In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus †
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Selection and Sequence Retrieval
2.2. Physicochemical Characterization of the Selected Protein
2.3. Functional Annotation of the Selected Protein
2.4. Secondary Structural Properties and Assessment
2.5. Three-Dimensional Structure Prediction and Validation of the Selected Protein
3. Results and Discussion
3.1. Protein Sequence Retrieval
3.2. Identification of the Physicochemical Properties of the Protein
3.3. Functional Annotation Anticipation of the Selected Protein
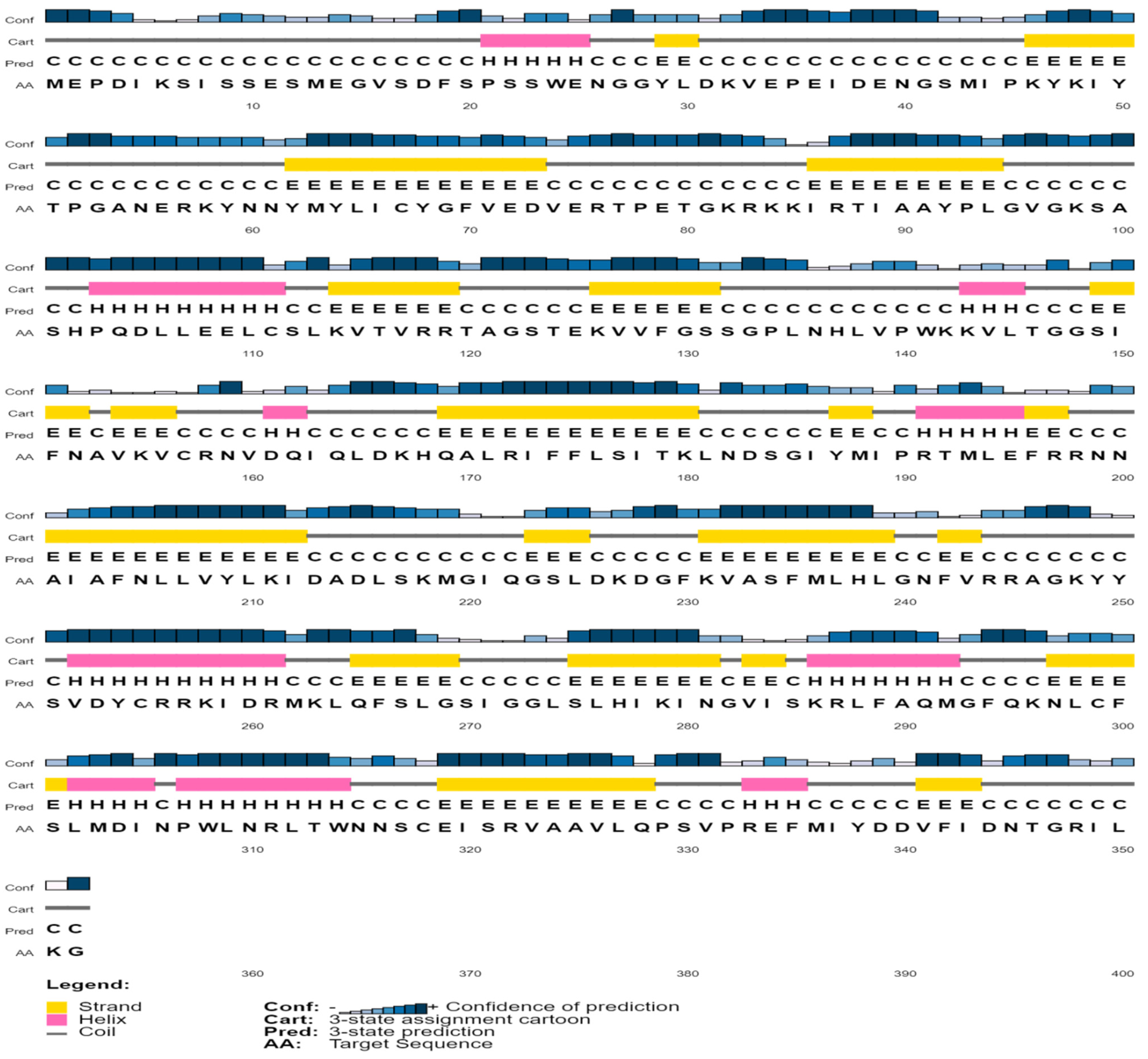
3.4. Secondary Structural Inquiry
3.5. Tertiary-Structure Anticipation and Validation of the Protein
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ang, B.S.P.; Lim, T.C.C.; Wang, L. Nipah Virus Infection. J. Clin. Microbiol. 2018, 56, 1–10. [Google Scholar] [CrossRef]
- Paul, L. Nipah virus in Kerala: A deadly Zoonosis. Clin. Microbiol. Infect. 2018, 24, 1113–1114. [Google Scholar] [CrossRef] [PubMed]
- Aditi; Shariff, M. Nipah virus infection: A review. Epidemiol. Infect. 2019, 147, e95. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Kaushik, S.; Kumar, R.; Yadav, J.P.; Kaushik, S. Emerging trends of Nipah virus: A review. Rev. Med. Virol. 2019, 29, e2010. [Google Scholar] [CrossRef] [PubMed]
- Soman Pillai, V.; Krishna, G.; Veettil, M.V. Nipah Virus: Past Outbreaks and Future Containment. Viruses 2020, 12, 465. [Google Scholar] [CrossRef]
- Lo, M.K.; Rota, P.A. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J. Clin. Virol. 2008, 43, 396–400. [Google Scholar] [CrossRef]
- Ternhag, A.; Penttinen, P. Nipah virus--another product from the Asian “virus factory”. Lakartidningen 2005, 102, 1046–1047. [Google Scholar]
- Choi, C. Nipah’s return. The lethal “flying fox” virus may spread between people. Sci. Am. 2004, 291, 21A–22A. [Google Scholar] [CrossRef]
- Singh, R.K.; Dhama, K.; Chakraborty, S.; Tiwari, R.; Natesan, S.; Khandia, R.; Munjal, A.; Vora, K.S.; Latheef, S.K.; Karthik, K.; et al. Nipah virus: Epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—A comprehensive review. Vet. Q. 2019, 39, 26–55. [Google Scholar] [CrossRef]
- Epstein, J.H.; Anthony, S.J.; Islam, A.; Kilpatrick, A.M.; Ali Khan, S.; Balkey, M.D.; Ross, N.; Smith, I.; Zambrana-Torrelio, C.; Tao, Y.; et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. USA 2020, 117, 29190–29201. [Google Scholar] [CrossRef]
- Yadav, P.D.; Shete, A.M.; Kumar, G.A.; Sarkale, P.; Sahay, R.R.; Radhakrishnan, C.; Lakra, R.; Pardeshi, P.; Gupta, N.; Gangakhedkar, R.R.; et al. Nipah Virus Sequences from Humans and Bats during Nipah Outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 2019, 25, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Sudeep, A.B.; Yadav, P.D.; Gokhale, M.D.; Balasubramanian, R.; Gupta, N.; Shete, A.; Jain, R.; Patil, S.; Sahay, R.R.; Nyayanit, D.A.; et al. Detection of Nipah virus in Pteropus medius in 2019 outbreak from Ernakulam district, Kerala, India. BMC Infect. Dis. 2021, 21, 162. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.D.; Raut, C.G.; Shete, A.M.; Mishra, A.C.; Towner, J.S.; Nichol, S.T.; Mourya, D.T. (Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am. J. Trop. Med. Hyg. 2012, 87, 576–578. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Aurine, N.; Dumont, C.; Fouret, J.; Ferren, M.; Mathieu, C.; Reynard, O.; Volchkov, V.E.; Legras-Lachuer, C.; Georges-Courbot, M.C.; et al. High Pathogenicity of Nipah Virus from Pteropus lylei Fruit Bats, Cambodia. Emerg. Infect. Dis. 2020, 26, 104–113. [Google Scholar] [CrossRef]
- Rathish, B.; Vaishnani, K. Nipah Virus. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Looi, L.M.; Chua, K.B. Lessons from the Nipah virus outbreak in Malaysia. Malays. J. Pathol. 2007, 29, 63–67. [Google Scholar]
- Lam, S.K.; Chua, K.B. Nipah virus encephalitis outbreak in Malaysia. Clin. Infect. Dis. 2002, 34 (Suppl. 2), S48–S51. [Google Scholar] [CrossRef]
- Singhai, M.; Jain, R.; Jain, S.; Bala, M.; Singh, S.; Goyal, R. Nipah Virus Disease: Recent Perspective and One Health Approach. Ann. Glob. Health 2021, 87, 102. [Google Scholar] [CrossRef]
- Gómez Román, R.; Wang, L.F.; Lee, B.; Halpin, K.; de Wit, E.; Broder, C.C.; Rahman, M.; Kristiansen, P.; Saville, M. Nipah@20: Lessons Learned from Another Virus with Pandemic Potential. mSphere 2020, 5, e00602-20. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.; Cerutti, L.; Hulo, N.; Gattiker, A.; Falquet, L.; Pagni, M.; Bairoch, A.; Bucher, P. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 2002, 3, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Madera, M.; Vogel, C.; Chothia, C.; Gough, J. The SUPERFAMILY database in 2007: Families and functions. Nucleic Acids Res. 2007, 35, D308–D313. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Deléage, G. ALIGNSEC: Viewing protein secondary structure predictions within large multiple sequence alignments. Bioinformatics 2017, 33, 3991–3992. [Google Scholar] [CrossRef]
- Moffat, L.; Jones, D.T. Increasing the accuracy of single sequence prediction methods using a deep semi-supervised learning framework. Bioinformatics 2021, 37, 3744–3751. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Gamage, D.G.; Gunaratne, A.; Periyannan, G.R.; Russell, T.G. Applicability of Instability Index for In vitro Protein Stability Prediction. Protein Pept. Lett. 2019, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Pihlasalo, S.; Auranen, L.; Hänninen, P.; Härmä, H. Method for estimation of protein isoelectric point. Anal. Chem. 2012, 84, 8253–8258. [Google Scholar] [CrossRef]
- Audain, E.; Ramos, Y.; Hermjakob, H.; Flower, D.R.; Perez-Riverol, Y. Accurate estimation of isoelectric point of protein and peptide based on amino acid sequences. Bioinformatics 2016, 32, 821–827. [Google Scholar] [CrossRef]
- Saikat, A.S.M. An In Silico Approach for Potential Natural Compounds as Inhibitors of Protein CDK1/Cks2. Chem. Proc. 2022, 8, 5. [Google Scholar]
- Wilkins, M.R.; Williams, K.L. Cross-species protein identification using amino acid composition, peptide mass fingerprinting, isoelectric point and molecular mass: A theoretical evaluation. J. Theor. Biol. 1997, 186, 7–15. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Khan, R.A.; Hossain, R.; Siyadatpanah, A.; Al-Khafaji, K.; Khalipha, A.B.R.; Dey, D.; Asha, U.H.; Biswas, P.; Saikat, A.S.M.; Chenari, H.A.; et al. Diterpenes/Diterpenoids and Their Derivatives as Potential Bioactive Leads against Dengue Virus: A Computational and Network Pharmacology Study. Molecules 2021, 26, 6821. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Dey, D.; Biswas, P.; Paul, P.; Mahmud, S.; Ema, T.I.; Khan, A.A.; Ahmed, S.Z.; Hasan, M.M.; Saikat, A.S.M.; Fatema, B.; et al. Natural flavonoids effectively block the CD81 receptor of hepatocytes and inhibit HCV infection: A computational drug development approach. Mol. Divers. 2022. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.T.; Jin, T.Y.; Zhang, Z.L.; Ye, Y.N.; Deng, Z.; Wang, J.; Guo, F.B. Quantitative elucidation of associations between nucleotide identity and physicochemical properties of amino acids and the functional insight. Comput. Struct. Biotechnol. J. 2021, 19, 4042–4048. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M.; Islam, R.; Mahmud, S.; Imran, M.A.S.; Alam, M.S.; Masud, M.H.; Uddin, M.E. Structural and Functional Annotation of Uncharacterized Protein NCGM946K2_146 of Mycobacterium Tuberculosis: An In-Silico Approach. Proceedings 2020, 66, 13. [Google Scholar]
- Saikat, A.S.M.; Uddin, M.E.; Ahmad, T.; Mahmud, S.; Imran, M.A.S.; Ahmed, S.; Alyami, S.A.; Moni, M.A. Structural and Functional Elucidation of IF-3 Protein of Chloroflexus aurantiacus Involved in Protein Biosynthesis: An In Silico Approach. BioMed Res. Int. 2021, 2021, 9050026. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.J.; Meng, G.; Winkler, D.C.; McGinnes, L.W.; Plevka, P.; Steven, A.C.; Morrison, T.G.; Rossmann, M.G. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. USA 2012, 109, 13996–14000. [Google Scholar] [CrossRef]
- Shtykova, E.V.; Petoukhov, M.V.; Dadinova, L.A.; Fedorova, N.V.; Tashkin, V.Y.; Timofeeva, T.A.; Ksenofontov, A.L.; Loshkarev, N.A.; Baratova, L.A.; Jeffries, C.M.; et al. Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH. J. Virol. 2019, 93, e01450-18. [Google Scholar] [CrossRef]
- Stollar, E.J.; Smith, D.P. Uncovering protein structure. Essays Biochem. 2020, 64, 649–680. [Google Scholar] [CrossRef]
- Heizinger, L.; Merkl, R. Evidence for the preferential reuse of sub-domain motifs in primordial protein folds. Proteins 2021, 89, 1167–1179. [Google Scholar] [CrossRef]
- Xie, J.; Lai, L. Protein topology and allostery. Curr. Opin. Struct. Biol. 2020, 62, 158–165. [Google Scholar] [CrossRef]
- Santhouse, J.R.; Rao, S.R.; Horne, W.S. Analysis of folded structure and folding thermodynamics in heterogeneous-backbone proteomimetics. Methods Enzymol. 2021, 656, 93–122. [Google Scholar] [PubMed]
- Vishwanath, S.; de Brevern, A.G.; Srinivasan, N. Same but not alike: Structure, flexibility and energetics of domains in multi-domain proteins are influenced by the presence of other domains. PLoS Comput. Biol. 2018, 14, e1006008. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, I.N.; Guarnera, E.; Zheng, Z. Basic units of protein structure, folding, and function. Prog. Biophys. Mol. Biol. 2017, 128, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Padjasek, M.; Kocyła, A.; Kluska, K.; Kerber, O.; Tran, J.B.; Krężel, A. Structural zinc binding sites shaped for greater works: Structure-function relations in classical zinc finger, hook and clasp domains. J. Inorg. Biochem. 2020, 204, 110955. [Google Scholar] [CrossRef]
- Zhang, G.J.; Ma, L.F.; Wang, X.Q.; Zhou, X.G. Secondary Structure and Contact Guided Differential Evolution for Protein Structure Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, D.; van Dijk, J.; Titulaer, W.; Lange, J.; Vriend, G.; Xue, L. The Future of Protein Secondary Structure Prediction Was Invented by Oleg Ptitsyn. Biomolecules 2020, 10, 910. [Google Scholar] [CrossRef]
- Wardah, W.; Khan, M.G.M.; Sharma, A.; Rashid, M.A. Protein secondary structure prediction using neural networks and deep learning: A review. Comput. Biol. Chem. 2019, 81, 1–8. [Google Scholar] [CrossRef]

| Protein Individualities | Protein Information |
|---|---|
| Locus | QBQ56721 |
| Amino acid | 352 aa |
| Definition | matrix protein [Nipah henipavirus] |
| Accession | QBQ56721 |
| Version | QBQ56721.1 |
| Source | Nipah henipavirus |
| Organism | Nipah henipavirus |
| FASTA sequence | >QBQ56721.1 matrix protein [Nipah henipavirus] MEPDIKSISSESMEGVSDFSPSSWENGGYLDKVEPEIDENGSMIPKYKIYTPGANERKYNNYMYLICYGF VEDVERTPETGKRKKIRTIAAYPLGVGKSASHPQDLLEELCSLKVTVRRTAGSTEKVVFGSSGPLNHLVP WKKVLTGGSIFNAVKVCRNVDQIQLDKHQALRIFFLSITKLNDSGIYMIPRTMLEFRRNNAIAFNLLVYL KIDADLSKMGIQGSLDKDGFKVASFMLHLGNFVRRAGKYYSVDYCRRKIDRMKLQFSLGSIGGLSLHIKI NGVISKRLFAQMGFQKNLCFSLMDINPWLNRLTWNNSCEISRVAAVLQPSVPREFMIYDDVFIDNTGRIL KG |
| Parameters | Value |
|---|---|
| Molecular weight | 39,847.16 |
| Theoretical pI | 9.31, 9.65 * |
| Total number of negatively charged residues (Asp + Glu) | 36 |
| Total number of positively charged residues (Arg + Lys) | 48 |
| Formula | C1787H2831N485O510S18 |
| Total number of atoms | 5631 |
| The estimated half-life | (a) 30 h (mammalian reticulocytes, in vitro). (b) >20 h (yeast, in vivo). (c) >10 h (Escherichia coli, in vivo). |
| Instability index (II) | 30.59 |
| Aliphatic index | 89.69 |
| Grand average of hydropathicity (GRAVY) | −0.212 |
| Amino Acids | Percentage (%) |
|---|---|
| Ala (A) | 5.2% |
| Arg (R) | 2.1% |
| Asn (N) | 9.4% |
| Asp (D) | 5.7% |
| Cys (C) | 0.2% |
| Gln (Q) | 5.2% |
| Glu (E) | 8.5% |
| Gly (G) | 4.8% |
| His (H) | 1.3% |
| Ile (I) | 6.3% |
| Leu (L) | 9.2% |
| Lys (K) | 9.1% |
| Met (M) | 2.0% |
| Phe (F) | 4.0% |
| Pro (P) | 3.0% |
| Ser (S) | 6.9% |
| Thr (T) | 6.9% |
| Trp (W) | 0.6% |
| Tyr (Y) | 3.0% |
| Val (V) | 6.7% |
| Secondary Structure Elements | Values (%) |
|---|---|
| Alpha helix (Hh) | 60 (17.05) |
| 310 helix (Gg) | 0 |
| Pi helix (Ii) | 0 |
| Beta bridge (Bb) | 0 |
| Extended strand (Ee) | 87 (24.72) |
| Beta turn (Tt) | 20 (5.64) |
| Bend region (Ss) | 0 |
| Random coil (Cc) | 185 (52.56) |
| Ambiguous states | 0 |
| Other states | 0 |
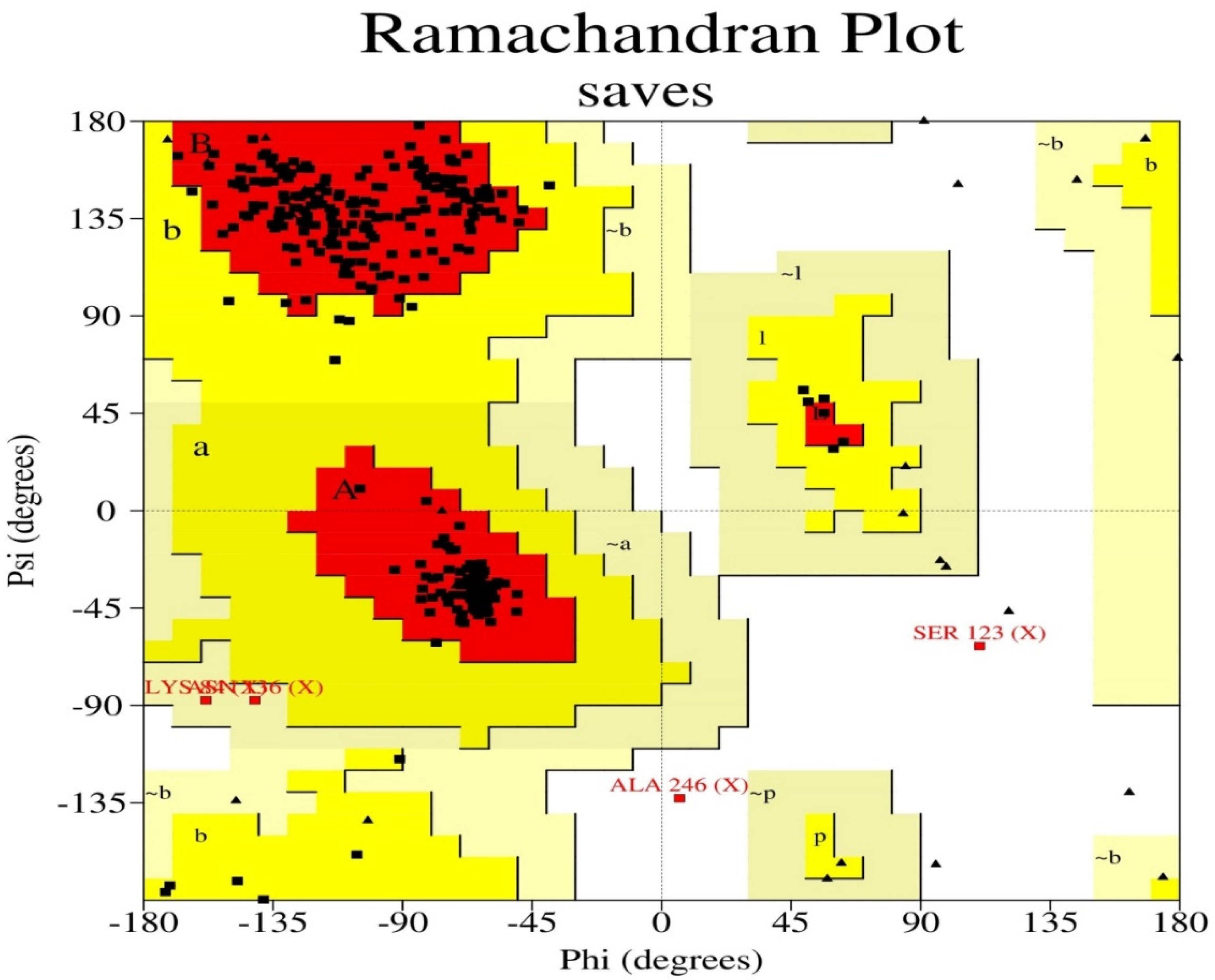
| Ramachandran Plot Statistics | Value (%) |
|---|---|
| Residues in the most favored regions (A, B, L) | 278 (92.4) |
| Residues in additional allowed regions (a, b, l, p) | 19 (6.3) |
| Residues in generously allowed regions (~a, ~b, ~l, ~p) | 2 (0.7) |
| Residues in disallowed regions | 2 (0.7) |
| Number of nonglycine and nonproline residues | 301 |
| Number of end residues (excl. Gly and Pro) | 1 |
| Number of glycine residues (shown as triangles) | 27 |
| Number of proline residues | 13 |
| Total number of residues | 342 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saikat, A.S.M.; Paul, A.K.; Dey, D.; Das, R.C.; Das, M.C. In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus . Chem. Proc. 2022, 12, 21. https://doi.org/10.3390/ecsoc-26-13522
Saikat ASM, Paul AK, Dey D, Das RC, Das MC. In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus . Chemistry Proceedings. 2022; 12(1):21. https://doi.org/10.3390/ecsoc-26-13522
Chicago/Turabian StyleSaikat, Abu Saim Mohammad, Apurbo Kumar Paul, Dipta Dey, Ranjit Chandra Das, and Madhab Chandra Das. 2022. "In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus " Chemistry Proceedings 12, no. 1: 21. https://doi.org/10.3390/ecsoc-26-13522
APA StyleSaikat, A. S. M., Paul, A. K., Dey, D., Das, R. C., & Das, M. C. (2022). In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus . Chemistry Proceedings, 12(1), 21. https://doi.org/10.3390/ecsoc-26-13522








