Machine Learning-Based Classification of Albanian Wines by Grape Variety, Using Phenolic Compound Dataset
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Description and Sample Selection
2.2. Data Cleaning and Preprocessing
2.3. Model Selection and Training
2.4. Validation Strategy and Performance Metrics
2.5. Feature Importance and Interpretation
3. Results and Discussions
3.1. Overview of Dataset and Analytical Scope
3.2. Exploratory Data Analysis
3.3. Benchmarking with International Wines
3.4. Phenolic Fingerprints and Classification Potential
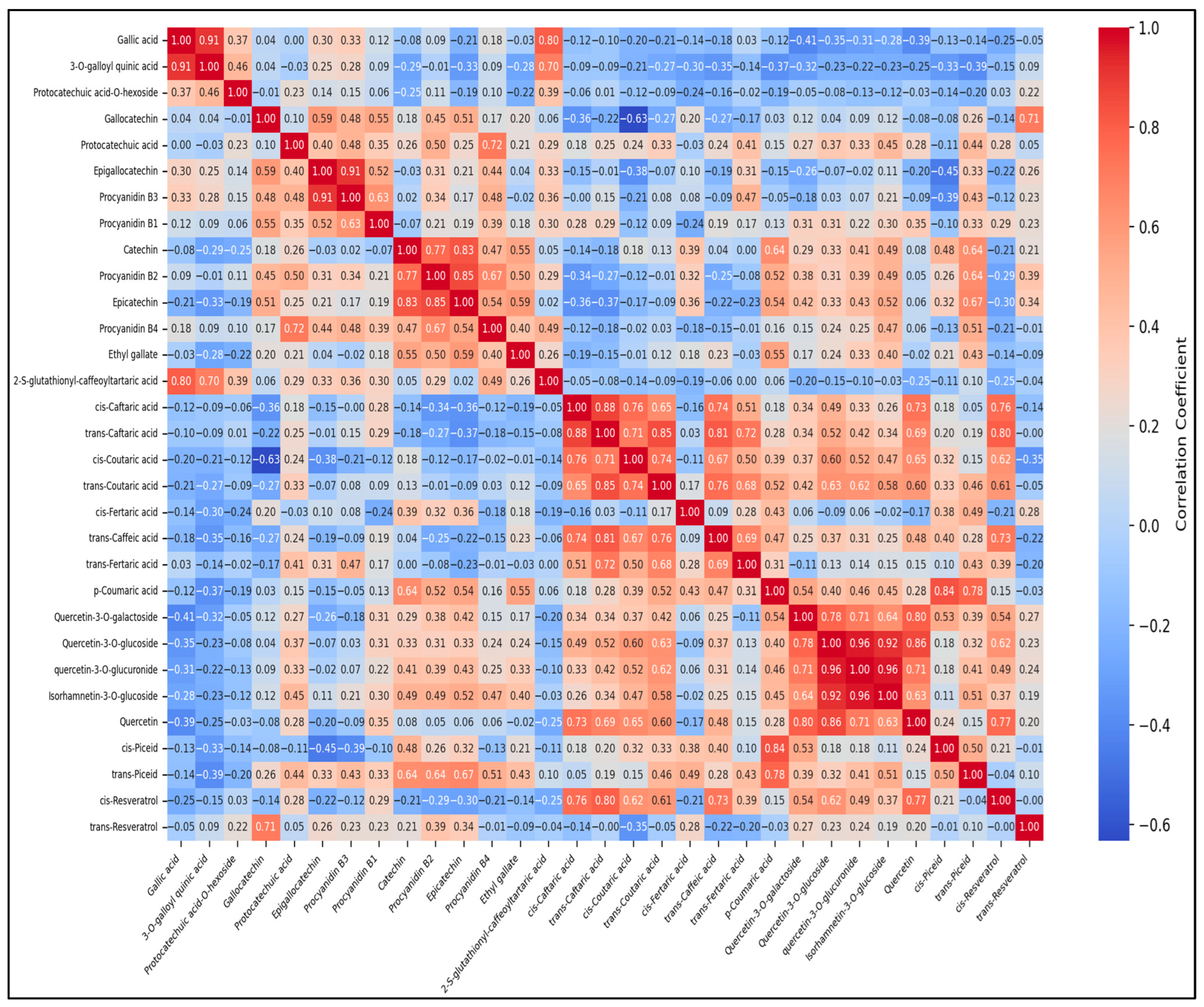
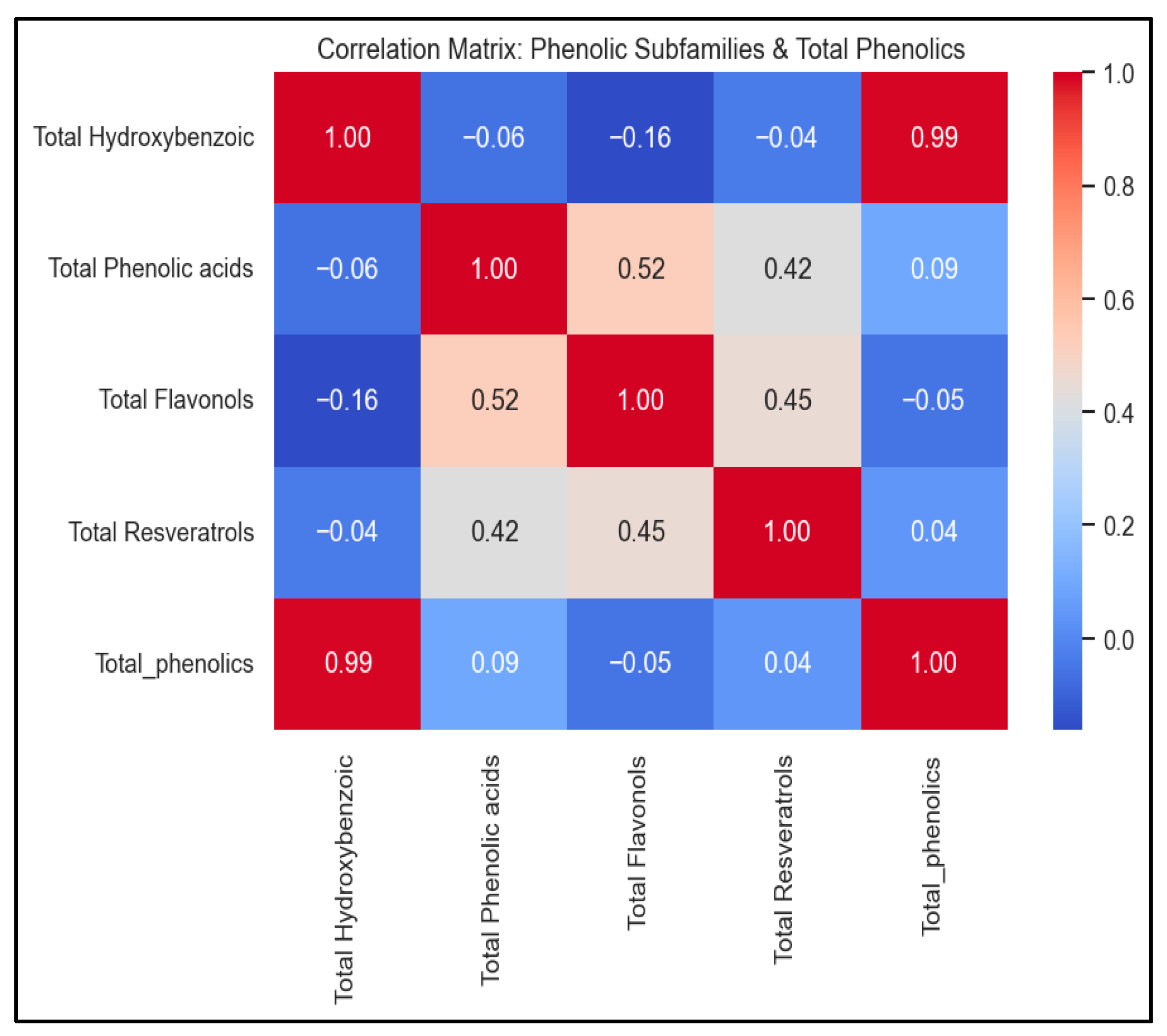
3.5. Correlation Analysis
3.6. Unsupervised Learning for Wine Typing
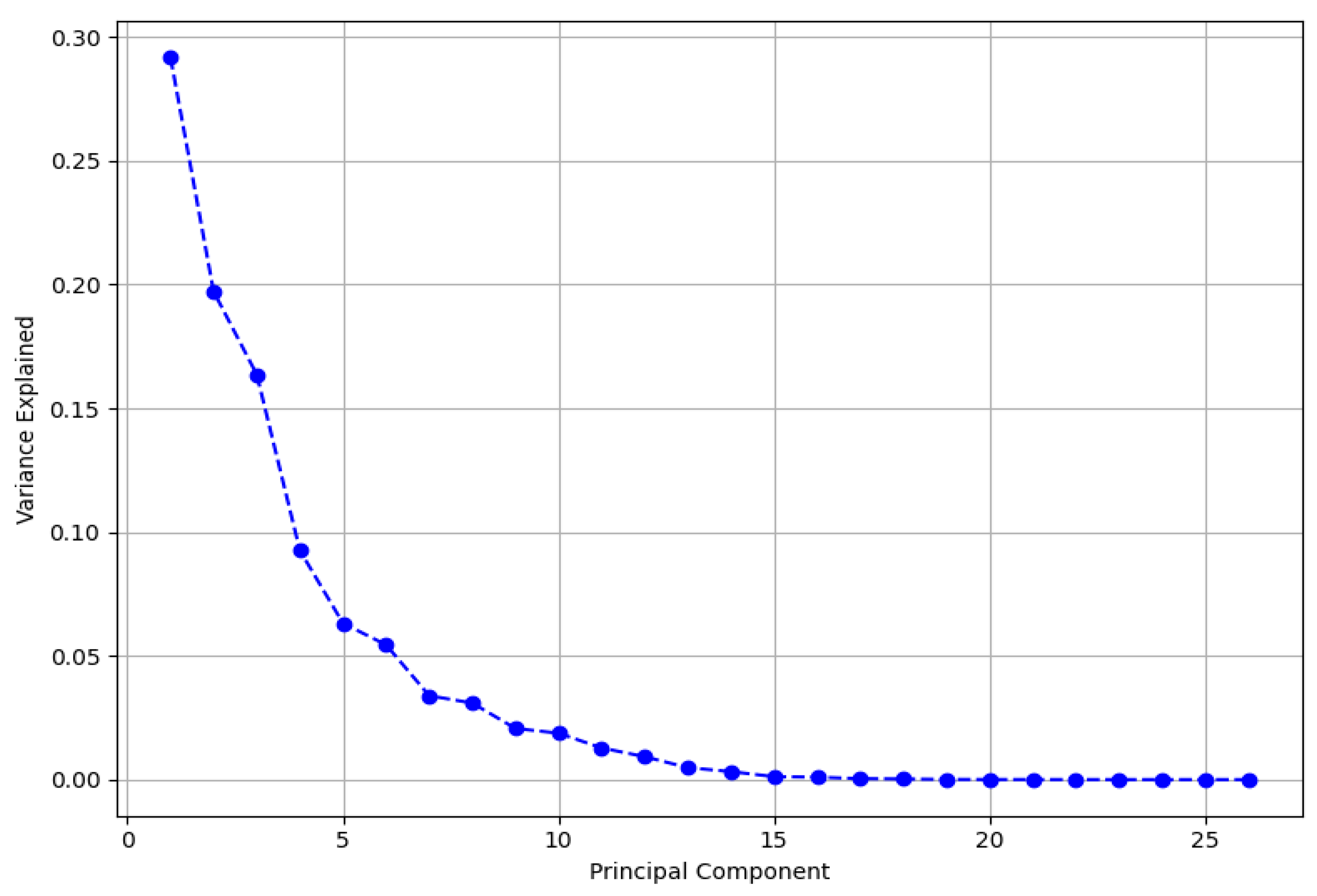
3.6.1. Principal Component Analysis (PCA)
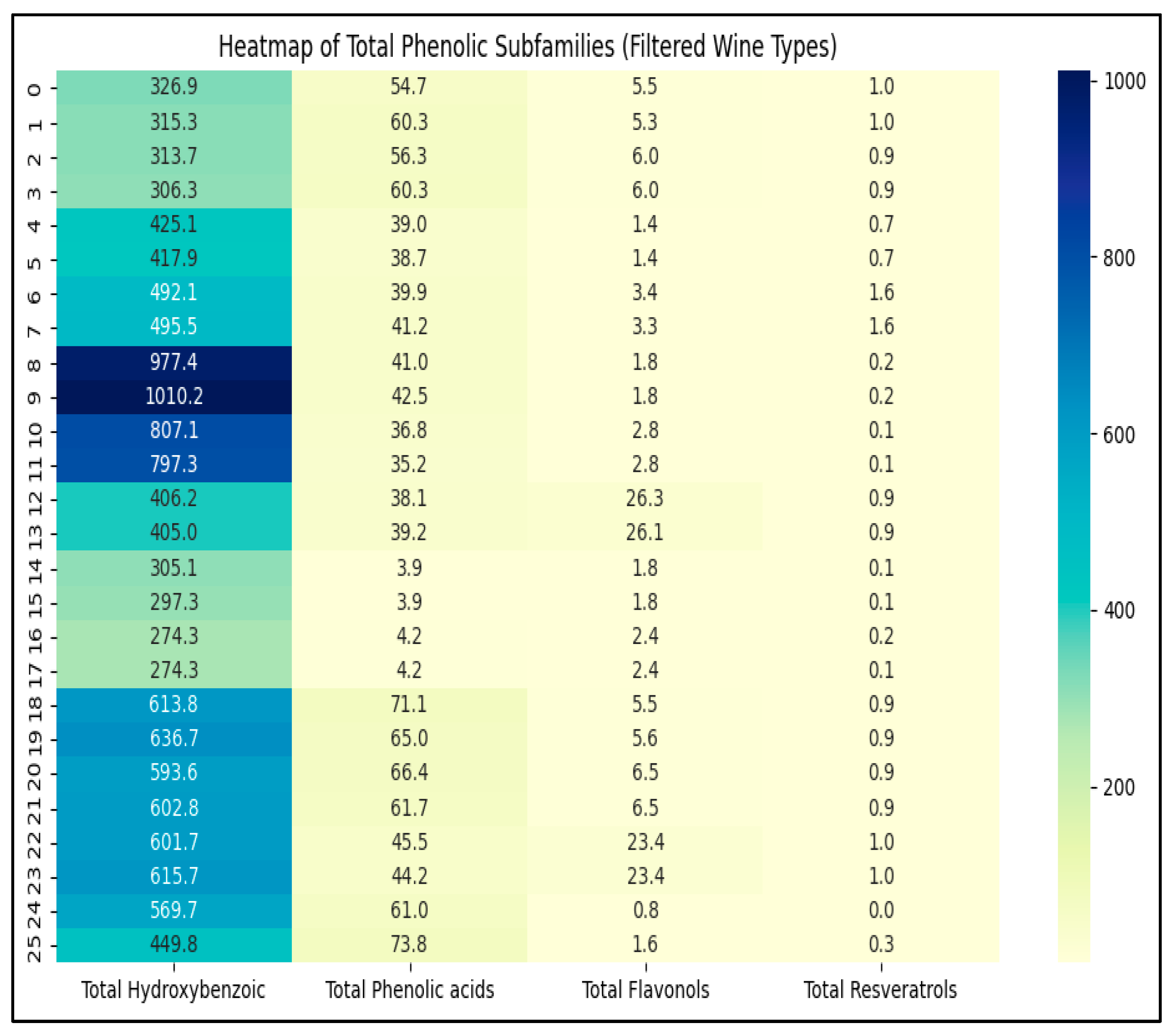
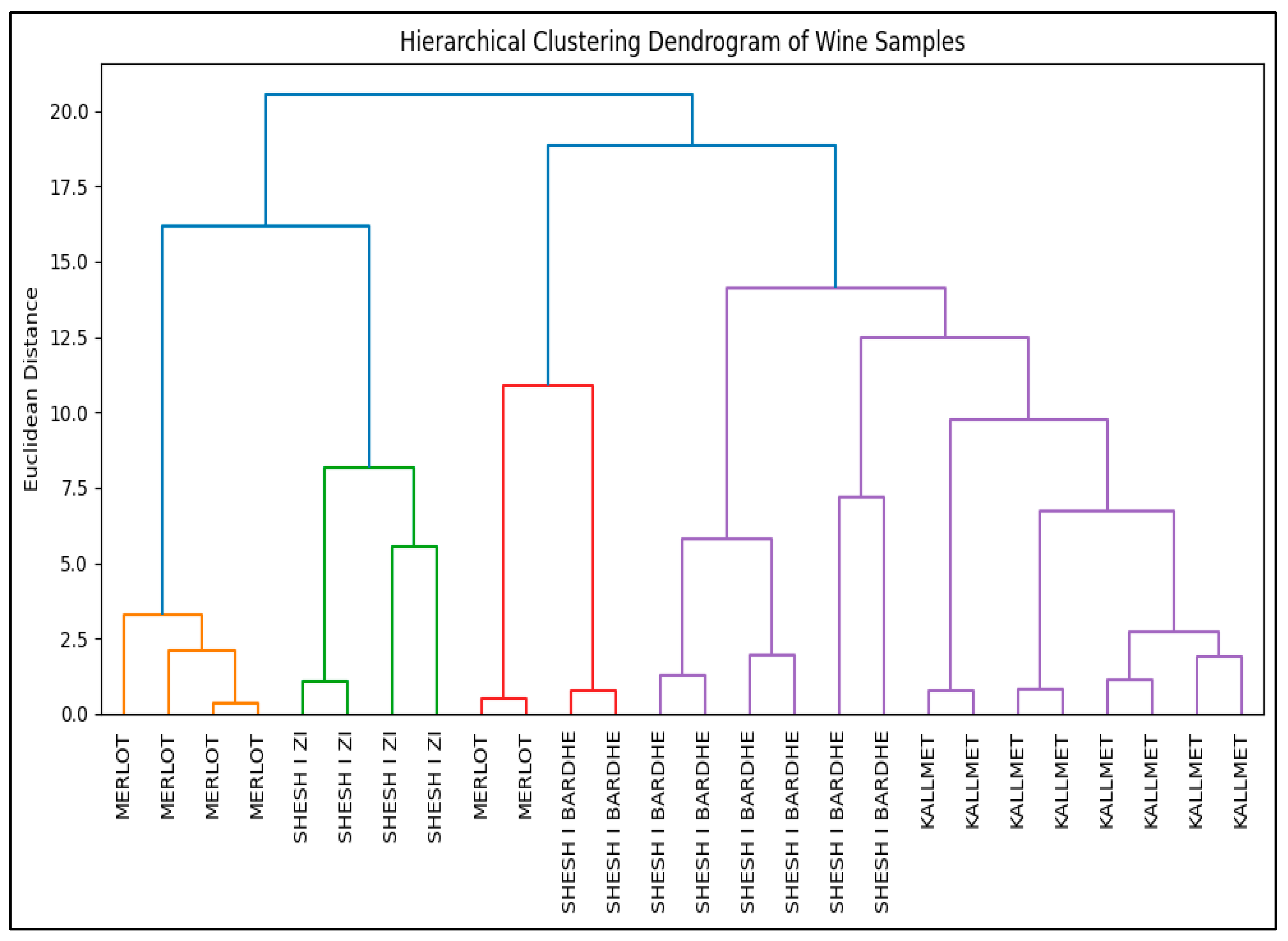
3.6.2. Phenolic Heatmap and Hierarchical Clustering
3.6.3. Interpretation of Compound Groupings
3.7. Supervised Learning Classification of Wine Varieties
3.7.1. Model Setup and Cross-Validation
- Random Forest.
- Support Vector Machine (SVM with RBF kernel).
- K-Nearest Neighbors (KNN).
- Logistic Regression.
- XGBoost.
3.7.2. Performance Metrics and Evaluation
3.7.3. Hyperparameter Optimization
- Random Forest: n_estimators = 50, max_depth = None, min_samples_split = 2-F1-macro = 0.9375.
- SVM: C = 1, kernel = ‘rbf’, gamma = ‘scale’-F1-macro = 1.0000.
- Logistic Regression: C = 0.1, penalty = ‘l2′, solver = ‘lbfgs’-F1-macro = 0.9020.
- KNN: n_neighbors = 7, weights = ‘distance’, metric = ‘manhattan’-F1-macro = 0.9270.
- XGBoost: learning_rate = 0.1, max_depth = 3, n_estimators = 50-F1-macro = 0.8931.
3.7.4. Final LOOCV Results and Permutation Testing
3.8. Model Interpretability and Phenolic Markers
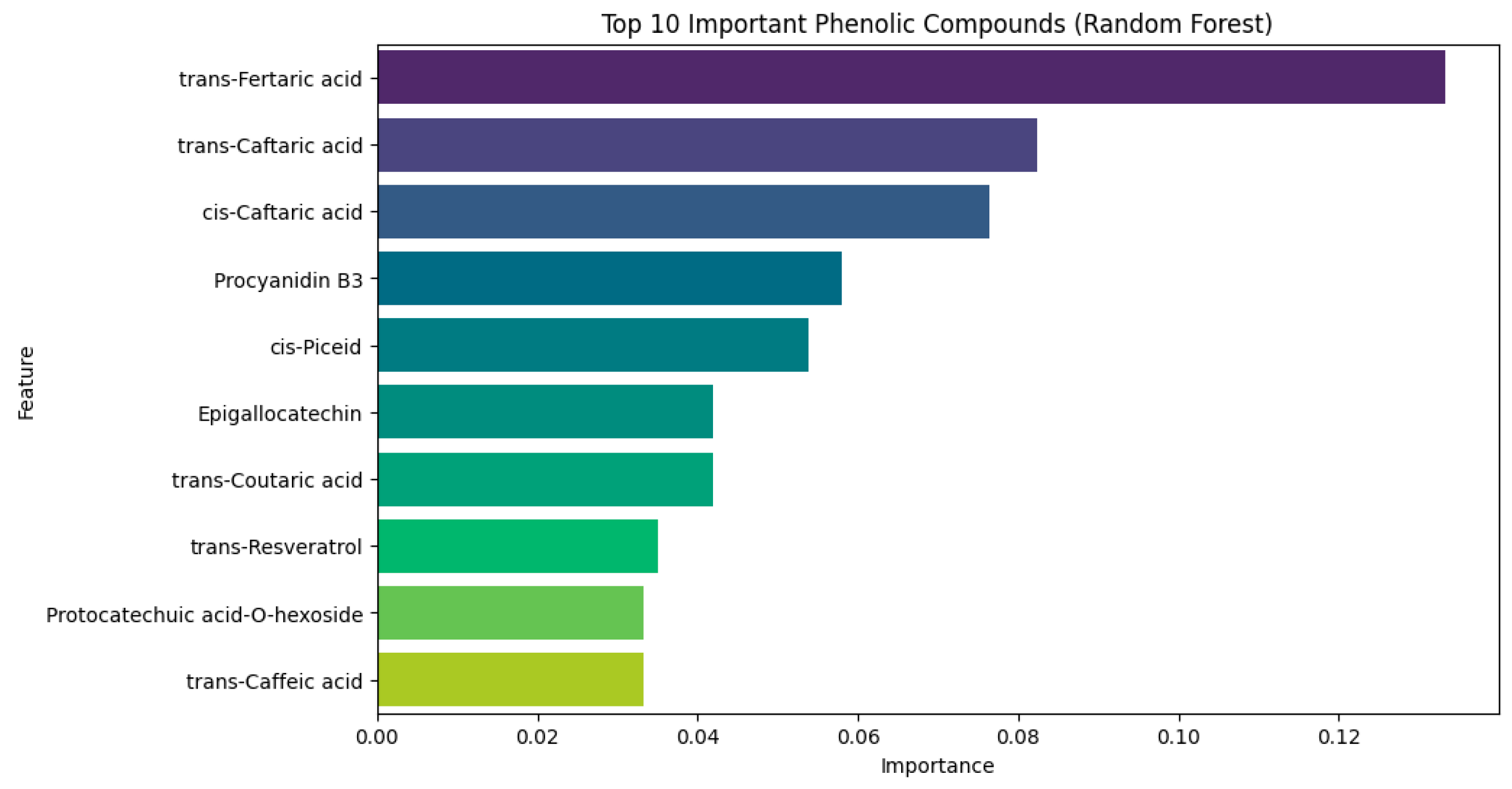
3.8.1. Feature Ranking
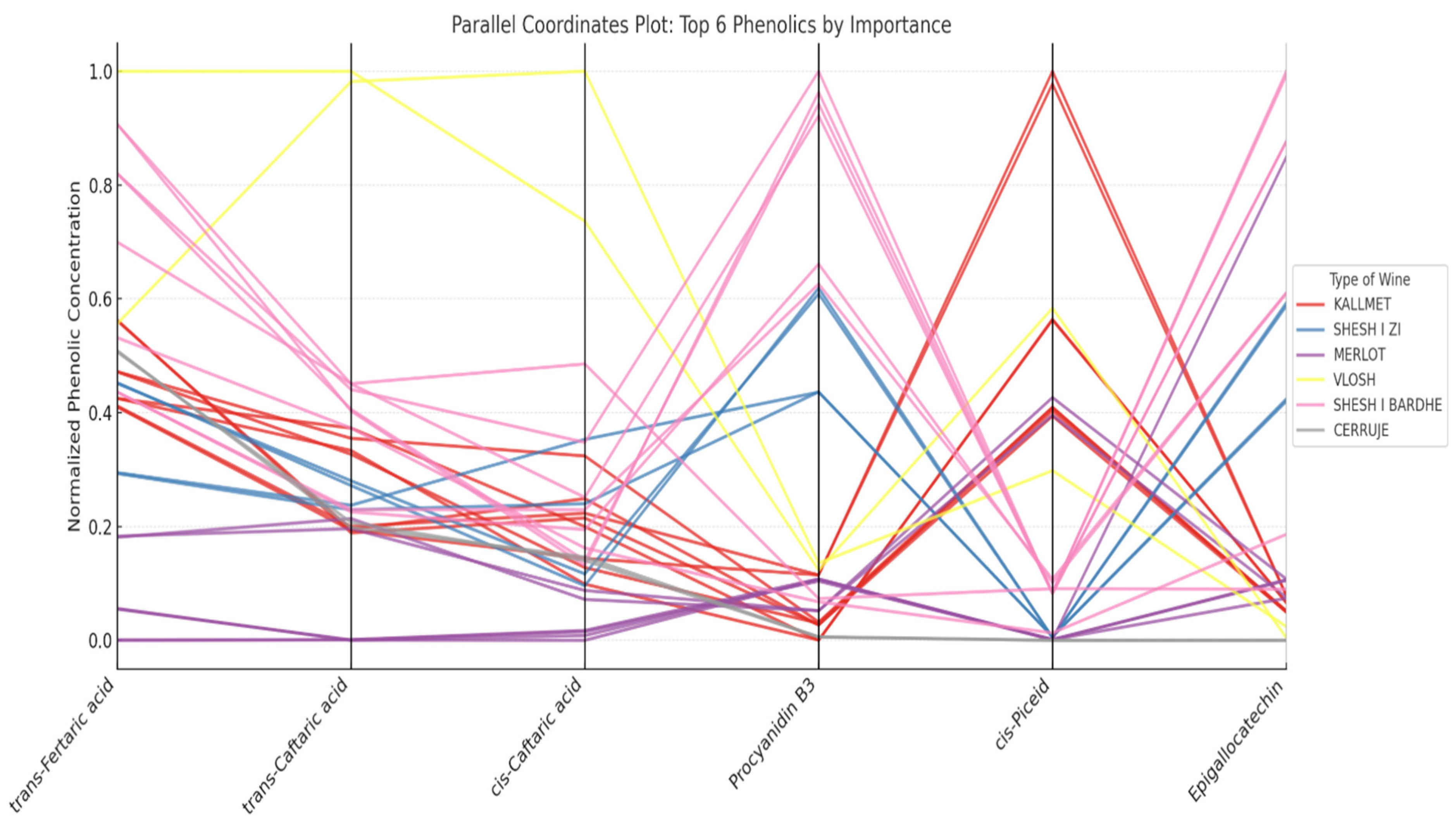
3.8.2. Parallel Coordinates Plot and Chemical Interpretation
3.8.3. Implications for Wine Typicity and Traceability
3.9. Further Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Topi, D.; Guclu, G.; Kelebek, H.; Selli, S. Olive oil production in Albania, chemical characterization, and authenticity. In Olive Oil-New Perspectives and Applications; Akram, M., Ed.; IntechOpen: Rijeka, Croatia, 2021; pp. 77–95. [Google Scholar] [CrossRef]
- Topi, D.; Topi, A.; Guclu, G.; Selli, S.; Uzlasir, T.; Kelebek, H. Targeted analysis for detecting phenolics and authentication of Albanian wines using LC-DAD/ESI–MS/MS combined with chemometric tools. Heliyon 2024, 10, e31127. [Google Scholar] [CrossRef]
- Topi, D.; Arapi, D.; Seiti, B. Vine Pruning Residues and Wine Fermentation By-Products: A Non-Exploited Source of Sustainable Agriculture, Albania Case. Resources 2025, 14, 29. [Google Scholar] [CrossRef]
- Topi, D.; Kelebek, H.; Shehi, G.; Guclu, G.; Selli, S. Phenolic Profiling of Merlot Wines from Albania: Influence of Geographical Origin and Vintage Assessed by LC-DAD-ESI-MS/MS. Analytica 2025, 6, 31. [Google Scholar] [CrossRef]
- Merkytė, V.; Longo, E.; Windisch, G.; Boselli, E. Phenolic Compounds as Markers of Wine Quality and Authenticity. Foods 2020, 9, 1785. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V. Wines. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M., Ed.; Elsevier: Oxord, UK, 2011; pp. 260–274. [Google Scholar] [CrossRef]
- Garrido, J.; Borges, F. Wine and grape polyphenols—A chemical perspective. Food Res. Int. 2013, 54, 1844–1858. [Google Scholar] [CrossRef]
- Kelebek, H.; Canbas, A.; Jourdes, M.; Teissedre, P.-L. HPLC-DAD-MS Determination of Colored and Colorless Phenolic Compounds in Kalecik Karasi Wines: Effect of Different Vineyard Locations. Anal. Lett. 2011, 44, 991–1008. [Google Scholar] [CrossRef]
- Gómez Gallego, M.A.; García-Carpintero, E.G.; Sánchez-Palomo, E.; González Viñas, M.A.; Hermosín-Gutiérrez, I. Evolution of the phenolic content, chromatic characteristics, and sensory properties during bottle storage of red single-cultivar wines from Castilla La Mancha region. Food Res. Int. 2013, 51, 554–563. [Google Scholar] [CrossRef]
- Bavaresco, L.; Lucini, L.; Busconi, M.; Flamini, R.; De Rosso, M. Wine Resveratrol: From the Ground Up. Nutrients. 2016, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Tiziana Lisanti, M.; Gambuti, A.; Vecchio, R.; Moio, L.; Frusciante, L.; Aversano, R.; Carputo, D. Wine varietal authentication based on phenolics, volatiles and DNA markers: State of the art, perspectives and drawbacks. Food Control 2017, 80, 1–10. [Google Scholar] [CrossRef]
- Tzachristas, A.; Pasvanka, K.; Calokerinos, A.; Proestos, C. Polyphenols: Natural antioxidants to be used as a quality tool in wine authenticity. Appl. Sci. 2020, 10, 5908. [Google Scholar] [CrossRef]
- Heras-Roger, J.; Díaz-Romero, C. From Vine to Wine: Coloured Phenolics as Fingerprints. Appl. Sci. 2025, 15, 1755. [Google Scholar] [CrossRef]
- Monagas, M.; Bartolomé, B.; Gómez-Cordovés, C. Updated knowledge about the presence of phenolic compounds in wine. Crit. Rev. Food Sci. Nutr. 2005, 45, 85–118. [Google Scholar] [CrossRef]
- Waterhouse, A.L.; Sacks, G.L.; Jeffery, D.W. Understanding Wine Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2016; p. 560. [Google Scholar]
- Hategan, A.R.; Pirnau, A.; Magdas, D.A. Applications of Machine Learning for Wine Recognition Based on 1H NMR Spectroscopy. Beverages 2025, 11, 45. [Google Scholar] [CrossRef]
- Sarlo, L.; Duroux, C.; Clément, Y.; Lanteri, P.; Rossetti, F.; David, O.; Tillement, A.; Gillet, P.; Hagège, A.; Laurent, D.; et al. Enhancing wine authentication: Leveraging 12,000+ international mineral wine profiles and artificial intelligence for accurate origin and variety prediction. OENO One 2024, 58. [Google Scholar] [CrossRef]
- Zaza, S.; Atemkeng, M.; Hamlomo, S. Wine feature importance and quality prediction: A comparative study of machine learning algorithms with unbalanced data. arXiv 2023, arXiv:2310.01584. [Google Scholar] [CrossRef]
- Aiello, G. An Artificial Intelligence-based tool to predict “unhealthy” wine and olive oil. J. Agric. Food Res. 2024, 16, 101179. [Google Scholar] [CrossRef]
- Llupa, J.; Gašić, U.; Brčeski, I.; Demertzis, P.; Tešević, V.; Topi, D. LC-MS/MS characterization of phenolic compounds in the quince (Cydonia oblonga Mill.) and sweet cherry (Prunus avium L.) fruit juices. Agric. For. 2022, 68, 193–205. [Google Scholar] [CrossRef]
- Topi, D.; Kelebek, H.; Güçlü, G.; Selli, S. LC DAD ESI MS/MS characterization of phenolic compounds in wines from Vitis vinifera’ Shesh i bardhë’ and ‘Vlosh’ cultivars. J. Food Process. Preserv. 2022, 46, e16157. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Topi, A.; Këlliçi, E.; Hudhra, D.; Topi, D. A machine learning-based classification of monocultivar olive oils—Specifically Kalinjot, Ulli i bardhë Tirana, and Mixan—Comparing their chemical composition. Edelweiss Appl. Sci. Technol. 2025, 9, 93–110. [Google Scholar] [CrossRef]
- Topi, D.; Topi, A.; Hudhra, D. Application of machine learning algorithmic models for the authentication of Albanian mono cultivar olive oils. J. Inf. Syst. Eng. Manag. 2025, 10, 486–507. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Tiwari, P.; Olejar, K.; Parr, W.; Kulasiri, D. A machine learning application in wine quality prediction. Mach. Learn. Appl. 2022, 8, 100261. [Google Scholar] [CrossRef]
- Gutiérrez-Escobar, R.; Aliaño-González, M.J.; Cantos-Villar, E. Wine Polyphenol Content and Its Influence on Wine Quality and Properties: A Review. Molecules 2021, 26, 718. [Google Scholar] [CrossRef]
- Piljac, J.; Martinez, S.; Valek, L.; Stipcevic, T.; Maletic, E. Influence of maceration time on the polyphenolic composition and antioxidant capacity of Plavac Mali wine. Food Technol. Biotechnol. 2005, 43, 219–225. [Google Scholar]
- Chira, K.; Pacella, N.; Jourdes, M.; Teissedre, P.-L. Chemical and sensory evaluation of Bordeaux wines (Cabernet-Sauvignon and Merlot) and correlation with wine age. Food Chem. 2011, 126, 1971–1977. [Google Scholar] [CrossRef]
- Branco, Z.; Baptista, F.; Paié Ribeiro, J.; Gouvinhas, I.; Barros, A.N. Impact of Winemaking Techniques on the Phenolic Composition and Antioxidant Properties of Touriga Nacional Wines. Molecules 2025, 30, 1601. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, Z.; Li, X. Comparison of phenolic compounds and antioxidant activities of red wines from different grape cultivars and vintages in China. J. Food Sci. 2012, 77, C614–C620. [Google Scholar]
- Santoro, V.; Di Renzo, G.C.; Carradori, S. Phenolic composition of international red wines: A comprehensive meta-analysis. Antioxidants 2020, 9, 200. [Google Scholar]
- Rapa, M.; Di Fabio, M.; Boccacci Mariani, M.; Giannetti, V. Characterization of Native Sicilian Wines by Phenolic Contents, Antioxidant Activity, and Chemometrics. Molecules 2025, 30, 534. [Google Scholar] [CrossRef]
- Ciucure, C.T.; Miricioiu, M.G.; Geana, E.I. Discrimination of Romanian Wines Based on Phenolic Composition and Identification of Potential Phenolic Biomarkers for Wine Authenticity and Traceability. Beverages 2025, 11, 44. [Google Scholar] [CrossRef]
- Clarke, S.; Bosman, G.; du Toit, W.; Aleixandre-Tudo, J.L. White wine phenolics: Current methods of analysis. J. Sci. Food Agric. 2023, 103, 7–25. [Google Scholar] [CrossRef]
- Di, S.; Yang, Y. Prediction of red wine quality using one-dimensional convolutional neural networks. arXiv 2023, arXiv:2208.14008. [Google Scholar] [CrossRef]
- Proestos, C.; Bakogiannis, A.; Komaitis, M. Determination of Phenolic Compounds in Wines. Int. J. Food Stud. 2012, 1, 33–41. [Google Scholar] [CrossRef]
- Stój, A.; Czernecki, T.; Domagała, D. Authentication of Polish Red Wines Produced from Zweigelt and Rondo Grape Varieties Based on Volatile Compounds Analysis in Combination with Machine Learning Algorithms: Hotrienol as a Marker of the Zweigelt Variety. Molecules 2023, 28, 1961. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Bengio, Y.; Grandvalet, Y. No Unbiased Estimator of the Variance of K-Fold Cross-Validation. J. Mach. Learn. Res. 2004, 5, 1089–1105. [Google Scholar]
- Berrar, D. Cross-validation. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 1, pp. 542–545. [Google Scholar] [CrossRef]
- Raschka, S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv 2020, arXiv:1811.12808. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Habas, P.A.; Zurada, J.M.; Lo, J.Y.; Baker, J.A.; Tourassi, G.D. Training neural network classifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Netw. 2008, 21, 427–436. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. Int. Jt. Conf. Artif. Intell. 1995, 14, 1137–1145. Available online: https://www.ijcai.org/Proceedings/95-2/Papers/016.pdf (accessed on 20 July 2025).
- Tsamardinos, I.; Greasidou, E.; Tsagris, M.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. arXiv 2017, arXiv:1708.07180. [Google Scholar] [CrossRef]
- Shan, G. Monte Carlo cross-validation for a study with a binary outcome and a limited sample size. BMC Med. Inform. Decis. Mak. 2022, 22, 270. [Google Scholar] [CrossRef]
- Du, J.-H.; Patil, P.; Roeder, K.; Kuchibhotla, A.K. Extrapolated cross-validation for randomized ensembles. arXiv 2023, arXiv:2302.13511. [Google Scholar] [CrossRef]
- Gorriz, J.M.; Martin Clemente, R.; Segovia, F.; Ramírez, J.; Ortiz, A.; Suckling, J. Is k-fold cross-validation the best model selection method for Machine Learning? arXiv 2024, arXiv:2401.16407. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Scikit-Learn Developers. Grid Search Documentation. 2023. Available online: https://scikit-learn.org/stable/modules/grid_search.html (accessed on 20 July 2025).
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and tuning strategies for random forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Eder, R.; Pajović Šćepanović, R.; Raičević, D.; Popović, T.; Korntheuer, K.; Wendelin, S.; Forneck, A.; Philipp, C. Effects of climatic conditions on phenolic content and antioxidant activity of Austrian and Montenegrin red wines. OENO One 2023, 57, 69–85. [Google Scholar] [CrossRef]
- Mollica, A.; Scioli, G.; Della Valle, A.; Cichelli, A.; Novellino, E.; Bauer, M.; Kamysz, W.; Llorent-Martínez, E.J.; Fernández-de Córdova, M.L.; Castillo-López, R.; et al. Phenolic analysis and in vitro biological activity of red wine, pomace and grape seed oil derived from Vitis vinifera L. cv. Montepulciano d’Abruzzo. Antioxidants 2021, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Arseni, A.; Crudu, S. The role of procyanidins in grapes and wines: Effects on quality and composition. J. Eng. Sci. 2025, 31, 175–192. [Google Scholar] [CrossRef]
- García Estévez, I.; Ramos Pineda, A.M.; Escribano Bailón, M.T. Interactions between wine phenolic compounds and human saliva in astringency perception: A review. Food Funct. 2018, 9, 1294–1309. [Google Scholar] [CrossRef]
- Lorrain, B.; Ky, I.; Pechamat, L.; Teissedre, P.-L. Evolution of Analysis of Polyphenols from Grapes, Wines, and Extracts. Molecules 2013, 18, 1076–1100. [Google Scholar] [CrossRef] [PubMed]
- Jordão, A.M.; Correia, A.C.; Martins, B.; Romão, A.; Oliveira, B. General physicochemical parameters, phenolic composition, and varietal aromatic potential of three red Vitis vinifera varieties “Merlot”, “Syrah”, and “Saborinho” cultivated on Pico Island—Azores Archipelago. Int. J. Plant Biol. 2024, 15, 1369–1390. [Google Scholar] [CrossRef]
- Ranaweera, R.K.; Gilmore, A.M.; Bastian, S.E.; Capone, D.L.; Jeffery, D.W. Spectrofluorometric analysis to trace the molecular fingerprint of wine during the winemaking process and recognise the blending percentage of different varietal wines. OENO One 2022, 56, 189–196. [Google Scholar] [CrossRef]
- Dahal, K.R.; Dahal, J.N.; Banjade, H.B.; Gaire, S.G. Prediction of Wine Quality Using Machine Learning Algorithms. Open J. Stat. 2021, 11, 278–289. [Google Scholar] [CrossRef]
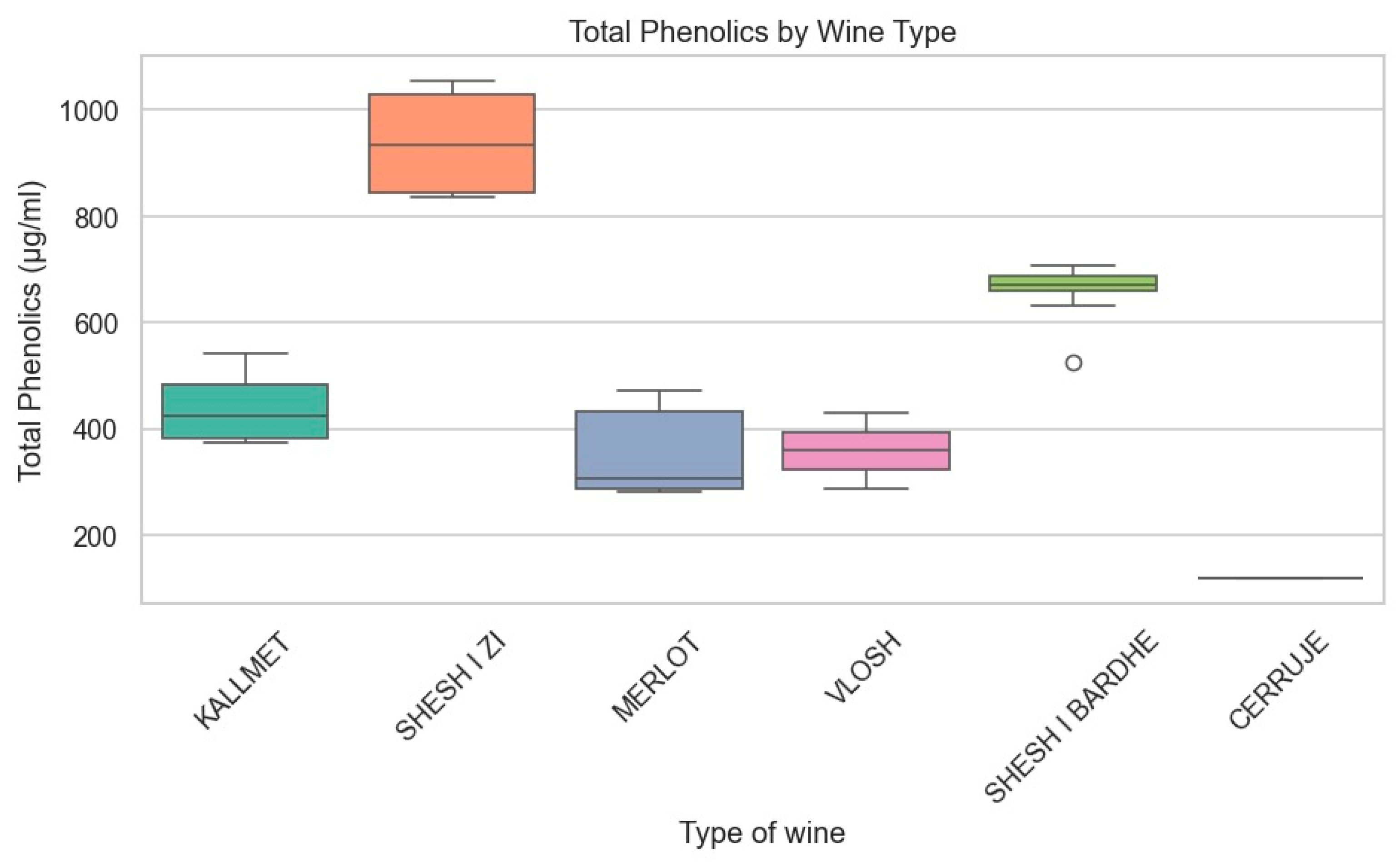
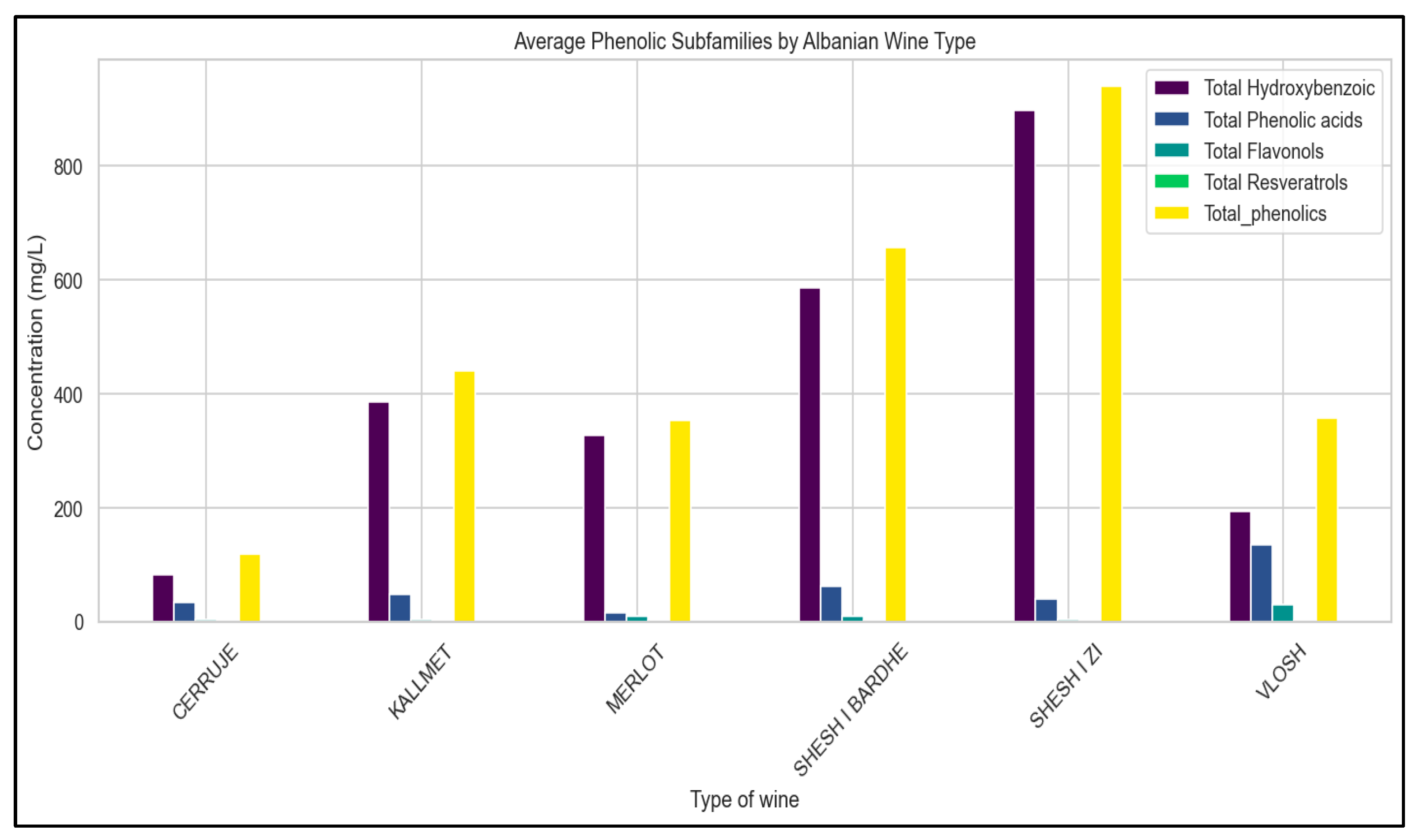


| Comparison | Phenolic Subclass | U Statistic | p-Value | Significance (α = 0.05) |
|---|---|---|---|---|
| Shesh i zi vs. Merlot | Hydroxybenzoic acids + flavanols | 0.0 | 0.0022 | Significant |
| Shesh i zi vs. Merlot | Total Phenolic acids | 0.0 | 0.0022 | Significant |
| Shesh i zi vs. Merlot | Total Flavonols | 0.0 | 0.0022 | Significant |
| Shesh i zi vs. Merlot | Total Resveratrols | 0.0 | 0.0022 | Significant |
| Shesh i zi vs. Kallmet | Hydroxybenzoic acids + flavanols | 0.0 | 0.0017 | Significant |
| Shesh i zi vs. Shesh i bardhë | Hydroxybenzoic acids + flavanols | 0.0 | 0.0017 | Significant |
| Shesh i bardhë vs. Cerruje | All subclasses | 0.0 | 0.0286 | Significant |
| Red vs. white wines | All subclasses | 0.0 | 0.0001 | Highly Significant |
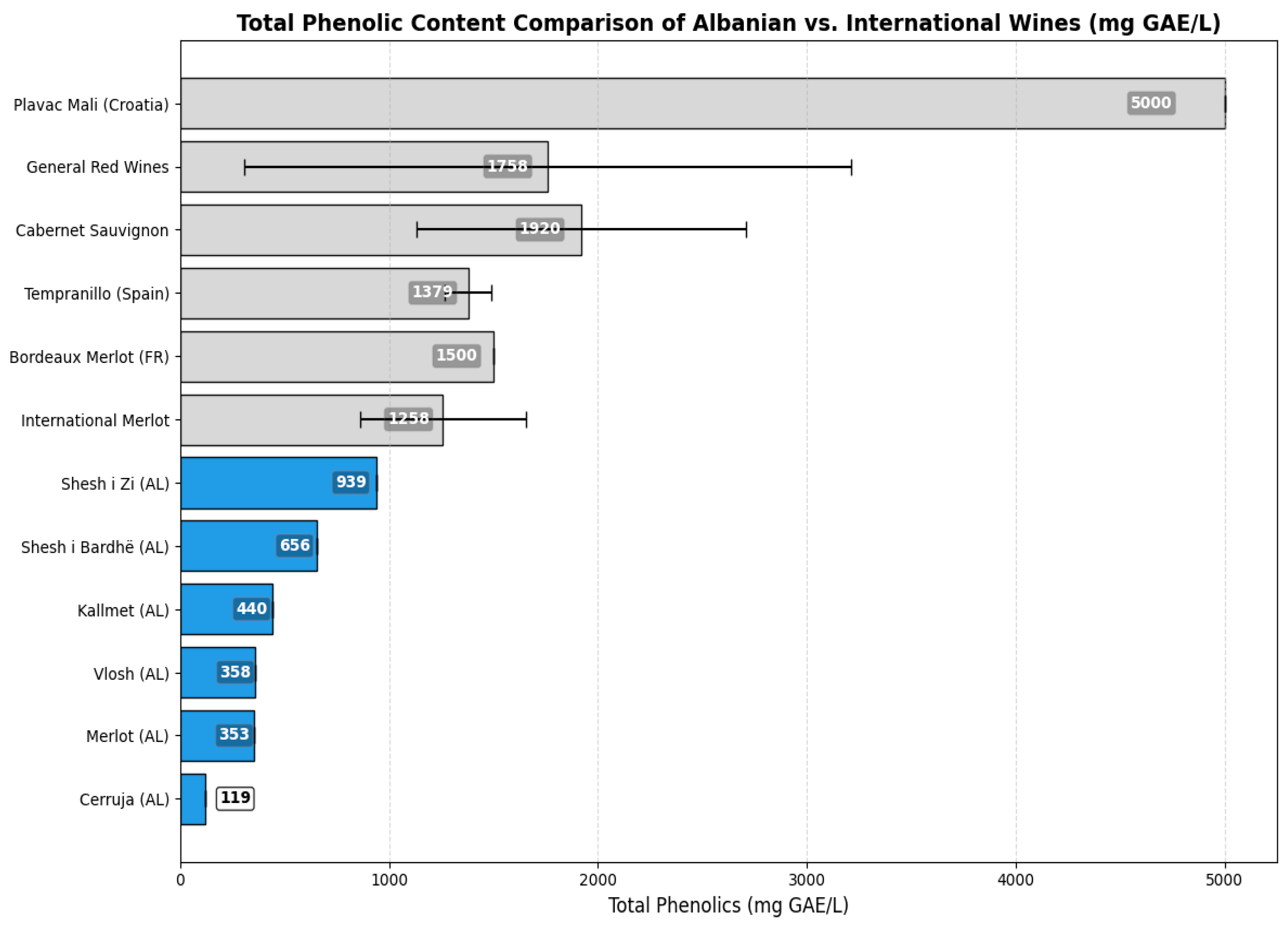
| Wine Type | Total Phenolics (mg GAE/L) | Citation | Interpretive Comment |
|---|---|---|---|
| Shesh i zi (AL) | 939 | wine dataset, 2017–2021 | A local Albanian red wine, Shesh i zi, exhibited strong phenolic intensity, reflecting native varietal richness and moderate oxidative stability. |
| Shesh i bardhë (AL) | 656 | wine dataset, 2017–2021 | The white variety Shesh i bardhë showed phenolic levels characteristic of light-colored wines, aligning with global white wine trends. |
| Vlosh (AL) | 358 | wine dataset, 2017–2021 | Vlosh, an autochthonous variety, demonstrated robust phenolic content, supporting its historic use in regional red blends. |
| Cerruje (AL) | 119 | wine dataset, 2017–2021 | Cerruja displayed moderate phenolic richness, possibly influenced by vintage and microclimate variations. |
| Merlot (AL) | 353 | wine dataset, 2017–2021 | Albanian Merlot samples had phenolic values in line with European counterparts, validating local vinification standards. |
| International Merlot | 860–1656 | [30] | Total phenol contents range (mg GAE/L) |
| General Red Wines | 305–3210 | [31] | Phenolic concentrations in global red wines are highly variable, spanning over an order of magnitude, and influenced by winemaking style and grape chemistry. |
| Plavac Mali (Croatia) | ~5000 | [27] | Plavac Mali stood out with ~5 g/L phenolics, attributed to thick grape skins and traditional extended maceration practices. |
| Bordeaux Merlot (FR) | ~1500 | [28] | Bordeaux-region Merlot wines showed intermediate total phenolics, shaped by climate and controlled fermentation processes. |
| Cabernet Sauvignon | 1129–2710 | [30] | Widely cultivated Cabernet Sauvignon exhibited some of the highest phenolic levels among commercial reds, often exceeding 2 g/L. |
| Model | ROC-AUC | F1-Score | Accuracy |
|---|---|---|---|
| Random Forest | 0.9907 | 0.9375 | 0.9259 |
| SVM | 1.0000 | 1.0000 | 1.0000 |
| KNN | 0.9360 | 0.5208 | 0.5370 |
| Logistic Regression | 1.0000 | 0.9020 | 0.8889 |
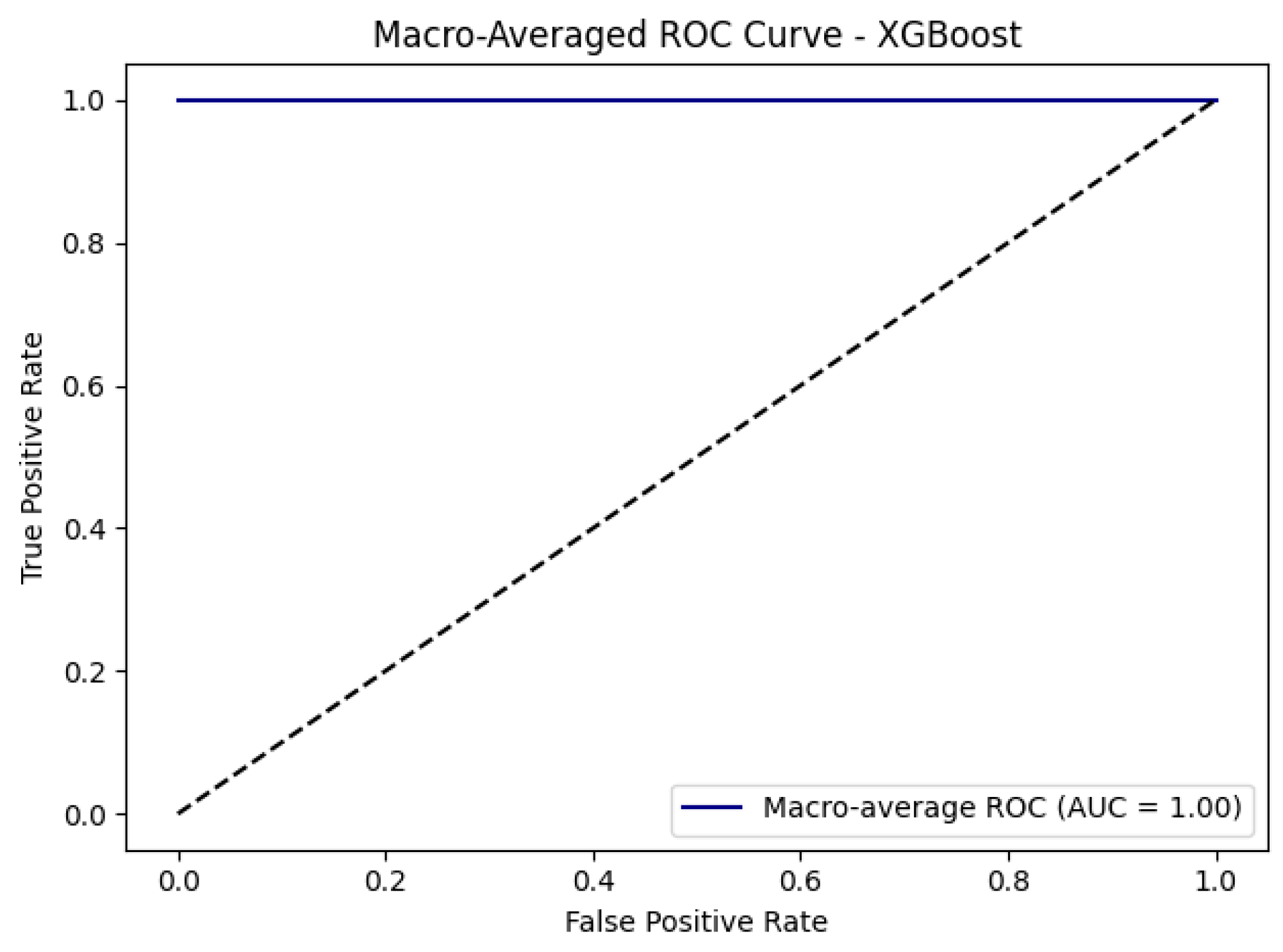
| XGBoost | 1.0000 | 0.8931 | 0.8843 |
| Model | LOOCV Accuracy | Permutation p-Value |
|---|---|---|
| SVM | 1.0000 | 0.0010 |
| Random Forest | 0.9615 | 0.0010 |
| Logistic Regression | 0.9615 | 0.0010 |
| KNN | 0.9231 | 0.0010 |
| XGBoost | 0.9231 | 0.0010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topi, A.; Kasaj, A.; Hudhra, D.; Kelebek, H.; Guclu, G.; Selli, S.; Topi, D. Machine Learning-Based Classification of Albanian Wines by Grape Variety, Using Phenolic Compound Dataset. Analytica 2025, 6, 43. https://doi.org/10.3390/analytica6040043
Topi A, Kasaj A, Hudhra D, Kelebek H, Guclu G, Selli S, Topi D. Machine Learning-Based Classification of Albanian Wines by Grape Variety, Using Phenolic Compound Dataset. Analytica. 2025; 6(4):43. https://doi.org/10.3390/analytica6040043
Chicago/Turabian StyleTopi, Ardiana, Agim Kasaj, Daniel Hudhra, Hasim Kelebek, Gamze Guclu, Serkan Selli, and Dritan Topi. 2025. "Machine Learning-Based Classification of Albanian Wines by Grape Variety, Using Phenolic Compound Dataset" Analytica 6, no. 4: 43. https://doi.org/10.3390/analytica6040043
APA StyleTopi, A., Kasaj, A., Hudhra, D., Kelebek, H., Guclu, G., Selli, S., & Topi, D. (2025). Machine Learning-Based Classification of Albanian Wines by Grape Variety, Using Phenolic Compound Dataset. Analytica, 6(4), 43. https://doi.org/10.3390/analytica6040043











