchenko, S.V.; O’Neill, D.P.; DiStasio, R.A., Jr.; et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef] [PubMed]
- Kinjo, J.; Nakano, D.; Fujioka, T.; Okabe, H. Screening of promising chemotherapeutic candidates from plants extracts. J. Nat. Med. 2016, 70, 335–360. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).