Predicting the Structure of Hydrogenase in Microalgae: The Case of Nannochloropsis salina
Abstract
1. Introduction
- FeS clusters are close one to each other to allow electron transfer among them;
- the terminal (active) FeS cluster (H-cluster) must interact with water molecules to transfer electrons to a polarized H-O bond; this step must be hindered to all except H-cluster;
- FeS clusters must resist, in the assembly, as much as possible to oxidation by reactive oxygen species formed by O2, to sustain hydrogen production as long as possible in the presence of a small amount of O2 formed at the OEC and diffusing in the chloroplast.
2. Materials and Methods
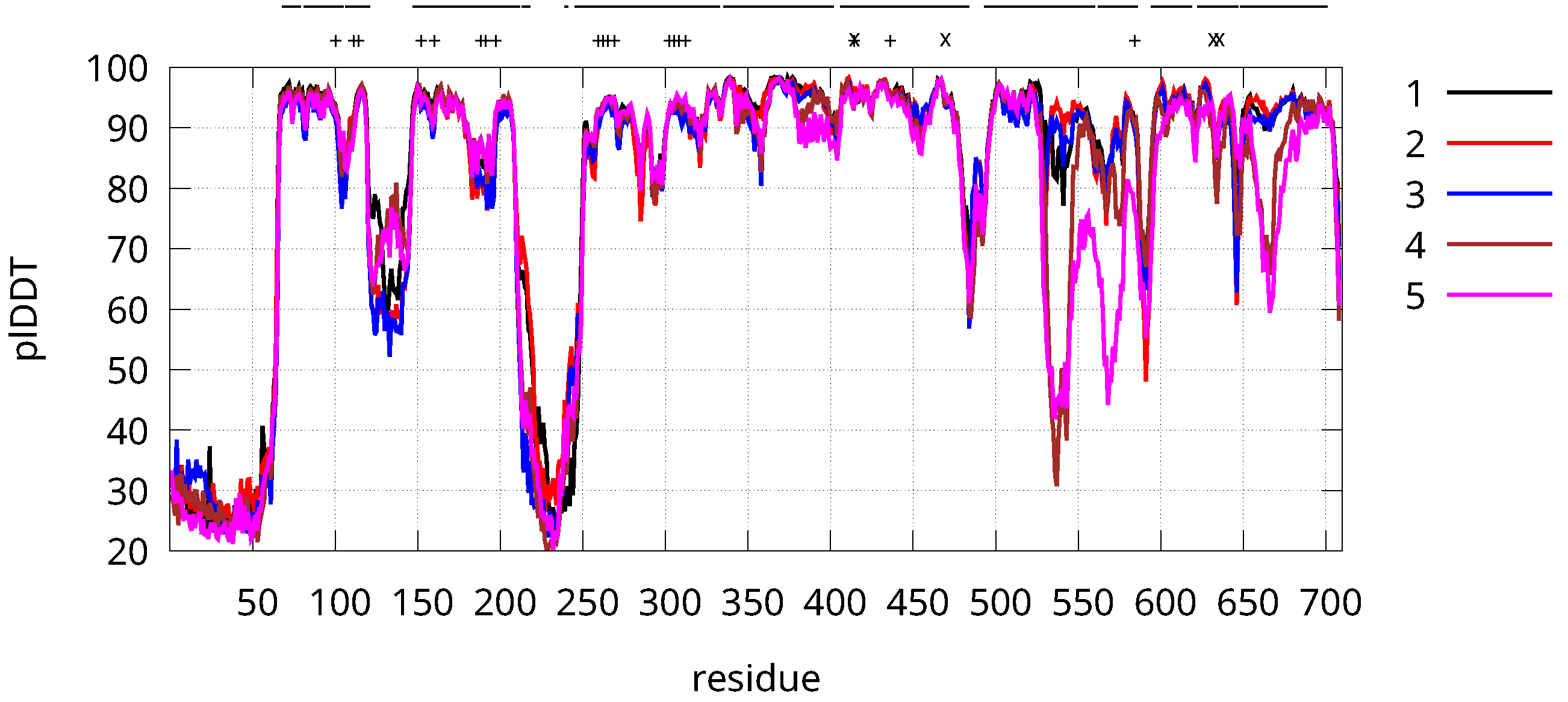
2.1. Structure Predictions from Sequence
2.2. Structure Refinement
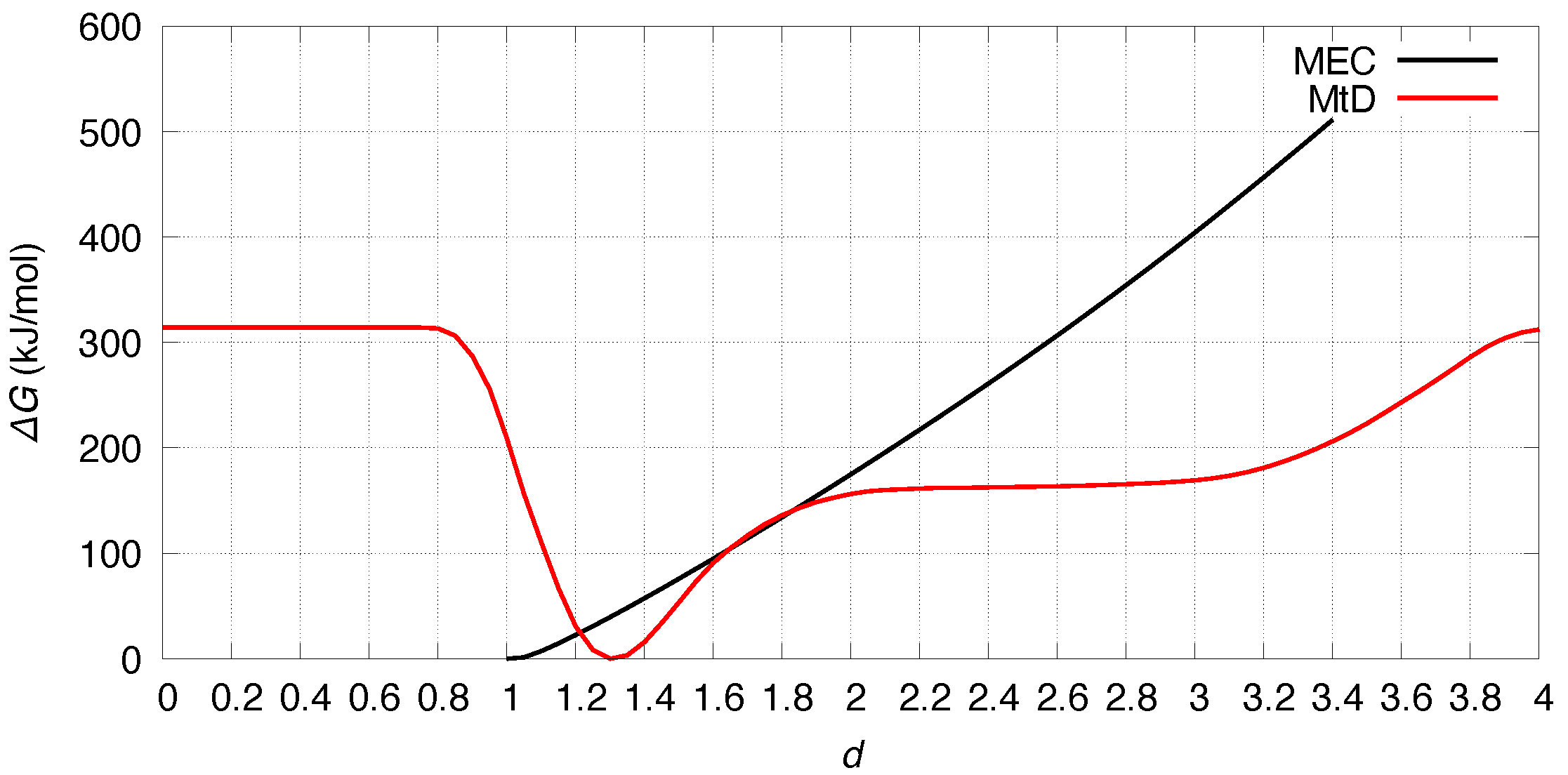
2.3. Free Energy
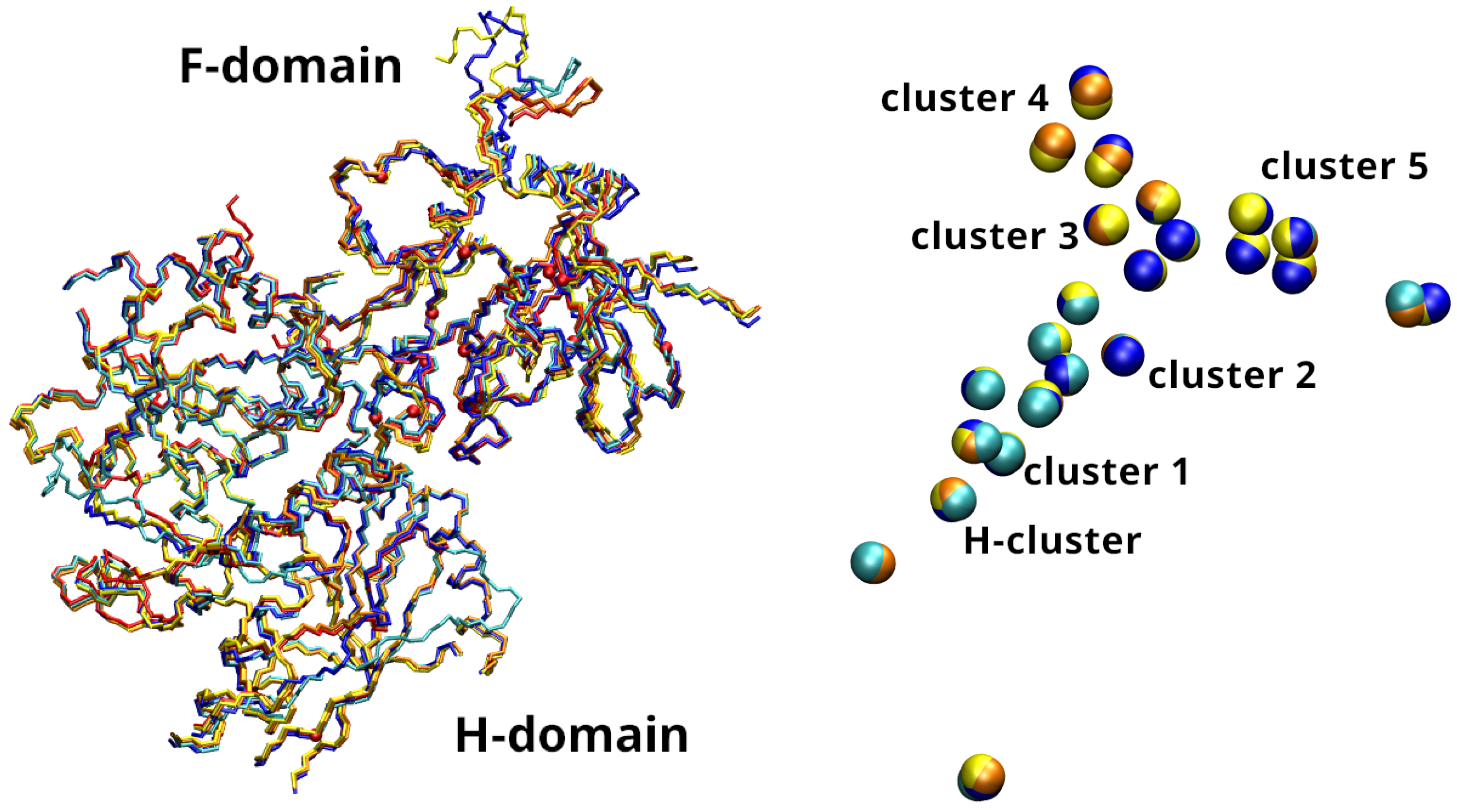
2.4. Analysis
3. Results
3.1. Structure Predictions from Sequence
3.2. Constraints on FeS Clusters Positions
| Cluster | Binding Residues |
|---|---|
| Fe2(adt)(CO)4(CN)2 (H-cluster) | C636 |
| Fe4S4 1 (H-cluster) | C415, C470, C632, C636 |
| Fe4S4 2 | C269, C302, C305, C308 |
| Fe4S4 3 | C259, C262, C265, C312 |
| Fe4S4 4 | H184, C188, C191, C197 |
| Fe2S2 5 | C100, C111, C114, C152 |
3.3. Free Energy Profiles
3.4. Solvent Access to Fe Atoms
3.5. Distal Iron Protection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MA | microalgae |
| MSA | multiple sequence alignment |
| OEC | oxygen evolving center |
| PS | photosystem |
| FeS | iron-sulfur |
| Hyd | [FeFe] hydrogenase |
| Ns | Nannochloropsis salina |
| CpI | [FeFe] hydrogenase of Clostridium pasteurianum, isoform I |
| Dd | Desulfovibrio desulfuricans |
| Cr | Chlamydomonas reinhardtii |
| Cb | Clostridium beijerinckii |
| MD | molecular dynamics |
| RMSD | root-mean square deviation |
| RMSF | root-mean square fluctuation |
References
- Megía, P.J.; Vizcaíno, A.J.; Calles, J.A.; Carrero, A. Hydrogen Production Technologies: From Fossil Fuels toward Renewable Sources. A Mini Review. Energy Fuels 2021, 35, 16403–16415. [Google Scholar] [CrossRef]
- Echim, S.M.; Budea, S. Use of Hydrogen Energy and Fuel Cells in Marine and Industrial Applications—Current Status. Hydrogen 2025, 6, 50. [Google Scholar] [CrossRef]
- Min Woon, J.; Shiong Khoo, K.; Akermi, M.; Alanazi, M.M.; Wei Lim, J.; Jing Chan, Y.; Sean Goh, P.; Silas Chidi, B.; Kee Lam, M.; Zaini, J.; et al. Reviewing biohydrogen production from microalgal cells through fundamental mechanisms, enzymes and factors that engendering new challenges and prospects. Fuel 2023, 346, 128312. [Google Scholar] [CrossRef]
- Ghirardi, M.L.; King, P.W.; Mulder, D.W.; Eckert, C.; Dubini, A.; Maness, P.C.; Yu, J. Hydrogen Production by Water Biophotolysis. In Microbial BioEnergy: Hydrogen Production; Zannoni, D., De Philippis, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 101–135. [Google Scholar] [CrossRef]
- Seibert, M.; Torzillo, G. (Eds.) Microalgal Hydrogen Production; Comprehensive Series in Photochemical & Photobiological Sciences; The Royal Society of Chemistry: Cambridge, UK, 2018. [Google Scholar] [CrossRef]
- Babu Pallam, R.; Ramamoorthy, N.K.; Rakshit, S.; Jaiswal, K.K.; Sarma, V.V. Microalgae for the Sustainable Production of Biohydrogen. In Clean Energy Transition-via-Biomass Resource Utilization: A Way to Mitigate Climate Change; Kumar, S., Sundaramurthy, S., Kumar, D., Chandel, A.K., Eds.; Springer Nature: Singapore, 2024; pp. 151–175. [Google Scholar] [CrossRef]
- Chandrasekhar, T.; Chandra Obul Reddy, P.; Swapna, B.; Veeranjaneya Reddy, L.; Anuprasanna, V.; Dakshayani, L.; Ramachandra Reddy, P.; Reddy, M.C. Algae: The game-changers in biohydrogen sector. Crit. Rev. Biotechnol. 2025, 45, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Lubitz, W.; Ogata, H.; Rüdiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef] [PubMed]
- Birrell, J.A.; Rodríguez-Maciá, P.; Reijerse, E.J.; Martini, M.A.; Lubitz, W. The catalytic cycle of [FeFe] hydrogenase: A tale of two sites. Coord. Chem. Rev. 2021, 449, 214191. [Google Scholar] [CrossRef]
- Artz, J.H.; Zadvornyy, O.A.; Mulder, D.W.; Keable, S.M.; Cohen, A.E.; Ratzloff, M.W.; Williams, S.G.; Ginovska, B.; Kumar, N.; Song, J.; et al. Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases. J. Am. Chem. Soc. 2020, 142, 1227–1235. [Google Scholar] [CrossRef]
- Sahin, S.; Brazard, J.; Zuchan, K.; Adachi, T.B.M.; Mühlenhoff, U.; Milton, R.D.; Stripp, S.T. Probing the ferredoxin:hydrogenase electron transfer complex by infrared difference spectroscopy. Chem. Sci. 2025, 16, 10465–10475. [Google Scholar] [CrossRef]
- Frowijn, L.S.; van Sark, W.G.J.H.M. Analysis of photon-driven solar-to-hydrogen production methods in the Netherlands. Sustain. Energy Techn. 2021, 48, 101631. [Google Scholar] [CrossRef]
- Touloupakis, E.; Faraloni, C.; Silva Benavides, A.M.; Torzillo, G. Recent Achievements in Microalgal Photobiological Hydrogen Production. Energies 2021, 14, 7170. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, M.; Yu, L.; Sun, H.; Liu, J. Nannochloropsis as an Emerging Algal Chassis for Light-Driven Synthesis of Lipids and High-Value Products. Mar. Drugs 2024, 22, 54. [Google Scholar] [CrossRef] [PubMed]
- Ramprakash, B.; Lindblad, P.; Eaton-Rye, J.J.; Incharoensakdi, A. Current strategies and future perspectives in biological hydrogen production: A review. Renew. Sustai. Energy Rev. 2022, 168, 112773. [Google Scholar] [CrossRef]
- Viviano, E.; Limongi, A.R. Chapter 13—Production of biofuels from microalgae. In Sustainable Industrial Processes Based on Microalgae; Lafarga, T., Acién, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2024; pp. 297–321. [Google Scholar] [CrossRef]
- Nicolet, Y.; Piras, C.; Legrand, P.; Hatchikian, C.E.; Fontecilla-Camps, J.C. Desulfovibrio desulfuricans iron hydrogenase: The structure shows unusual coordination to an active site Fe binuclear center. Structure 1999, 7, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Mulder, D.W.; Boyd, E.S.; Sarma, R.; Lange, R.K.; Endrizzi, J.A.; Broderick, J.B.; Peters, J.W. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA Delta EFG. Nature 2010, 465, 248–251. [Google Scholar] [CrossRef]
- Swanson, K.D.; Ratzloff, M.W.; Mulder, D.W.; Artz, J.H.; Ghose, S.; Hoffman, A.; White, S.; Zadvornyy, O.A.; Broderick, J.B.; Bothner, B.; et al. [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster. J. Am. Chem. Soc. 2015, 137, 1809–1816. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Botticelli, S.; La Penna, G.; Nobili, G.; Rossi, G.; Stellato, F.; Morante, S. Modelling Protein Plasticity: The Example of Frataxin and Its Variants. Molecules 2022, 27, 1955. [Google Scholar] [CrossRef]
- Nobili, G.; Botticelli, S.; La Penna, G.; Morante, S.; Rossi, G.; Salina, G. Probing protein stability: Towards a computational atomistic, reliable, affordable, and improvable model. Front. Mol. Biosci. 2023, 10, 1122269. [Google Scholar] [CrossRef]
- Botticelli, S.; La Penna, G.; Minicozzi, V.; Stellato, F.; Morante, S.; Rossi, G.; Faraloni, C. Predicting the Structure of Enzymes with Metal Cofactors: The Example of [FeFe] Hydrogenases. Int. J. Mol. Sci. 2024, 25, 3663. [Google Scholar] [CrossRef]
- Tamayo-Ordoñez, Y.d.J.; Ayil-Gutiérrez, B.A.; Moreno-Davila, I.M.M.; Tamayo-Ordoñez, F.A.; Córdova-Quiroz, A.V.; Poot-Poot, W.A.; Damas-Damas, S.; Villanueva-Alonzo, H.d.J.; Tamayo-Ordoñez, M.C. Bioinformatic analysis and relative expression of hyd and fdx during H2 production in microalgae. Phycol. Res. 2023, 71, 37–55. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucl. Acids Res. 2023, 52, D368–D375. [Google Scholar] [CrossRef]
- Chang, C.H.; Kim, K. Density Functional Theory Calculation of Bonding and Charge Parameters for Molecular Dynamics Studies on [FeFe] Hydrogenases. J. Chem. Theory Comput. 2009, 5, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi1 and chi2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N-log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Hardy, D.J.; Stone, J.E.; Tajkhorshid, E.; Kohlmeyer, A. TopoGromacs: Automated Topology Conversion from CHARMM to GROMACS within VMD. J. Chem. Inform. Model. 2016, 56, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Hošek, P.; Kříž, P.; Toulcová, D.; Spiwok, V. Multisystem altruistic metadynamics- Well-tempered variant. J. Chem. Phys. 2017, 146, 125103. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Branduardi, D.; Bussi, G.; Camilloni, C.; Provasi, D.; Raiteri, P.; Donadio, D.; Marinelli, F.; Pietrucci, F.; Broglia, R.A.; et al. PLUMED: A Portable Plugin for Free-Energy Calculations with Molecular Dynamics. J. Comput. Phys. 2009, 180, 1961–1972. [Google Scholar] [CrossRef]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New feathers for an old bird. Comp. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef]
- Bonomi, M.; Bussi, G.; Camilloni, C.; Tribello, G.A.; Banás̆, P.; Barducci, A.; Bernetti, M.; Bolhuis, P.G.; Bottaro, S.; Branduardi, D.; et al. Promoting transparency and reproducibility in enhanced molecular simulations. Nat. Methods 2019, 16, 670–673. [Google Scholar] [CrossRef]
- Rumpel, S.; Siebel, J.F.; Diallo, M.; Farès, C.; Reijerse, E.J.; Lubitz, W. Structural Insight into the Complex of Ferredoxin and [FeFe] Hydrogenase from Chlamydomonas reinhardtii. ChemBioChem 2015, 16, 1663–1669. [Google Scholar] [CrossRef]
- Fukuyama, K. Structure and Function of Plant-Type Ferredoxins. Photosynth. Res. 2004, 81, 289–301. [Google Scholar] [CrossRef]
- Pandey, A.S.; Harris, T.V.; Giles, L.J.; Peters, J.W.; Szilagyi, R.K. Dithiomethylether as a Ligand in the Hydrogenase H-Cluster. J. Am. Chem. Soc. 2008, 130, 4533–4540. [Google Scholar] [CrossRef]
- Winkler, M.; Duan, J.; Rutz, A.; Felbek, C.; Scholtysek, L.; Lampret, O.; Jaenecke, J.; Apfel, U.P.; Gilardi, G.; Valetti, F.; et al. A safety cap protects hydrogenase from oxygen attack. Nat. Commun. 2021, 12, 756. [Google Scholar] [CrossRef]
- Rodríguez-Maciá, P.; Galle, L.M.; Bjornsson, R.; Lorent, C.; Zebger, I.; Yoda, Y.; Cramer, S.P.; DeBeer, S.; Span, I.; Birrell, J.A. Caught in the Hinact: Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O2-stable State of [FeFe] Hydrogenase. Angew. Chem. Int. Ed. 2020, 59, 16786–16794. [Google Scholar] [CrossRef]
- Duan, J.; Rutz, A.; Kawamoto, A.; Naskar, S.; Edenharter, K.; Leimkühler, S.; Hofmann, E.; Happe, T.; Kurisu, G. Structural determinants of oxygen resistance and Zn2+-mediated stability of the [FeFe]-hydrogenase from Clostridium beijerinckii. Proc. Natl. Acad. Sci. USA 2025, 122, e2416233122. [Google Scholar] [CrossRef]
- Koo, J.; Shiigi, S.; Rohovie, M.; Mehta, K.; Swartz, J.R. Characterization of [FeFe] Hydrogenase O2 Sensitivity Using a New, Physiological Approach. J. Biol. Chem. 2016, 291, 21563–21570. [Google Scholar] [CrossRef]
- Ghirardi, M.L.; Togasaki, R.K.; Seibert, M. Oxygen sensitivity of algal H2 production. Appl. Biochem. Biotechnol. 1997, 63, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Bingham, A.S.; Smith, P.R.; Swartz, J.R. Evolution of an [FeFe] hydrogenase with decreased oxygen sensitivity. Int. J. Hydrogen Energy 2012, 37, 2965–2976. [Google Scholar] [CrossRef]
- Morra, S.; Arizzi, M.; Valetti, F.; Gilardi, G. Oxygen Stability in the New [FeFe]-Hydrogenase from Clostridium beijerinckii SM10 (CbA5H). Biochemistry 2016, 55, 5897–5900. [Google Scholar] [CrossRef] [PubMed]
- Morra, S. Fantastic [FeFe]-Hydrogenases and Where to Find Them. Front. Microbiol. 2022, 13, 853626. [Google Scholar] [CrossRef]
- Schumann, C.; Fernández Méndez, J.; Berggren, G.; Lindblad, P. Novel concepts and engineering strategies for heterologous expression of efficient hydrogenases in photosynthetic microorganisms. Front. Microbiol. 2023, 14, 1179607. [Google Scholar] [CrossRef]
- Jespersen, M.; Greening, C.; Ernst, L.; Man Leung, P.; Shafaat, H.S.; Grinter, R. Diverse lineages and adaptations of oxygen-adapted hydrogenases. Trends Biochem. Sci. 2025, 50, 596–609. [Google Scholar] [CrossRef]
- Stripp, S.T.; Goldet, G.; Brandmayr, C.; Sanganas, O.; Vincent, K.A.; Haumann, M.; Armstrong, F.A.; Happe, T. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. USA 2009, 106, 17331–17336. [Google Scholar] [CrossRef]
- Cohen, J.; Kim, K.; King, P.; Seibert, M.; Schulten, K. Finding Gas Diffusion Pathways in Proteins: Application to O2 and H2 Transport in CpI [FeFe]-Hydrogenase and the Role of Packing Defects. Structure 2005, 13, 1321–1329. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, C.K.; Uddin, S.; Stripp, S.T.; Engelbrecht, V.; Winkler, M.; Leimkühler, S.; Brocks, C.; Duan, J.; Schäfer, L.V.; et al. Protein Dynamics Affect O2-Stability of Group B [FeFe]-Hydrogenase from Thermosediminibacter oceani. J. Am. Chem. Soc. 2025, 147, 15170–15180. [Google Scholar] [CrossRef]
- Crack, J.C.; Green, J.; Cheesman, M.R.; Le Brun, N.E.; Thomson, A.J. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. USA 2007, 104, 2092–2097. [Google Scholar] [CrossRef] [PubMed]
- Outten, F.W. Iron-sulfur clusters as oxygen-responsive molecular switches. Nat. Chem. Biol. 2007, 3, 206–207. [Google Scholar] [CrossRef] [PubMed]
- Crack, J.C.; Green, J.; Thomson, A.J.; Le Brun, N.E. Iron–Sulfur Clusters as Biological Sensors: The Chemistry of Reactions with Molecular Oxygen and Nitric Oxide. Acc. Chem. Res. 2014, 47, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, A.; Pandelia, M.E.; Gärtner, W.; Lubitz, W. [Fe4S4]- and [Fe3S4]-cluster formation in synthetic peptides. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1414–1422. [Google Scholar] [CrossRef]
- Pizzanelli, S.; Pitzalis, E.; Botticelli, S.; Machetti, F.; Faraloni, C.; La Penna, G. Electron spin resonance in microalgae whole-cells to monitor hydrogen production. J. Biol. Inorg. Chem. 2025, 30, 229–240. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Touloupakis, E.; Faraloni, C.; Silva Benavides, A.M.; Masojídek, J.; Torzillo, G. Sustained photobiological hydrogen production by Chlorella vulgaris without nutrient starvation. Int. J. Hydrogen Energy 2021, 46, 3684–3694. [Google Scholar] [CrossRef]
- Torzillo, G.; Scoma, A.; Faraloni, C.; Ena, A.; Johanningmeier, U. Increased hydrogen photoproduction by means of a sulfur-deprived Chlamydomonas reinhardtii D1 protein mutant. Int. J. Hydrogen Energy 2009, 34, 4529–4536. [Google Scholar] [CrossRef]

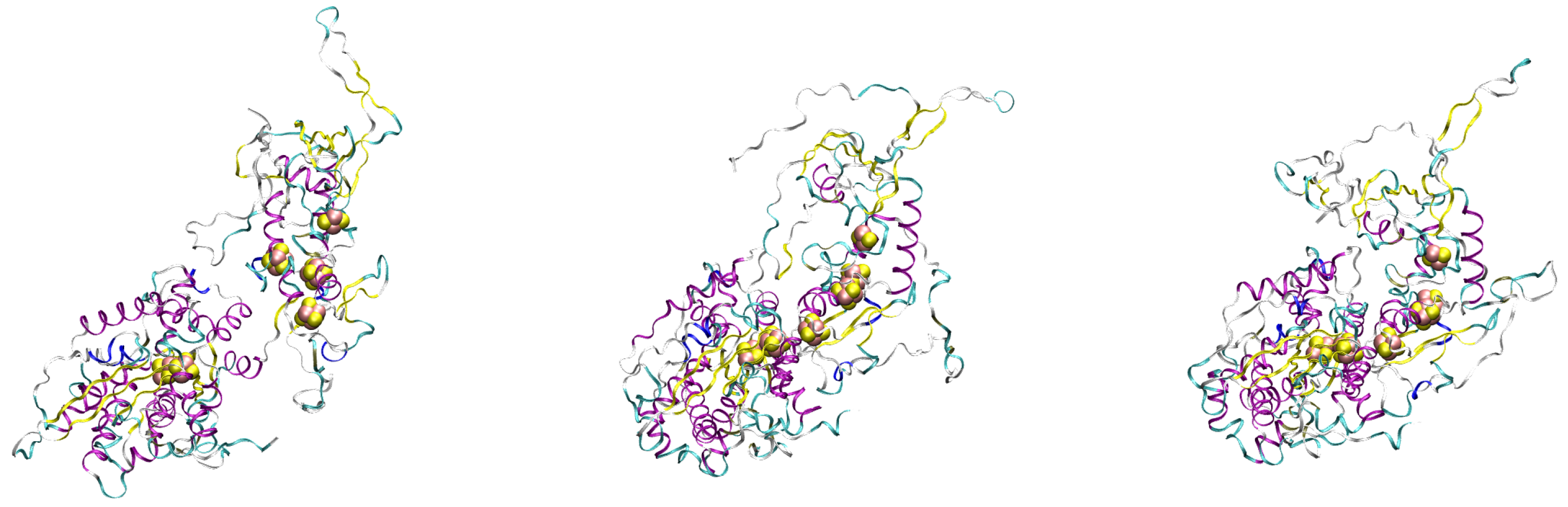
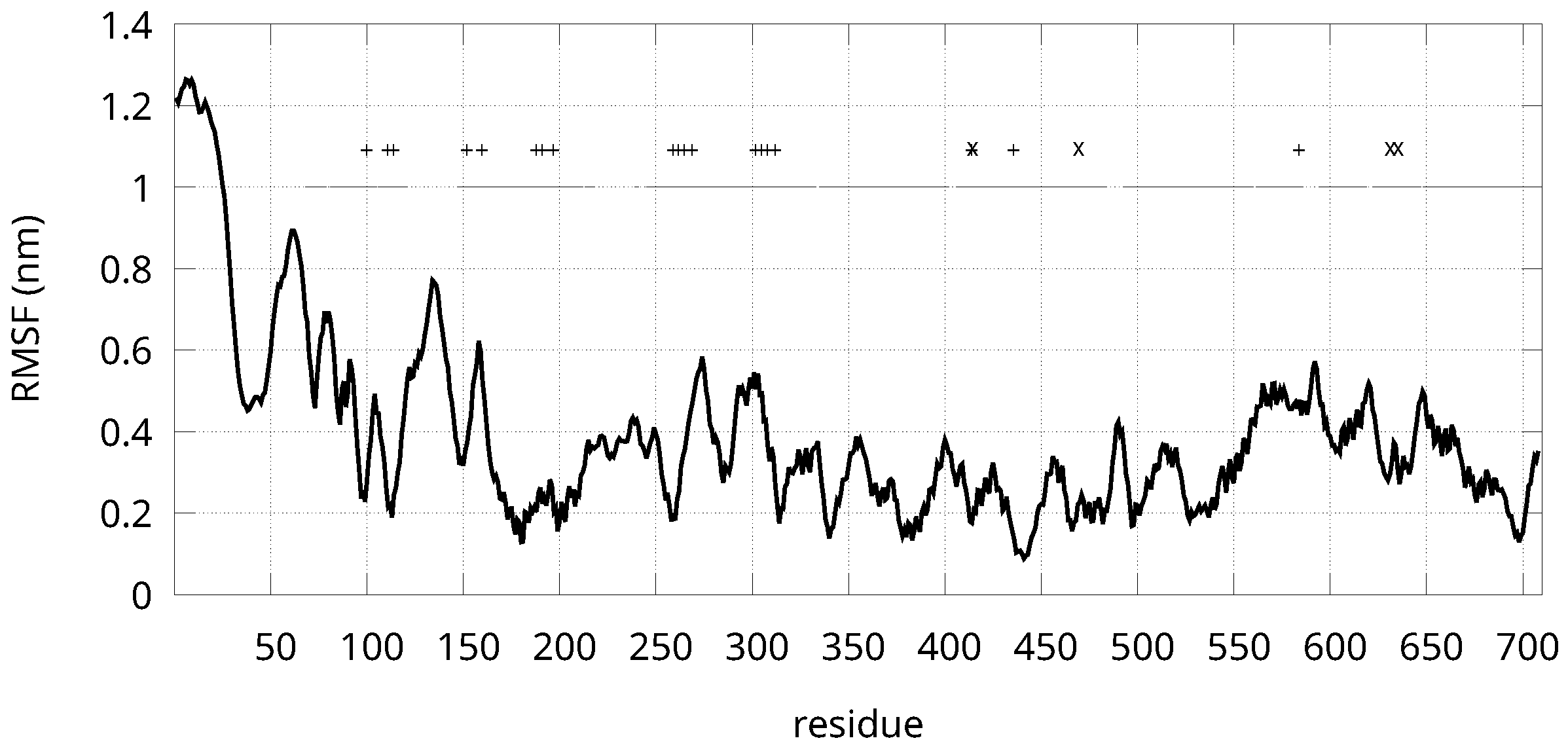
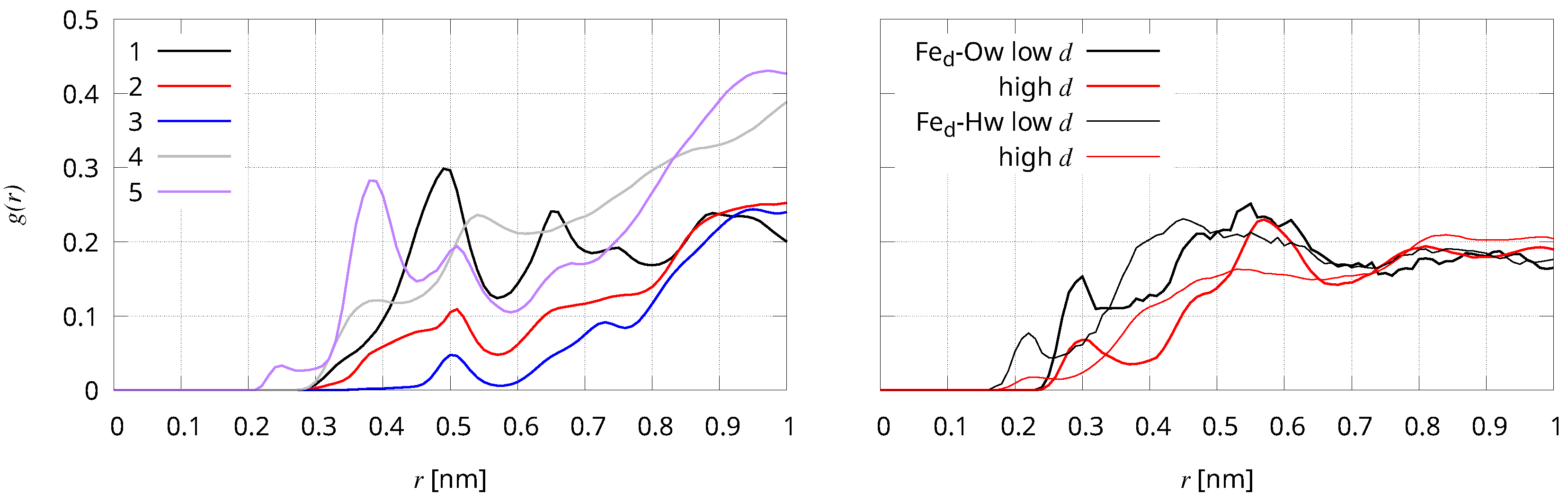
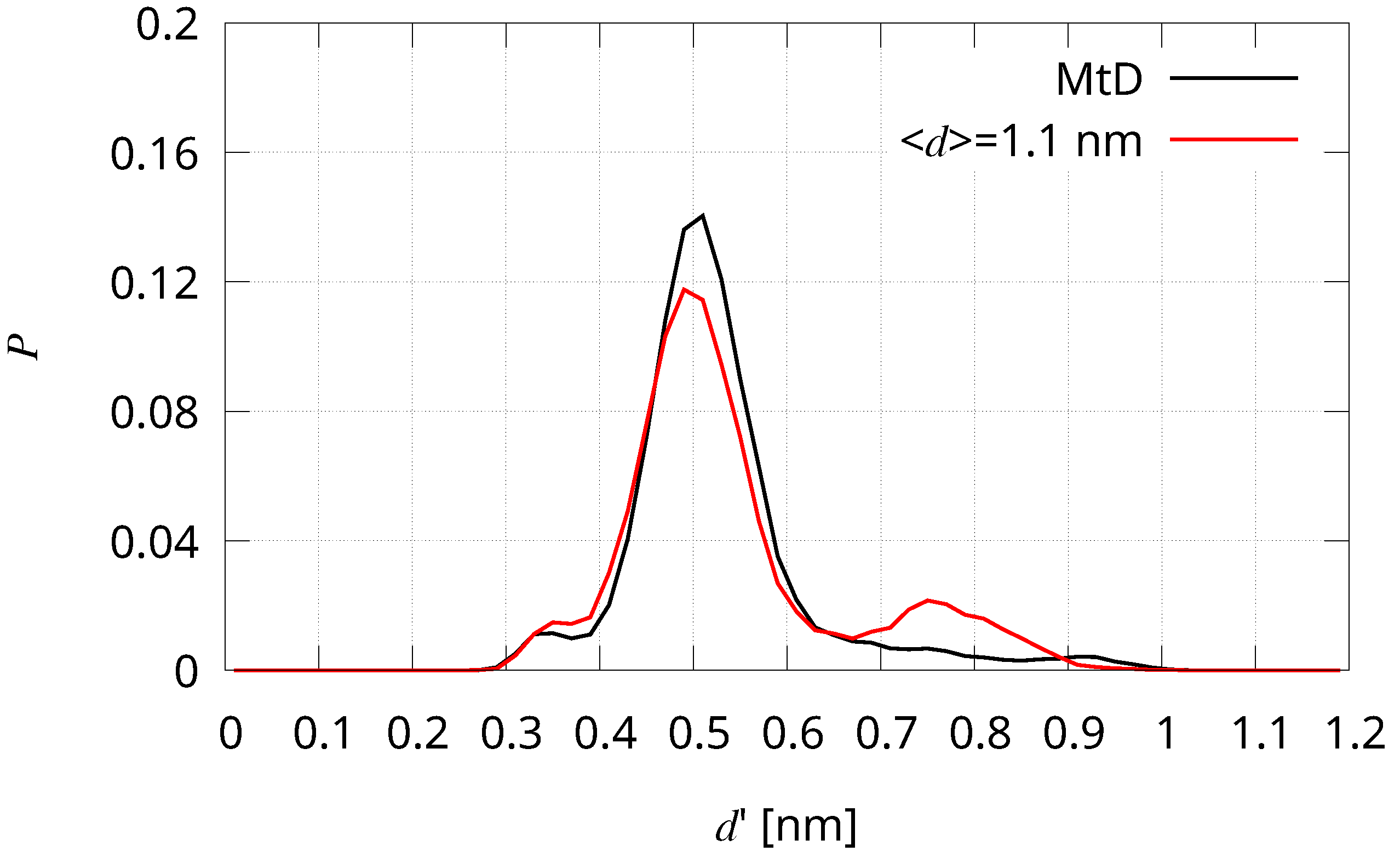
| Sample | Selection Rule |
|---|---|
| 1 | Maximal MEC modulation weight using the constraint 0.95 nm |
| 2 | Center of the maximally populated cluster in MtD |
| (using a RMSD cut-off of 0.3 nm) obtained for configurations with 0.95 nm | |
| 3 | Minimal distances between FeS clusters in MtD |
| Sample | CpI | 1 | 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster | 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 | 2 | 3 | 4 | 5 |
| 1 | 1.15 | 2.09 | 2.68 | 2.94 | 0.91 | 2.0 | 2.9 | 3.1 | 0.94 | 2.0 | 2.8 | 2.8 |
| 2 | - | 1.23 | 2.20 | 2.25 | - | 1.2 | 2.3 | 2.3 | - | 1.2 | 2.1 | 2.2 |
| 3 | - | - | 1.22 | 1.37 | - | - | 1.3 | 1.3 | - | - | 1.2 | 1.4 |
| 4 | - | - | - | 1.92 | - | - | - | 1.9 | - | - | - | 1.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Botticelli, S.; Faraloni, C.; La Penna, G. Predicting the Structure of Hydrogenase in Microalgae: The Case of Nannochloropsis salina. Hydrogen 2025, 6, 77. https://doi.org/10.3390/hydrogen6040077
Botticelli S, Faraloni C, La Penna G. Predicting the Structure of Hydrogenase in Microalgae: The Case of Nannochloropsis salina. Hydrogen. 2025; 6(4):77. https://doi.org/10.3390/hydrogen6040077
Chicago/Turabian StyleBotticelli, Simone, Cecilia Faraloni, and Giovanni La Penna. 2025. "Predicting the Structure of Hydrogenase in Microalgae: The Case of Nannochloropsis salina" Hydrogen 6, no. 4: 77. https://doi.org/10.3390/hydrogen6040077
APA StyleBotticelli, S., Faraloni, C., & La Penna, G. (2025). Predicting the Structure of Hydrogenase in Microalgae: The Case of Nannochloropsis salina. Hydrogen, 6(4), 77. https://doi.org/10.3390/hydrogen6040077







