Abstract
The main priority in leishmaniasis-endemic countries is to find safer and more accessible treatments for this neglected disease. In this study, we focus on a drug repositioning strategy using molecular docking. New molecular entities (NMEs) approved by the FDA from 2019 to the present were analyzed. The therapeutic target was the sterol 14-alpha demethylase from Leishmania infantum. Of the 125 NMEs tested, 16 demonstrated greater affinity in virtual screening than the co-crystallized inhibitor (fluconazole). This approach offers a promising method for identifying new uses for existing drugs and provides a rapid way to discover safer treatments for leishmaniasis.
1. Introduction
Considered the clinical form of leishmaniasis with the worst prognosis if left untreated, visceral leishmaniasis (VL) is caused by species of the genus Leishmania, such as Leishmania infantum in the New World and Leishmania donovani in the Old World [1,2]. This disease is endemic in around 60 countries worldwide, including 13 countries in the Americas, with a total of 1834 cases and a fatality rate of 9.38% in 2022 [1,3].
Chemotherapy for leishmaniasis remains inadequate regarding safety and accessibility to this day. The most commonly used drugs for this disease are amphotericin B, pentavalent antimonials, and miltefosine, whose adverse effects may include cardiotoxicity, pancreatitis, ototoxicity, and nephrotoxicity [4]. Furthermore, this disease remains directly linked to poverty, considering the limited access of certain populations to adequate healthcare services [4].
The new molecular entities (NMEs) approved by the U.S. Food and Drug Administration (FDA) consist of molecules that have passed safety and efficacy filters, and they represent both a new treatment option and a major advance in healthcare systems. The approval process of NMEs takes an average of 12 years of evaluation and around $1 billion in costs before the drug can enter the market [5,6]. It is important to note that some drugs characterized as NMEs contain active moieties closely related to those in products previously approved by the FDA, or they are pre-approved chemical entities that now constitute a combination product [6]. None of the NMEs approved since 2019 have been related to the treatment of leishmaniasis, which suggests that, with a drug repositioning approach, new uses for drugs that have already passed safety and efficacy filters, or important chemical entities, could be rapidly identified for leishmaniasis.
The enzyme sterol 14-alpha demethylase (CYP51) catalyzes the removal of the 14-alpha methyl group from sterol precursors, a key reaction to synthesizing the ergosterol necessary for the cell membrane of protozoa and fungi or synthesizing cholesterol needed for animal cell membranes [7,8]. This enzyme in trypanosomatids, such as Leishmania, has been targeted for inhibition using chemical entities that inhibit its orthologs in fungi, such as azoles including ketoconazole, itraconazole, and fluconazole. However, azoles have low efficacy against VL in humans and animal models [7]. Although the difference between the amino acid sequences of CYP51 found in Leishmania and fungi is around 25%, CYP51 enzymes preserve their conserved catalytic function, maintaining the enigmatic aspects of the effectiveness of azoles between Leishmania and fungi [9].
In this study, we report which of the NMEs approved by the FDA from 2019 to date inhibit L. infantum CYP51 in silico via molecular docking, finding that 16 out of the 125 evaluated chemical entities bind with greater affinity than the co-crystallized ligand (fluconazole) and interact with the same key residues involved in the inhibition of the enzyme.
2. Materials and Methods
The data for Novel drug approvals by the FDA from 2019 to 2024 were obtained from publicly accessible databases (“https://www.fda.gov/drugs/development-approval-process-drugs/novel-drug-approvals-fda (accessed on 24 May 2024)” [6]). Since these novel drugs may contain pre-approved chemical entities that now constitute a combination product, all chemical entities of the drug were considered. Only chemical entities were used, and medications with antibodies or peptides were not evaluated in this study. The SMILE code for all chemical entities was obtained from PubChem (“https://pubchem.ncbi.nlm.nih.gov/ (accessed on 24 May 2024)” [10]), and the 3D structures were generated using OMEGA software v5.0.0.3 [11], using the “pose” mode that produces conformers for the OEDocking suite, obtaining a total of up to 64 possible conformers for each of 125 NMEs selected.
A crystalline structure of L. infantum CYP51 (RCSB Protein Data Bank accession code: 3L4D) was used as the molecular target [9,12]. This enzyme is in complex with fluconazole, which was used to define the area for molecular docking, and the heme group which serves as a cofactor for this enzyme was kept in the binding site. The OEDocking suite was used to prepare the molecular target, determining the location and shape of the active site with the “make_receptor” program v4.3.0.3 [13,14].
A molecular docking was performed between L. infantum CYP51, fluconazole, and multiples conformers of 125 NMEs using the OEDocking suite, with FRED software v4.3.0.3 (Chemgauss4) [13,15]. The score for the best 4 conformers of each molecule was evaluated. This software was chosen based on the predictive capacity obtained in previous studies [16].
The analysis of interactions between the molecular target and molecules was analyzed using PyMOL v2.5.0 [17] and Discovery Studio Visualizer v21.1.0.20298 [18].
3. Results and Discussion
Figure 1 shows the residues involved in the interaction of CYP51 from L. infantum with the co-crystallized ligand fluconazole. Additionally, a heme cofactor is observed, which directly interacts with fluconazole.
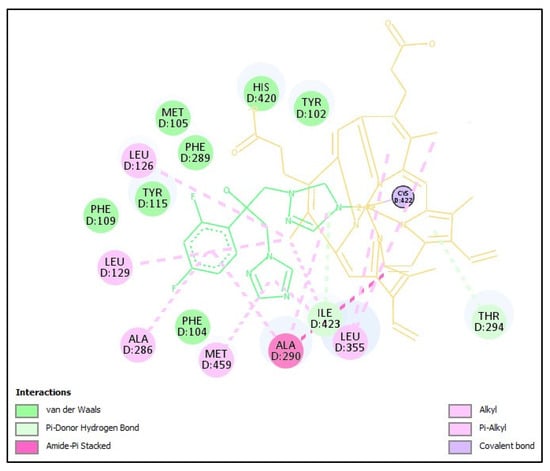
Figure 1.
Interaction of CYP51 from L. infantum with the crystallized ligand fluconazole. Fluconazole (in green) interacts mainly with the heme group (yellow) and protein residues such as ALA290, LEU355, ALA286, MET357, PHE289, TYR102, MET105, PHE109, THR294, and TYR115. Figure generated with Discovery Studio Visualizer v21.1.0.20298.
The binding cavity of CYP51 is largely shaped by residues unique to its phylum [7]. Interestingly, L. infantum CYP51 features a plant-specific phenylalanine at position 104, which alters its substrate preference. Despite this difference, the rest of the amino acids in the binding cavity are conserved as in other CYP51 enzymes from trypanosomatids, with the majority being nonpolar amino acids such as methionine, leucine, and alanine [9]. Inhibitors can fit into this space by conforming to the structure of the binding site, effectively locking the enzyme in an inactive, substrate-free state. Therefore, the greater the ability of an inhibitor to stabilize this inactive form, the stronger its inhibitory effect is expected to be [7,9].
Azole-based inhibitors, such as fluconazole, have limited potency against L. infantum CYP51. Although these inhibitors can displace the substrate in the enzyme’s active site, a high molar excess is often required to achieve inhibition [7,9]. Furthermore, azoles do not completely block sterol biosynthesis, resulting in the accumulation of intermediate sterols that can still function as partial membrane components [7]. All of the above suggests that new strategies targeting specific aspects of sterol biosynthesis are needed, with the potential to discover stronger inhibitors than azoles.
Table 1 shows the 16 NMEs whose free binding energy was higher than the co-crystallized ligand fluconazole. The scoring functions of FRED software do not use partial charges, giving rise to a direct comparison between the affinity energies obtained for the conformers of each molecule [13,15].

Table 1.
FRED Chemgauss4 score and interactions with CYP51 for each NME found better than fluconazole.
The molecular docking results reveal compounds such as drospirenone, brexanolone, ganaxolone, and zuranolone, which have tetracyclic structures similar to the sterols substrates of CYP51, like eburicol, lanosterol, and obtusifoliol [9]. However, these molecules also exhibit additional properties, including anxiolytic, anticonvulsant, and sedative effects [19,20,21]. Drospirenone is frequently used in oral contraceptives in combination with another estrogen identified in the virtual screening performed, estetrol [22].
Figure 2a shows the overlay of co-crystallized fluconazole (PDB: 3L4D) and fluconazole after molecular docking (highlighted in green), with the same orientation towards the heme group and a RMSD of 1.241. This result can be taken as a good indicator of the molecular docking protocol applied. It should be noted that the heme group interacts with all the best NMEs found in the virtual screening performed as shown in Table 1 and Figure 2b. Unlike fluconazole, the identified NMEs could permanently keep the enzyme in a substrate-free state. The therapeutic potential of these compounds can be determined through further evaluation in cellular and animal models.
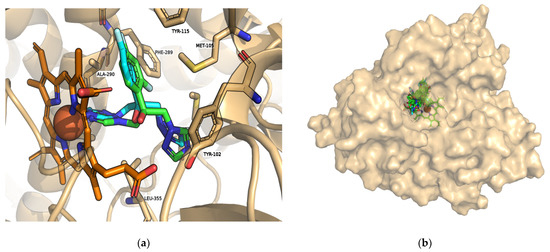
Figure 2.
Interactions with the heme group and substrate binding cavity of L. infantum CYP51. (a) Overlap of the crystalline fluconazole structure (cyan) and the pose obtained through molecular docking (highlighted in green); (b) fluconazole and all NMEs interact with the heme group (orange) in the same location. Figures generated with PyMOL v2.5.0.
It is important to highlight the high ranking of perfluorohexyloctane in the virtual screening performed. This molecule appears to be safe when used as an excipient in drug delivery emulsions, and its surfactant properties make it a promising candidate for enhancing the solubility of lipophilic drugs [23,24]. Additionally, its potential impact on L. infantum CYP51 could facilitate synergistic therapies. By using perfluorohexyloctane as an emulsifier for a leishmanicidal drug, it may be possible to combine the effects of both the emulsifier and the drug, targeting CYP51 alone or addressing multiple targets that include this enzyme.
Author Contributions
Conceptualization, J.D.G.; methodology, J.D.G.; software, J.D.G. and M.L.; validation, J.D.G. and M.L.; formal analysis, M.L.; investigation, J.D.G.; data curation, J.D.G.; writing—original draft preparation, J.D.G.; writing—review and editing, M.L.; visualization, J.D.G.; supervision, M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef] [PubMed]
- Leite, J.C.; Gonçalves, A.A.M.; de Oliveira, D.S.; Resende, L.A.; Boas, D.F.V.; Ribeiro, H.S.; Pereira, D.F.; Da Silva, A.V.; Mariano, R.M.; Reis, P.C.; et al. Transmission-Blocking Vaccines for Canine Visceral Leishmaniasis: New Progress and Yet New Challenges. Vaccines 2023, 11, 1565. [Google Scholar] [CrossRef] [PubMed]
- Organización Panamericana de la Salud. Available online: https://iris.paho.org/handle/10665.2/59156 (accessed on 12 August 2024).
- Shmueli, M.; Ben-Shimol, S. Review of Leishmaniasis Treatment: Can We See the Forest through the Trees? Pharmacy 2024, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Khurana, G.; Rohilla, A.; Deep, A. Drug Development Process and Novel Drugs Approved by FDA for 2017–18. Appl. Clin. Res. Clin. Trials Regul. Aff. 2018, 5, 80–98. [Google Scholar] [CrossRef]
- Novel Drug Approvals at FDA. Available online: https://www.fda.gov/drugs/development-approval-process-drugs/novel-drug-approvals-fda (accessed on 24 May 2024).
- Lepesheva, G.I.; Friggeri, L.; Waterman, M.R. CYP51 as Drug Targets for Fungi and Protozoan Parasites: Past, Present and Future. Parasitology 2018, 145, 1820–1836. [Google Scholar] [CrossRef] [PubMed]
- Van Rhijn, N.; Bromley, M.; Richardson, M.; Bowyer, P. CYP51 Paralogue Structure Is Associated with Intrinsic Azole Resistance in Fungi. MBio 2021, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, T.Y.; Wawrzak, Z.; Liu, J.; Nes, W.D.; Waterman, M.R.; Lepesheva, G.I. Substrate Preferences and Catalytic Parameters Determined by Structural Characteristics of Sterol 14α-Demethylase (CYP51) from Leishmania infantum. J. Biol. Chem. 2011, 286, 26838–26848. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High-Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- McGann, M. FRED and HYBRID Docking Performance on Standardized Datasets. J. Comput. Aided Mol. Des. 2012, 26, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Kelley, B.P.; Brown, S.P.; Warren, G.L.; Muchmore, S.W. POSIT: Flexible Shape-Guided Docking for Pose Prediction. J. Chem. Inf. Model. 2015, 55, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Guarimata, J.D.; Alcívar, C.; Lavecchia, M.; Poveda, A. Molecular Docking for the Development of Alternative Therapies against Leishmaniasis. Chem. Proc. 2023, 14, 82. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.5.0 Schrödinger, LLC. Available online: https://storage.googleapis.com/pymol-storage/installers/index.html (accessed on 24 May 2024).
- BIOVIA Discovery Studio-Dassault Systèmes. Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (accessed on 20 June 2024).
- Riebel, M.; Brunner, L.-M.; Nothdurfter, C.; Wein, S.; Schwarzbach, J.; Liere, P.; Schumacher, M.; Rupprecht, R. Neurosteroids and translocator protein 18 kDa (TSPO) ligands as novel treatment options in depression. In European Archives of Psychiatry and Clinical Neuroscience; Springer: Berlin/Heidelberg, Germany, 2024; Volume 2024, pp. 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Xu, H.; Ye, L.; Li, C.; Zhu, H.; Jiang, W.; Wang, W.; Yang, H.; Yang, Y.; Wang, Y.; et al. Synthesis and evaluation of neuroactive steroids with novel pharmacophore at C-21 let identify a compound with advantageous PK profile and higher EC(50) and E(max) as PAM on GABAA receptor. Eur. J. Med. Chem. 2024, 276, 116602. [Google Scholar] [CrossRef] [PubMed]
- Battipaglia, C.; Feliciello, L.; Genazzani, A.D.; Facchinetti, F.; Grandi, G. Combined Oral Contraceptive with Estetrol Plus Drospirenone: From Pharmacokinetics to Clinical Applications. Expert Opin. Drug Metab. Toxicol. 2023, 19, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Gérard, C.; Foidart, J.M. Estetrol: From Preclinical to Clinical Pharmacology and Advances in the Understanding of the Molecular Mechanism of Action. Drugs R D 2023, 23, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Tsagogiorgas, C.; Anger, F.; Beck, G.; Breedijk, A.; Yard, B.; Hoeger, S. Impact of Different Emulsifiers on Biocompatibility and Inflammatory Potential of Perfluorohexyloctane (F6H8) Emulsions for New Intravenous Drug Delivery Systems. Drug Des. Dev. Ther. 2019, 13, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Habbe, K.J.; Frings, A.; Saad, A.; Geerling, G. The Influence of a Mineral Oil Cationic Nanoemulsion or Perfluorohexyloctane on the Tear Film Lipid Layer and Higher Order Aberrations. PLoS ONE 2023, 18, e0279977. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).