Abstract
The control of insect pests and the need for increased food production due to the world population growth, together with environmental issues associated with synthetic pesticides, has stimulated the development of new and “greener” alternatives, based on natural compounds. Eugenol is a natural compound that is the major component of clove oil. It has demonstrated antimicrobial and antioxidant activity, being also a powerful insecticide. Recently, new eugenol derivatives have been developed, with some molecules displaying increased insecticide activity. One of the difficulties associated with the rational development of new eugenol derivatives with enhanced insecticidal activity lies in the lack of knowledge of the specific protein target responsible for its activity and to the binding conformation of these molecules. Here, we report the application of an integrated molecular modeling—inverted virtual screening protocol of a collection of eugenol derivatives with confirmed insecticide activity against a molecular library of protein targets typically associated with the insecticide activity of natural compounds. The protocol included six different scoring functions from popular docking software alternatives. The results consistently show a marked preference for interaction of the eugenol derivatives with the odorant binding proteins (OBPs) in insect species. Interestingly, OBPs have been regarded as promising targets in the insect periphery nerve system for environmental-friendly approaches in insect pest management. The present results provide clues for the rational development of new eugenol derivatives as bioinsecticides targeting OBPs.
1. Introduction
The increase in population has caused a strain in agriculture due to rising demand and decreased land availability. Crops need to become resistant to damage and disease and the use of pesticides, fungicides, and herbicides has allowed for crop protection and long-term storage [1,2]. However, when these chemicals are used extensively or incorrectly, they become hazardous to the environment and to human health [3]. Thus, the search for new natural, safe, and ecofriendly alternatives, i.e., biopesticides, is being encouraged. Plants, animals, and bacteria produce metabolites that can exhibit insecticidal activity. Using those metabolites to protect crops may lead to higher specificity, lower toxicity, and even a decrease in pest resistance [4].
Eugenol, the major component of clove oil, has established insecticidal and antimicrobial activity against a variety of targets [5,6,7]. Consequently, the search for new eugenol derivatives with higher efficiency was boosted to find additional alternatives to known insecticides. However, there is still significant work to be done to find out their correct binding conformation and, most importantly, their exact target(s) and mechanism(s) of action. This is precisely where computational chemistry can provide valuable insight.
In this study, it is presented the application of an integrated molecular modeling—an inverted virtual screening protocol for the identification of potential protein targets for a series of eugenol derivatives with confirmed insecticide activity. The protocol included the study of protein targets typically associated with the insecticide activity and included six different scoring functions from popular docking software alternatives.
2. Methods
A search on Scopus was performed for papers describing virtual screening (VS) studies, involving targets and molecules with insecticidal/herbicide activity. The selection criteria placed relevance of the target and year of publication. In the eighteen studies found, fourteen targets were identified (listed in Table 1).

Table 1.
List of targets selected for the inverted virtual screening study.
Eugenol and eleven derivatives (Figure 1 EU1–EU3e) were selected as new potential insecticides. These molecules have been previously synthesized and validated experimentally with good insecticidal activity.
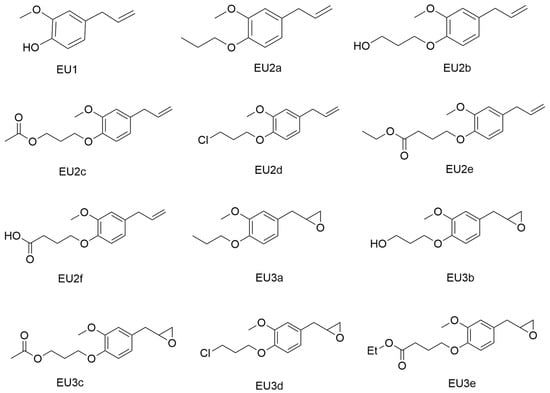
Figure 1.
Eugenol and derivatives used in this study.
Each Protein Databank (PDB) structure was prepared for docking using the AutoDock Vina plugin for PyMOL [26]. Crystallographic waters and cofactors were removed. The ligands were extracted and saved in separate files to be used for the re-docking and as a reference site for the docking coordinates. When there were no crystallographic ligands present, a selection based on the most important active site residues was made. Re-docking was used to evaluate the ability of the docking software to reproduce the geometry and orientation of the crystallographic pose as well as the quality of the docking protocol, and to optimize the docking protocol.
The docking programs/scoring functions used were GOLD [27] (PLP, ASP, ChemScore, and GoldScore scoring functions), AutoDock Vina [28], and LeDock [29]. With each docking program/scoring function, the protocol was optimized for each protein target, to minimize the rmsd in the docking predictions of the reference ligand in redocking, by comparison with the crystallographic structure of the corresponding complex.
The optimized parameters for each program/scoring function were: Vina—docking box position, docking box dimension, exhaustiveness; LeDock—docking box position, docking box dimension; GOLD (PLP, ASP, ChemScore, GoldScore)—binding pocket center, docking region radius, search efficiency, number of runs. The final optimized conditions were used for the subsequent stages. Eugenol and derivatives were prepared for docking using Datawarrior [30] and OpenBabel [31] and were docked into each structure with the optimized protocol with all the six scoring functions. A ranked list was prepared based on the average scores of each target.
3. Results and Discussion
Table 2 presents the average scores obtained for of all the eugenol derivatives for each potential target with each scoring function. The score for all of the GOLD scoring functions is dimensionless, and the higher the score, the better the binding affinity. Vina and LeDock scoring functions, on the other hand, use a metric that is a more precise approximation of binding free energy, so a more negative value means better affinity.

Table 2.
Average eugenol derivate scores obtained for all PDB structures with the six different scoring functions.
Overall, the results show good consistency, with odorant binding proteins, acetylcholinesterases, octopamine receptors, and chitinases yielding better scores. On the other hand, targets, such as voltage-gated sodium channels, sterol carrier protein-2 (HaSCP-2), and N-Acetylglucosamine-1-phosphate uridyltransferase (GlmU), are consistently presenting lower scores for all scoring functions.
The structure with the best score was selected for each potential target and they were ranked from the best target to worst, according to the predictions of the different docking programs/scoring functions. The results are listed in Table 3. Globally, considering the results obtained with the several scoring functions, odorant binding proteins are the target with the highest affinity towards eugenol derivatives, followed closely by acetylcholinesterase, chitinases, and octopamine receptors. Enan in 2001 [5] suggested that the insecticidal activity of eugenol was mediated by octopamine receptors. Our study implies that there might be other targets involved as well, as the binding affinity of eugenol derivates was higher for odorant binding proteins (OBPs) and acetylcholinesterase.

Table 3.
Ranking of targets obtained with the different docking programs/scoring functions.
Some variations between the predictions of different scoring functions exists. For example, for the PLP and ChemScore scoring function, odorant binding proteins, and acetylcholinesterase come in first and second as preferable targets for eugenol derivates. However, for ASP and Vina, the preferable target is the acetylcholinesterase, and for both Vina and LeDock, odorant binding proteins are the second preferable targets. The discrepancy is even higher for GoldScore, with odorant binding proteins coming in third place and octopamine receptors presenting the highest binding affinity for eugenol derivates. This may be explained by the own nature of each scoring function, as they consider different aspects of protein-ligand binding.
The consistency of the results was visually confirmed by the analysis of the corresponding poses. The hypothesis formed is that eugenol and eugenol derivatives can be used as repellents because they can bind to odorant binding proteins or be used as pesticides, inhibiting insect acetylcholinesterase. As observed in Figure 2, they are very different targets, both in size and in function.
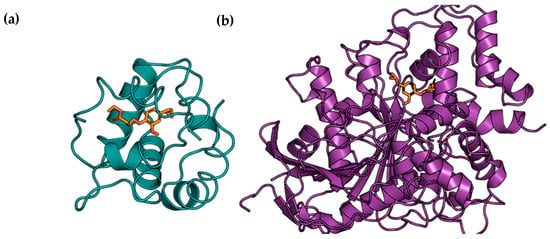
Figure 2.
Docking-predicted binding mode of EU3e to OBPs (a) and docking-predicted binding modes of EU3e to Acetylcholinesterase (b) with PLP scoring function.
Odorant binding proteins (OBPs) are a large family of insect proteins that are crucial for species survival and reproduction, as they use pheromones, plant volatiles, and other odorant molecules to mate, find food, and avoid predators [32]. OBPs are present in a variety of organisms, are highly expressed and highly divergent in sequence. They do, however, present a few common features, such as their small size and the presence of six conserved cysteines [33]. These features also make them good targets for rapid screenings. There is not enough consensus regarding the specificity of these proteins and further studies must be performed to better understand the sensitivity of OBPs [2].
Acetylcholinesterase (AChE) is one of the most common targets of synthetic pesticides, such as organophosphates and carbamate [34], and has been a target of reference for over 50 years. This enzyme is a serine hydrolase and is responsible for regulating the levels of acetylcholine in a variety of organisms, from mammals to insects [35]. Due to its extensive “attack”, some pests have become resistant to organophosphates, and the search for new and effective alternatives is currently being promoted [36].
Interestingly, during a search in the Protein Data Bank for eugenol, a structure of an odorant binding protein was found complexed with eugenol found. It is an OBP of Apis mellifera (PDB: 3S0E) that exhibits high affinity for eugenol [37]. This reinforces the proposed theory that eugenol and derivatives can, in fact, bind to OBPs and could potentially work as repellents. Still, additional computational and experimental studies need to be performed to further optimize and develop this hypothesis.
4. Conclusions
In the present study, we report the application of an integrated molecular modeling—an inverted virtual screening protocol of a collection of eugenol derivatives in order to find possible protein targets in which they present insecticidal activity.
First, we explored the literature for other virtual screening studies performed on known targets to minimize the candidate pool. Of 18 studies found, 14 targets were selected to continue the study. After careful optimization of the VS protocol, the eugenol derivatives were docked into each target with six different scoring functions (PLP, ASP, ChemScore, GoldScore, Vina and LeDock). The consistency of the scores was evaluated and a ranked list of most likely targets was created.
Eugenol derivates showed an increased binding affinity for odorant binding proteins and acetylcholinesterases. Since there is, already in the PDB database, a structure of an OBP bound to eugenol not considered in the VS, it reinforces the proposal that eugenol derivatives can potentially be used as repellents.
This work presents a simple approach for the application of inverted virtual screening in identification of possible targets for new insecticides.
Acknowledgments
This research was funded by COMPETE 2020 program, co-financed by the FEDER and the European Union, PTDC/ASP-AGR/30154/2017 (POCI-01-0145-FEDER-030154) and by UCIBIO (UIDB/04378/2020).
References
- Liu, X.; Cao, A.; Yan, D.; Ouyang, C.; Wang, Q.; Li, Y. Overview of mechanisms and uses of biopesticides. Int. J. Pest Manag. 2019, 67, 1–8. [Google Scholar] [CrossRef]
- Venthur, H.; Zhou, J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.A. Pesticides and Human Health. In Pestic. Mod. World—Eff. Pestic. Expo; InTech: Hong Kong, 2011. [Google Scholar]
- Amoabeng, B.W.; Johnson, A.C.; Gurr, G.M. Natural enemy enhancement and botanical insecticide source: a review of dual use companion plants. Appl. Entomol. Zool. 2019, 54, 1–19. [Google Scholar] [CrossRef]
- Enan, E. Insecticidal activity of essential oils: octopaminergic sites of action. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001, 130, 325–337. [Google Scholar] [CrossRef]
- Liska, A.; Rozman, V.; Kalinovic, I.; Ivezic, M.; Balicevir, R. Contact and fumigant activity of 1,8-cineole, eugenol and camphor against Tribolium castaneum (Herbst). Julius-Kühn-Archiv 2010, 425, 716. [Google Scholar]
- Wang, C.; Zhang, J.; Chen, H.; Fan, Y.; Shi, Z. Antifungal activity of eugenol against Botrytis cinerea. Trop. Plant Pathol. 2010, 35, 137–143. [Google Scholar] [CrossRef]
- Hu, X.; Yin, B.; Cappelle, K.; Swevers, L.; Smagghe, G.; Yang, X.; Zhang, L. Identification of novel agonists and antagonists of the ecdysone receptor by virtual screening. J. Mol. Graph. Model. 2018, 81, 77–85. [Google Scholar] [CrossRef]
- Harada, T.; Nakagawa, Y.; Ogura, T.; Yamada, Y.; Ohe, T.; Miyagawa, H. Virtual Screening for Ligands of the Insect Molting Hormone Receptor. J. Chem. Inf. Model. 2011, 51, 296–305. [Google Scholar] [CrossRef]
- Dong, Y.; Jiang, X.; Liu, T.; Ling, Y.; Yang, Q.; Zhang, L.; He, X. Structure-Based Virtual Screening, Compound Synthesis, and Bioassay for the Design of Chitinase Inhibitors. J. Agric. Food Chem. 2018, 66, 3351–3357. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Yao, Y.; Wang, J.; Li, Y.; Li, G.; Wang, Y. Identification of Novel Potential β-N-Acetyl-D-Hexosaminidase Inhibitors by Virtual Screening, Molecular Dynamics Simulation and MM-PBSA Calculations. Int. J. Mol. Sci. 2012, 13, 4545–4563. [Google Scholar] [CrossRef]
- Dong, L.; Shen, S.; Xu, Y.; Wang, L.; Yang, Q.; Zhang, J.; Lu, H. Identification of novel insect β-N-acetylhexosaminidase OfHex1 inhibitors based on virtual screening, biological evaluation, and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2021, 39, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Lin, D.; Zhang, Q.; Zhang, J.; Yu, Z. Structure-based virtual screening of novel inhibitors of the uridyltransferase activity of Xanthomonas oryzae pv. oryzae GlmU. Eur. J. Med. Chem. 2012, 53, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.; Costa, J.; Silva, R.; da Costa, G.; Rodrigues, A.; Rabelo, É.; Souto, R.; Taft, C.; Silva, C.; Rosa, J.; et al. Identification of Potential Inhibitors from Pyriproxyfen with Insecticidal Activity by Virtual Screening. Pharmaceuticals 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Riva, C.; Suzanne, P.; Charpentier, G.; Dulin, F.; Halm-Lemeille, M.-P.; Sopkova-de Oliveira Santos, J. In silico chemical library screening and experimental validation of novel compounds with potential varroacide activities. Pestic. Biochem. Physiol. 2019, 160, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Fattouch, S.; Raboudi-Fattouch, F.; Ponce, J.V.G.; Forment, J.V.; Lukovic, D.; Marzouki, N.; Vidal, D.R. Concentration dependent effects of commonly used pesticides on activation versus inhibition of the quince (Cydonia Oblonga) polyphenol oxidase. Food Chem. Toxicol. 2010, 48, 957–963. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, Y.-X.; Kang, T.; Sun, Y.-N.; Li, J.-Z.; Ye, F. Identification of novel inhibitors of p-hydroxyphenylpyruvate dioxygenase using receptor-based virtual screening. J. Taiwan Inst. Chem. Eng. 2019, 103, 33–43. [Google Scholar] [CrossRef]
- Liu, X.-H.; Chen, P.-Q.; Wang, B.-L.; Dong, W.-L.; Li, Y.-H.; Xie, X.-Q.; Li, Z.-M. High Throughput Receptor-Based Virtual Screening Under ZINC Database, Synthesis, and Biological Evaluation of Ketol-Acid Reductoisomerase Inhibitors. Chem. Biol. Drug Des. 2010, 75, 228–232. [Google Scholar] [CrossRef]
- Shen, H.; Li, Z.; Jiang, Y.; Pan, X.; Wu, J.; Cristofori-Armstrong, B.; Smith, J.J.; Chin, Y.K.Y.; Lei, J.; Zhou, Q.; et al. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 2018, 362, 1–8. [Google Scholar] [CrossRef]
- Offermann, L.R.; Chan, S.L.; Osinski, T.; Tan, Y.W.; Chew, F.T.; Sivaraman, J.; Mok, Y.-K.; Minor, W.; Chruszcz, M. The major cockroach allergen Bla g 4 binds tyramine and octopamine. Mol. Immunol. 2014, 60, 86–94. [Google Scholar] [CrossRef]
- Cai, J.; Du, X.; Wang, C.; Lin, J.; Du, X. Identification of Potential Helicoverpa armigera (Lepidoptera: Noctuidae) Sterol Carrier Protein-2 Inhibitors Through High-Throughput Virtual Screening. J. Econ. Entomol. 2017, 110, 1779–1784. [Google Scholar] [CrossRef]
- Joshi, T.; Joshi, T.; Sharma, P.; Chandra, S.; Pande, V. Molecular docking and molecular dynamics simulation approach to screen natural compounds for inhibition of Xanthomonas oryzae pv. Oryzae by targeting peptide deformylase. J. Biomol. Struct. Dyn. 2021, 39, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Correy, G.J.; Zaidman, D.; Harmelin, A.; Carvalho, S.; Mabbitt, P.D.; Calaora, V.; James, P.J.; Kotze, A.C.; Jackson, C.J.; London, N. Overcoming insecticide resistance through computational inhibitor design. Proc. Natl. Acad. Sci. USA 2019, 116, 21012–21021. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, J.D.; Ha, T.S.; Jones, D.N.M.; Smith, D.P. Activation of Pheromone-Sensitive Neurons Is Mediated by Conformational Activation of Pheromone-Binding Protein. Cell 2008, 133, 1255–1265. [Google Scholar] [CrossRef]
- Oliferenko, P.V.; Oliferenko, A.A.; Poda, G.I.; Osolodkin, D.I.; Pillai, G.G.; Bernier, U.R.; Tsikolia, M.; Agramonte, N.M.; Clark, G.G.; Linthicum, K.J.; et al. Promising Aedes aegypti Repellent Chemotypes Identified through Integrated QSAR, Virtual Screening, Synthesis, and Bioassay. PLoS ONE 2013, 8, e64547. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided. Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Liu, N.; Xu, Z. Using LeDock as a docking tool for computational drug design. IOP Conf. Ser. Earth Environ. Sci. 2019, 218, 012143. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Brito, N.F.; Moreira, M.F.; Melo, A.C.A. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.S.; Xiao, S.; Carlson, J.R. The diverse small proteins called odorant-binding proteins. Open Biol. 2018, 8, 180208. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Luo, J.; Zhou, Y.; Xiao, H.; He, K.; Yin, C.; Xu, J.; Li, F. ACE: An efficient and sensitive tool to detect insecticide resistance-associated mutations in insect acetylcholinesterase from RNA-Seq data. BMC Bioinform. 2017, 18, 330. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.-P.; Brimijoin, S.; Ragsdale, D.W.; Zhu, K.Y.; Suranyi, R. Novel and Viable Acetylcholinesterase Target Site for Developing Effective and Environmentally Safe Insecticides. Curr. Drug Targets 2012, 13, 471–482. [Google Scholar] [CrossRef]
- Pang, Y. Insect Acetylcholinesterase as a Target for Effective and Environmentally Safe Insecticides. In Target Recept. Control Insect Pests Part II; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 435–494. [Google Scholar]
- Spinelli, S.; Lagarde, A.; Iovinella, I.; Legrand, P.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 2012, 42, 41–50. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).