New 1,2,4-Triazole Potential Inhibitors of Mycobacterial Imidazoleglycerol-Phosphate Dehydratase (IGPD) †
Abstract
:1. Introduction
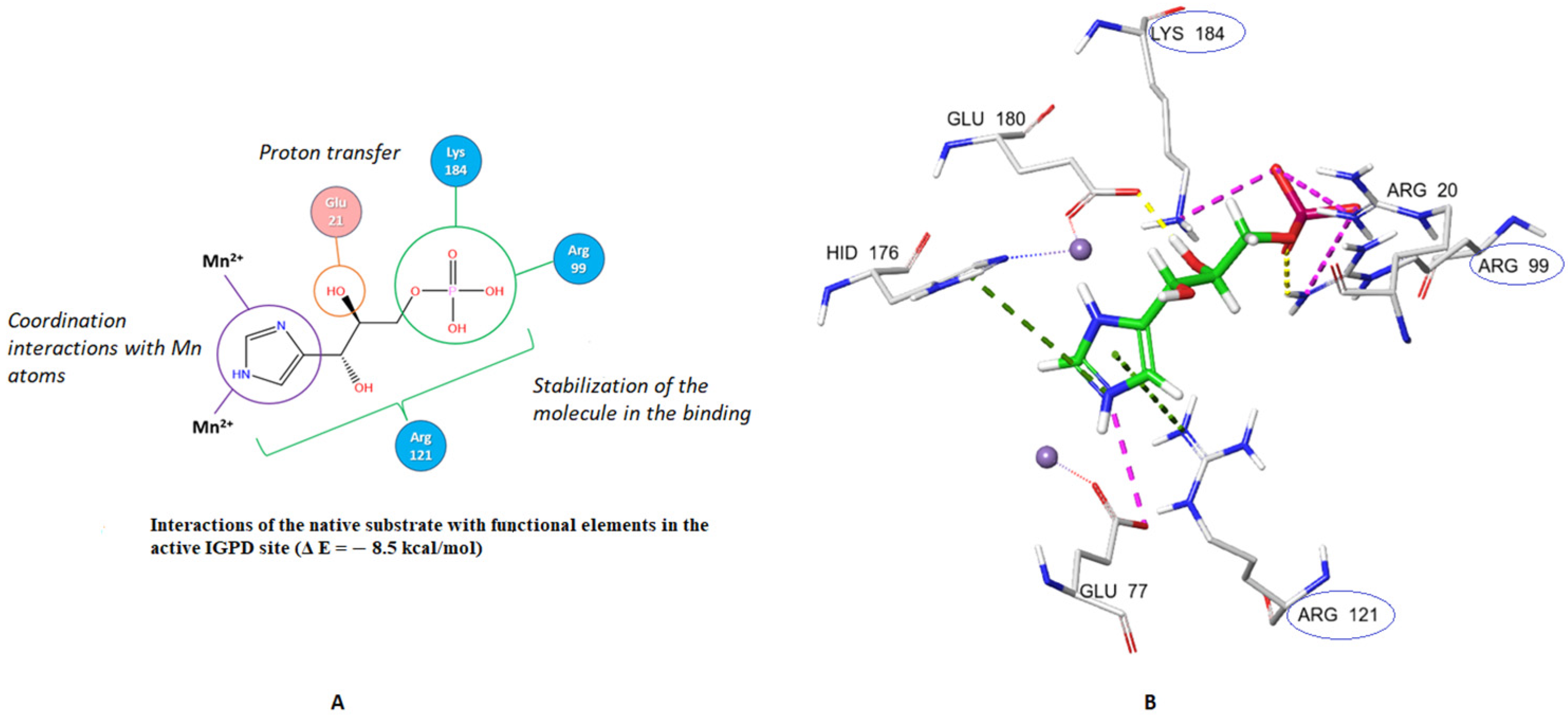
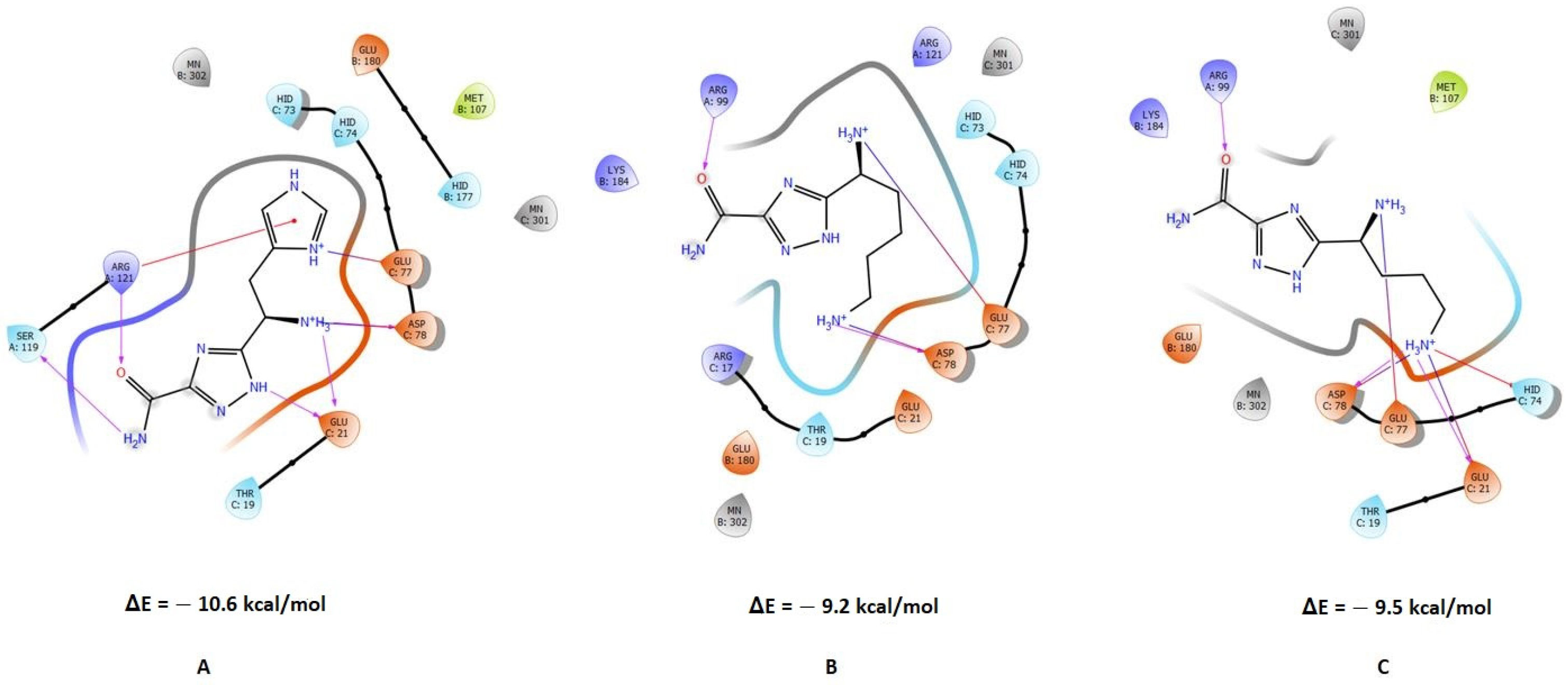
2. Results and Discussion
2.1. Design and In Silico Studies
2.2. Synthetic Section
2.3. Biology Section
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Synthesis
4.3. In Silico Studies
4.4. Antibacterial Activity
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agüero, F.; Al-Lazikani, B.; Aslett, M.; Berriman, M.; Buckner, F.S.; Campbell, R.K.; Carmona, S.; Carruthers, I.M.; Chan, A.W.E.; Chen, F.; et al. Genomic-scale prioritization of drug targets: The TDR Targets database. Nat. Rev. Drug Discov. 2008, 7, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.S.; Bhole, R.P.; Khedekar, P.B.; Chikhale, R.V. Mycobacterium enoyl acyl carrier protein reductase (InhA): A key target for antitubercular drug discovery. Bioorg. Chem. 2021, 115, 105242. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.J.; Carroll, P.; Lewis, D.G.; Dunford, A.J.; Seward, H.E.; Neeli, R.; Cheesman, M.R.; Marsollier, L.; Douglas, P.; Smith, W.E.; et al. Characterization of active site structure in CYP121. A cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J. Biol. Chem. 2008, 283, 33406–33416. [Google Scholar] [CrossRef] [PubMed]
- Lunardi, J.; Nunes, J.; Bizarro, C.; Basso, L.; Santos, D.; Machado, P. Targeting the Histidine Pathway in Mycobacterium tuberculosis. Curr. Top. Med. Chem. 2013, 13, 2866–2884. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, T.; Xiao, J.; Huang, G. Antibacterial activity study of 1,2,4-triazole derivatives. Eur. J. Med. Chem. 2019, 173, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Drozd, M.; Ginalska, G.; Pachuta-Stec, A.; Pitucha, M. New Application of 1,2,4-Triazole Derivatives as Antitubercular Agents. Structure, In Vitro Screening and Docking Studies. Molecules 2020, 25, 6033. [Google Scholar] [CrossRef] [PubMed]
- Simons, C.; Alshabani, L.A.; Willcocks, S.J.; Srithiran, G.; Bhakta, S.; Estrada, D.F. Synthesis, biological evaluation and computational studies of pyrazole derivatives as Mycobacterium tuberculosis CYP121A1 inhibitors. RSC Med. Chem. 2022, 13, 1350–1360. [Google Scholar]
- Kumar, D.; Jha, B.; Bhatia, I.; Ashraf, A.; Dwivedy, A.; Biswal, B.K. Characterization of a triazole scaffold compound as an inhibitor of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase. Proteins Struct. Funct. Bioinform. 2022, 90, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Hilton, J.L.; Kearney, P.C.; Ames, B.N. Mode of action of the herbicide, 3-amino-1,2,4-triazole(amitrole): Inhibition of an enzyme of histidine biosynthesis. Arch. Biochem. Biophys. 1965, 112, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Ahangar, M.S.; Vyas, R.; Nasir, N.; Biswal, B.K. Structures of native, substrate-bound and inhibited forms of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase. Acta Crystallogr. Sect. D—Biol. Crystallogr. 2013, 69, 2461–2467. [Google Scholar] [CrossRef] [PubMed]
- Bisson, C.; Britton, K.L.; Sedelnikova, S.E.; Rodgers, H.F.; Eadsforth, T.C.; Viner, R.C.; Hawkes, T.R.; Baker, P.J.; Rice, D.W. Crystal structures reveal that the reaction mechanism of imidazoleglycerol-phosphate dehydratase is controlled by switching Mn(II) coordination. Structure 2015, 23, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Ahangar, M.S.; Khandokar, Y.; Nasir, N.; Vyas, R.; Biswal, B.K. HisB from Mycobacterium tuberculosis: Cloning, overexpression in Mycobacterium smegmatis, purification, crystallization and preliminary X-ray crystallographic analysis. Acta Crystallogr. Sect. F—Struct. Biol. Cryst. Commun. 2011, 67, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Glynn, S.E.; Baker, P.J.; Sedelnikova, S.E.; Davies, C.L.; Eadsforth, T.C.; Levy, C.W.; Rodgers, H.F.; Blackburn, G.; Hawkes, T.R.; Viner, R.; et al. Structure and Mechanism of Imidazoleglycerol-Phosphate Dehydratase. Structure 2005, 13, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Borg, S.; Etenne-Bouhtou, G.; Luthman, K. Synthesis of 1,2,4-Oxadiazole-, 1,3,4-Oxadiazole-, and 1,2,4-Triazole-Derived Dipeptidomimetics. J. Org. Chem. 1995, 60, 3112–3120. [Google Scholar] [CrossRef]
- Lelovic, N.; Mitachi, K.; Yang, J.; Lemieux, M.R.; Ji, Y.; Kurosu, M. Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J. Antibiot. 2020, 73, 780–789. [Google Scholar] [CrossRef] [PubMed]


| Compounds Structures | Time, h | Inhibition Growth M. tuberculosis, % * |
|---|---|---|
| 10 mM | ||
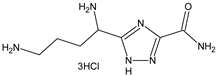 | 24 | 34 ± 3 |
| 48 | 50 ± 4 | |
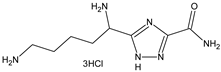 | 24 | 30 ± 4 |
| 48 | 36 ± 5 | |
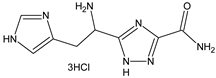 | 24 | 44 ± 2 |
| 48 | 74 ± 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oleynik, E.; Pyankina, E.; Mitina, E.; Mikhina, E.; Matveev, A. New 1,2,4-Triazole Potential Inhibitors of Mycobacterial Imidazoleglycerol-Phosphate Dehydratase (IGPD). Chem. Proc. 2024, 16, 67. https://doi.org/10.3390/ecsoc-28-20178
Oleynik E, Pyankina E, Mitina E, Mikhina E, Matveev A. New 1,2,4-Triazole Potential Inhibitors of Mycobacterial Imidazoleglycerol-Phosphate Dehydratase (IGPD). Chemistry Proceedings. 2024; 16(1):67. https://doi.org/10.3390/ecsoc-28-20178
Chicago/Turabian StyleOleynik, Eugenia, Ekaterina Pyankina, Ekaterina Mitina, Ekaterina Mikhina, and Andrey Matveev. 2024. "New 1,2,4-Triazole Potential Inhibitors of Mycobacterial Imidazoleglycerol-Phosphate Dehydratase (IGPD)" Chemistry Proceedings 16, no. 1: 67. https://doi.org/10.3390/ecsoc-28-20178
APA StyleOleynik, E., Pyankina, E., Mitina, E., Mikhina, E., & Matveev, A. (2024). New 1,2,4-Triazole Potential Inhibitors of Mycobacterial Imidazoleglycerol-Phosphate Dehydratase (IGPD). Chemistry Proceedings, 16(1), 67. https://doi.org/10.3390/ecsoc-28-20178






