Abstract
In recent decades, sustainable forest management has been increasingly recognized, promoting the diffusion of silvicultural practices aimed at considering all components of the forest system. Deadwood is an important component of the forest ecosystem. It plays a fundamental role in providing nutrients and habitats for a wide variety of saprotrophic and heterotrophic organisms and significantly contributes to soil formation and carbon storage. Deadwood is inhabited by a plethora of organisms from various kingdoms that have evolved the ability to utilize decaying organic matter. This community, consisting of both eukaryotic and prokaryotic species, can be defined as “necrobiome”. Through the interactions between its various members, the necrobiome influences the decay rates of deadwood and plays a crucial role in the balance between organic matter decomposition, carbon sequestration, and gas exchanges (e.g., CO2) with the atmosphere. The present work aims to provide an overview of the biodiversity and role of the microbial communities that inhabit deadwood and their possible involvement in greenhouse gas (CO2, N2O, and CH4) emissions.
1. Introduction
Forest ecosystems host a large portion of terrestrial biodiversity and play a crucial role in the biogeochemical cycles of the elements, influencing the climate and providing ecosystem services for the well-being of society. Therefore, forest management actions for forest ecosystem preservation represent an important tool for biodiversity conservation and ecosystem functioning [1]. Sustainable forest management (SFM) has become a central guiding principle to preserve forest ecosystems’ biodiversity and functionality, allowing the spread of management practices aimed at considering all forest components. Since the 1990s, the role and importance of deadwood in forest ecosystems have been recognized at a political level. Consequently, deadwood volume has become one of the most important indicators of forest biodiversity belonging to the Pan-European qualitative indicators for SFM [2], and it has been included within the list of the five carbon pools provided by the Intergovernmental Panel on Climate Change [3].
According to the Global Forest Resources Assessment 2005, forest deadwood can be defined as all non-living woody biomass not contained in the litter, either standing, lying on the ground, or in the soil [4] (Figure 1). Deadwood includes several components such as snags, standing dead trees (including high stumps), lying dead trunks, fallen branches, fallen twigs, and stumps [5].

Figure 1.
Deadwood: standing and lying trunks, fallen branches, and coarse wood debris.
In recent decades, the perception of the importance of deadwood in forest ecosystems has gradually changed. Consequently, forest management actions aimed at regulating the amount of deadwood in forests have been modified. In the past, non-living woody biomass present in forests was perceived negatively for health reasons, such as pests, insect attacks, [6] or greater risk of fires [7], and deadwood was generally removed during silvicultural operations. Nowadays, deadwood is recognized as an essential structural and functional component of forests [8], with a central role in sustaining forest biodiversity and delivering ecosystem services (ESs). It functions as a habitat for numerous plant and animal species (Figure 2), an essential substrate for numerous insects [9] and fungi [10], a key factor in nutrient cycling [11], a fundamental element in the geomorphological and soil hydrological processes [12], and a valuable forest carbon pool contributing to climate change mitigation [13,14].

Figure 2.
Deadwood provides a favorable environment for the regeneration of plant species and natural shelters for animals.
The management of forests for timber and bioenergy production while maintaining and/or improving other ecosystem services has been one of the most significant research challenges since the concept of ESs was developed [15]. The amount of deadwood in a forest depends on a set of natural and anthropogenic variables: forest type, stage of forest development, local geo-climatic situation, and the kind and frequency of anthropogenic disturbances. In particular, the qualitative and quantitative presence of deadwood in a forest is influenced by both forest systems (coppice or high forest) and management intensity [16,17]. Silvicultural strategies play a crucial role in maintaining or increasing the volume of deadwood in forests. Thus, ad hoc solutions and the well-designed planning of different silvicultural actions may be fundamental to achieving a balance between forest productive functions and deadwood environmental benefits [17].
In forest ecosystems, most biodiversity, even at the intraspecific level, is found among bacteria, fungi, protozoa, lichens, and arthropods [18]. These organisms live mainly in the soil, but they are also abundant in the necromass (deadwood, litter, animal carcasses), where they are key actors in decomposition processes [19]. Two types of organisms, which depend on deadwood presence in the forest ecosystem, can be distinguished: (i) directly dependent organisms that use the deadwood as a substrate for germination, as a power source, or as a nesting site, and (ii) indirectly dependent organisms that occasionally find shelter in the coarse woody debris or in standing dead trees at either an early or advanced level of decomposition [20].
Considering the central role of deadwood in the biological diversity and functionality of forest ecosystems, studies concerning its typology and abundance, and the mechanisms involved in its decomposition, are of increasing interest.
2. The Necrobiome of Deadwood
The term necrobiome was originally used to describe “the community of species (e.g., prokaryotic and eukaryotic) associated with decomposing remains of heterotrophic biomass, including animal carrion and human corpses” [21]. This term, initially focusing mainly on vertebrate carrions, was later expanded by Benbow et al. [22] to include other forms of necromass, such as leaves, wood, and dung. Therefore, the term necrobiome can be associated with vegetable and animal necromass decomposition. Some aspects of the decomposing action of the necrobiome are common to all types of necromass, such as tissue disintegration, microbial activity, and the release and recycling of nutrients within the ecosystem. Conversely, other aspects are characteristic of each different form of necromass, such as community dynamics, decomposition rates, and specific decomposing taxa [22].
Deadwood is an important source of organic matter consisting of simple sugars, organic acids, and complex structural biopolymers such as cellulose, hemicellulose, and lignin. It is a crucial factor in nutrient recycling in forest soil [19]. In fact, through decomposition, the structural polymers of wood cell walls are demolished and transferred to the soil, making the nutrients contained therein available for soil microorganisms and absorption by plant roots [14].
The refractoriness of the lignin–cellulosic complexes is the main driver controlling the decay rate of deadwood. Only a limited number of fungi and prokaryotes possess the capability to decompose the lignin–cellulosic complexes, as they have developed the enzymes necessary to break them down [23]. The action of pedofauna, consisting mainly of invertebrates such as xylophagous insects (Figure 3), is fundamental in exposing a larger surface of the wood to microorganism attack. Through the chewing apparatus, the pedofauna fragment deadwood, favoring the mixing of organic molecules and soil mineral components [24].

Figure 3.
The arthropods inhabiting deadwood contribute to its fragmentation.
Unicellular protozoa can also significantly contribute to mineralization processes and the recycling of the nutrients contained in deadwood [25].
The deadwood necrobiome is composed of organisms belonging to different kingdoms that have evolved to use decaying organic matter as a nutrient resource or as a habitat, interacting with each other through relationships that can be synergistic, antagonistic, and/or neutral [26]. Through these interactions, the necrobiome influences the deadwood decay rates and plays a crucial role in the balance between organic matter decomposition, carbon sequestration, and gas exchanges with the atmosphere (mainly carbon dioxide; CO2) [27,28].
The necrobiome colonizes plant tissues immediately after or before their death and may have an internal (endonecrotic community) or external (epinecrotic community) origin [22]. Climate and the quality of the woody substrates (density, pH, moisture content, total lignin, and cellulose contents) are the driving forces of deadwood decomposition, as they strongly influence the structure of the necrobiome and the speed of the decomposition process [29,30]. As the woody material decays, its structure and chemical composition gradually change, inducing a succession of microbial communities, as species are progressively replaced by other species that are better adapted to the new habitat [31,32,33,34,35,36]. The incorporation on the soil surface organic layer of woody material at various stages of decomposition can have consequences on the chemical and biological properties of forest soil [37].
While it remains largely unclear which factors influence each group of organisms and to what extent, there is no doubt that deadwood offers a wide range of niches for many specialized organisms [38] and that its decomposition largely contributes to sustaining forest biodiversity [39].
2.1. Fungi
Fungi, in particular Basidiomycetes (Figure 4) and Ascomycetes, have always been considered pioneering microorganisms in the wood decomposition process [33]. Thanks to their ability to secrete various enzymes that attack the structural biopolymers of wood, fungi can readily colonize deadwood and modulate the availability of nutritional resources for their growth [23,33]. Furthermore, they promptly contribute to opening the way in the recalcitrant substrate for colonization by other microorganisms, including bacteria [40]. Several authors found that Ascomycetes are more abundant than Basidiomycetes, especially in the early phases of deadwood decomposition [28,33,41,42].

Figure 4.
Deadwood as a substrate for the growth of Basidiomycete fungi.
Endophytes are generally the first colonizing fungi [42]. Outside taxa preferentially take over in the later stages [43,44]. Fungi use two main colonization strategies: the dispersion of spores in the atmosphere and the invasion of mycelial filaments from the surrounding soil [45]. Following these stochastic events, the ability of certain fungal groups to degrade different substrates leads to temporary changes in the chemical composition of deadwood. As the state of decay progresses, a greater number of ecological niches become available to support the colonization of a highly diversified microbial community. As the wood decomposes, a greater variety of substrates gradually becomes available, requiring a greater metabolic diversity within the active bacterial and fungal communities [36,42].
Fungal taxa exhibit specific preferences for wood at a definite stage of decay [46]. Based on the different degradation strategies they operate, fungi are commonly classified into soft rot fungi, white rot fungi, and brown rot fungi. The soft rot fungi are mostly Ascomycetes and Deuteromycetes and are among the first colonizers [47,48]. They can degrade cellulose and hemicellulose, creating localized gaps that allow access to the necromass for other decomposing microorganisms. The term soft rot was originally proposed by Savory in 1954 [49] to distinguish the decay caused by these fungi, which attack only the cellulose, making the surface of the wood very soft, from the action of the white and brown rot fungi, which, conversely, destroy the wood [50]. White rot fungi and brown rot fungi are mainly taxonomically classified within the subdivision of Basidiomycetes [51,52]. White rot fungi secrete a plethora of extracellular oxidative enzymes capable of degrading all cell wall biopolymers [14,53]. They significantly reduce the lignin content of deadwood thanks to the action of oxidative metalloenzymes, such as laccase and manganese peroxidase [23,54,55]. Some species preferentially remove the lignin, leaving white degraded areas mainly consisting of cellulose, while other species concurrently degrade lignin and cellulose [56]. After a first phase dominated by soft rot and white rot fungi, the decay process is predominantly driven by brown rot fungi, which can attack hemicellulose and cellulose, causing a relative increase in lignin [48,52]. They are not able to mineralize lignin into CO2 and do not secrete peroxidase, but they apply a singular mechanism that uses hydroxyl radicals, produced by the non-enzymatic Fenton reaction, as an oxidizing agent [23,52]. Therefore, the lignin is modified while maintaining its polymeric structure [52]. Despite having limited ligninolytic activity, brown rot fungi are good competitors and are abundant in the late stages of deadwood decomposition [31,52,57].
Hypoxylon frangiforme and Lopadostoma turgidum were found almost exclusively on deadwood at early stages of decomposition while Trechispora farinacea and Phanerochaete velutina were more frequently isolated from deadwood at late decay stages [58]. A key player in Norway spruce decomposition is Guehomyces pullulans, which is very efficient in conquering the habitat in the early stages of decomposition and, successively, as the decay progresses, providing its biomass to feed other species [42]. The white rot fungus Phlebia radiata proved to be an efficient early colonizer of deadwood, contributing to wood decomposition by the secretion of oxidative and carbohydrate-active enzymes [23]. In Norway spruce deadwood, these enzyme activities were associated with the emission of volatile organic compounds (VOCs), such as methyl-3-furoate [23]. Fungal VOCs are considered important signaling molecules in hyphal interspecific interactions.
Several species, such as Eutypa spinosa and Fomes fomentarius, have been identified as endophytes latently present in the living sapwood of the European beech [59]. They can utilize the available nutritional resources early, before the entry of secondary and late colonizers who may be stronger competitors [60]. Other examples of late colonizers are Mycena haematopus and Pluteus spp. [57]. The species Resinicium bicolor (Hymenochaetales), Fomitopsis pinicola (Polyporales), and Heterobasidion spp. (Russulales) have been found in the deadwood of conifers (Picea abies) in temperate and boreal forests [61,62,63]. Trametes versicolor (Polyporales) and members of Xylariales have frequently been found in European temperate beech and oak forests. Resinicium spp. have been considered functional and structural key members of the necrobiome and are abundant in deadwood of both deciduous and coniferous species [64].
Furthermore, Mucoromycota were found in a significant percentage (11%) in Norway spruce deadwood blocks, almost exclusively referring to the genus Mucor [42]. Although this phylum is not considered a typical deadwood decomposer, it is presumed to be involved in facilitating the breakdown of complex sugars [57].
2.2. Bacteria
Bacteria are also involved in the deadwood decomposition process [65] (Figure 5). They colonize deadwood in early decay stages growing on easily degradable substrates such as sugars, organic acids, pectin, and easily accessible cellulose. In recent years, it has been shown that even bacteria can degrade wood’s structural biopolymers, including lignin, and catabolize secondary products deriving from lignin’s incomplete degradation by fungi [66,67]. However, their role in cellulose and lignin decomposition is lesser than that of fungi. Tláskal et al. [65] found that more than 91% of the transcripts involved in the degradation of structural wood biopolymers were of fungal origin, while only 7% were assigned to bacteria. By using a gene-centric approach, selecting 60 enzyme-encoding genes putatively involved in lignin depolymerization and the metabolism of lignin-derived aromatic compounds, Díaz-García et al. [67] predicted that some species within the Pseudomonadaceae family could possess broad and relevant ligninolytic activity.
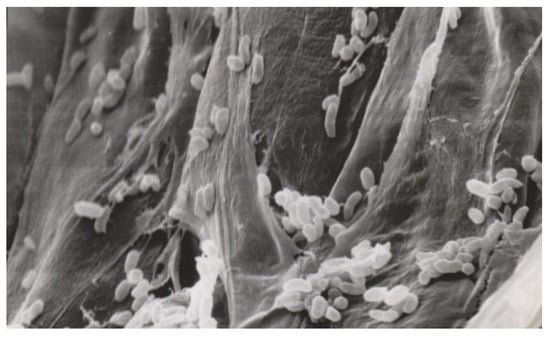
Figure 5.
Bacterial cells colonize dead plant tissue (scanning electron microscope photo, 2620× magnification).
In 1971, Greaves [68] first conceptualized a functional classification of bacteria inhabiting deadwood: (i) bacteria that affect permeability but do not significantly alter the strength of the wood material, (ii) bacteria that affect the wood strength properties by attacking wood structures, (iii) bacteria that act as synergistic members of the total microflora, contributing to the ultimate deadwood breakdown, and (iv) “passive” bacteria, which can act as antagonists to other bacteria. Bacterial pectinases play a key role in increasing wood permeability [69].
Based mainly on electron microscope ultrastructural observations, three main types of bacterial decay, clearly different from those operated by fungi, have been recognized: erosion, tunneling, and cavitation [56]. The erosion bacteria degrade the cellulose and hemicellulose of the secondary walls, producing deep channels parallel to the cell-wall microfibrils and leaving lignin residues. The tunneling bacteria produce tiny tunnels in the secondary walls and medium lamellae. The cavitation bacteria form small diamond-shaped or irregular cavities in the secondary wall [56]. While tunneling bacteria appear to require the presence of oxygen for their activity, erosion bacteria can tolerate conditions of extremely low oxygen levels [70].
Most deadwood saproxylic bacteria belong to the Proteobacteria, Actinobacteria, and Acidobacteria phyla [71]. They participate in the whole deadwood decay, being able to use a wide variety of more or less labile substrates and interacting with fungi through complex synergistic and competitive relationships [72,73,74]. Bradyrhizobium and Caulobacter were found to be the most frequent proteobacterial species in deciduous temperate mixed forest ecosystems [28]. The presence of Bradyrhizobium could potentially contribute to N-enrichment, thanks to its nitrogen (N2)-fixing activity [28].
Deadwood is a favorable environment for the growth of actinobacteria. Many actinobacterial species can secrete cellulases and hydrolytic enzymes that degrade cellulose and hemicellulose [72]. Furthermore, they are presumably also involved in lignin degradation [75], although their role is not yet clear. Lynd et al. [76] retain the actinobacteria as early colonizers of deadwood; thanks to their cellulolytic action, they contribute to increasing the permeability of water and the humidity of the wood, thus favoring fungal colonization [77]. On the other hand, in the more advanced stages of decomposition, a greater metabolic specialization is hypothesized for actinobacteria, thus confirming their limited ability to degrade lignin [78]. Pastorelli et al. [36] found a greater number of actinobacterial taxa in the early stages of deadwood decay than in the more advanced phases. This finding suggests a greater involvement of this bacterial group in the degradation of more labile structural compounds, such as hemicellulose and cellulose, compared to more recalcitrant compounds, such as lignin.
Although the cultivation of the acidobacterial group has proved to be challenging, some members belonging to subdivision 1 have been isolated from deadwood colonized by the white rot fungus Hypholoma fasciculare [73]. Subdivision 1 hosts acidobacteria preferring a moderately acidic pH range [79] which are, therefore, presumably well adapted to the deadwood environment. By using an Illumina MiSeq platform, Lee et al. [28] identified Terriglobus as the most cosmopolitan species in the acidobacterial deadwood community. Acidobacteria have been described as abundant in the forest soil methylotrophic community [80]. However, their physiology and role in deadwood decomposition are still poorly known.
A significant portion of the deadwood prokaryotic necrobiome consists of bacteria capable of using reduced carbon substrates without CC bonding, such as methanol, a by-product of the lignin decomposition operated by fungi [65]. Vorob’ev et al. [81] identified the presence of methylotrophic microorganisms in association with the H. fascicular fungus, in decaying beech wood. The obligate methanotroph Methyloferula sp. was found in association with Amelanchier arborea deadwood [28]. The group of methylotrophic bacteria also includes members with the ability to oxidize methane (CH4), the so-called methanotrophic bacteria. In a mesocosm experiment performed on Pinus nigra deadwood fragments, methanotrophic bacteria increased with increasing decay class and CH4 consumption, suggesting relatively greater involvement of this microbial group as decomposition progresses [36]. Mäkipää et al. [82] found methanotrophs as the main group within the N2-fixing prokaryotic community in spruce deadwood. They assumed the presence of synergistic interactions between methanotrophs and fungi, with the former providing ammonium (NH4+) to the fungi in return for the methanol produced by the latter [83].
Nitrogen-fixing bacteria also significantly increase as decomposition progresses [36,84]. Using a meta-transcriptomic approach to analyze the microbiome associated with European beech deadwood decay, Tláskal et al. [65] found that N2-fixation is one of the dominant processes of the N cycle occurring in deadwood, second only to the incorporation of NH4+ into organic molecules. On the contrary, the respiratory pathway (denitrification) that reduces nitrates (NO3−) and nitrites (NO2−) seems to be considerably less important in deadwood than in soil, and the transcripts related to the nitrification process were absent. Denitrifying bacteria are abundant and widespread in forest soils [27]. Denitrification is a stepwise process that involves several enzymes (reductases) and results in the conversion of dissolved NO3− and NO2− to molecular nitrogen (N2), passing through the production of nitrous oxide (N2O) [85]. Not all denitrifying bacteria harbor the complete battery of genes encoding for all the reductases of the denitrification process. Some denitrifying species have a truncated metabolic pathway [85]. Pastorelli et al. [36] quantified two key denitrification genes, the nirK, and nosZ genes, and N2O potential emission from black pine deadwood. The obtained results suggested that the deadwood necrobiome may host a diversity of species that could drive denitrification towards a complete reduction in nitrate up to the release of N2, thus lowering N2O emissions.
The amoA gene involved in NH4+ oxidation, the first step of the nitrification pathway, was detected only at low levels both in the bacterial and archaeal communities of deadwood [36,65]. Little is known about the role of this group of prokaryotes in wood decomposition. They are also involved in N2O emissions [86], but their contribution to deadwood appears potentially less relevant than that of denitrifying bacteria.
2.3. Archaea
Little is known about the metabolic activities of the prokaryotes belonging to the Archaea domain and their involvement in deadwood degradation. Enzymes such as cellulase and xylanase have been discovered in extremophilic archaea [87], but it is not known whether these enzymes may also be present in temperate archaea [88].
Numerous members of this domain can produce CH4 as a metabolic by-product. The CH4 emission is the result of the activity of a consortium of microorganisms, where simple C-compounds are produced by the degradative and/or fermentative activity of other microorganisms and used as terminal electron acceptors by methanogenic archaea [89]. The colonization of living tree tissue by methanogenic archaea was documented as early as the 1970s [90]. The early stages of decomposition showed the highest methanogenic activity [91]. This finding suggested that methanogenesis is fueled by non-structural labile C substrates, most abundant in less-decayed wood [91]. Fungi break down cell structural biopolymers, generating by-products that other wood microorganisms may use to produce CO2 and H2, primary substrates for the methanogenesis process [92]. However, non-methanogenic archaea have also been found in decaying wood, indicating archaea as integral and dynamic members of the plant necrobiome [88].
To date, Thaumarchaeota have been found as prominent members of the archaea community of forest necromass, highlighting the versatility and cosmopolitan nature of this phylum in the natural environment [88]. However, it is easier to assume that CH4 evolution occurs mainly as a result of symbiotic interactions between methanogenic archaea and xylophagous insects, protozoa, or fungi inhabiting the deadwood [92,93,94]. In a mesocosm experiment conducted on P. nigra deadwood, Pastorelli et al. [36] found the presence of Methanobrevibacter strongly correlated with the high production of CH4 (Figure 6). Methanobrevibacter is a strictly anaerobic species that generally live as a symbiote of protozoa or are attached to the intestinal epithelium of both lower and upper termites. Furthermore, a great abundance of Methanobrevibacter has also been found within the cells of Spirotrichonympha leidyi, a flagellate of the parabasalid group that lives in the termite intestine [94].
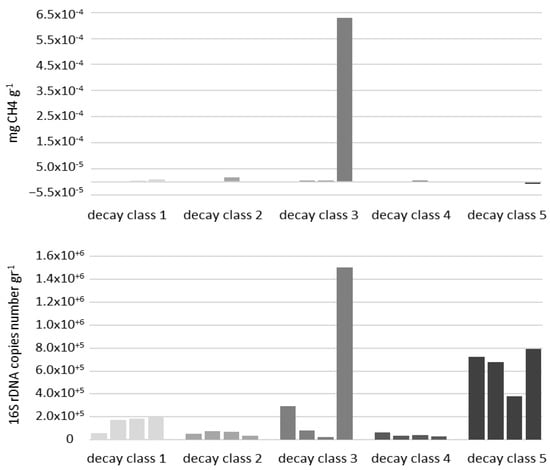
Figure 6.
Maximum values of CH4 production and number of methanogenic archaeal 16S rDNA gene copies resulting from a mesocosm experiment consisting of the cores of five different decay classes from black pine (Pinus nigra J.F. Arnold) lying deadwood. The sequence analysis conducted on the sample with high CH4 emission showed a great abundance of Methanobrevibacter [36].
2.4. Ciliate
Ciliophora is one of the most abundant phyla of the protozoan community in soil, but ciliates can also inhabit mosses, lichens, litter, and deadwood [95,96]. Ciliates are important members of the ecosystem trophic network since they prey on microorganisms and are preyed upon by other protozoa and metazoans, thus playing a very central role in nutrient recycling [95]. Understanding ciliate functions and diversity is essential to extending our knowledge about the nutrient cycle in forest ecosystems. However, most ecological studies on this group of protozoa have been conducted on the soil, and reliable data on their abundance and taxonomic composition in deadwood are still scarce.
Different ciliate species have specific food preferences and tolerate specific microclimates and abiotic conditions [96]. Ciliates are highly adaptable to environmental changes and, thanks to their ability to develop inactive forms (cysts), they can survive adverse conditions [97], such as wet–dry alternations. Pastorelli et al. [98] showed that, like the other members of the necrobiome, the composition of the ciliate community varies as deadwood decays, becoming more and more homogeneous (Figure 7). The early stages of deadwood decomposition are characterized by great variability in deadwood quality, probably due to stochastic events that led to colonization by bacterial and fungal taxa with different degrading capacities [45] and ciliate taxa with different food preferences. As the decomposition progresses, the bacterial, fungal, and ciliate taxa are mainly selected according to deterministic mechanisms [99] controlled by the metabolic processes involved in the degradation of complex wood residues and by the palatability of the degrading microorganisms [19].
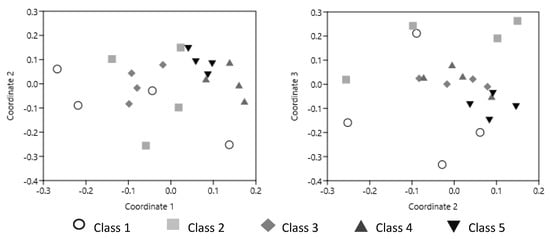
Figure 7.
nMDS ordination plots of ciliate 18S rDNA gene obtained by PCR-DGGE from black pine deadwood at different decay classes [98].
Ciliates are ubiquitous and can be easily dispersed in the air [97]. They are susceptible to a wide range of environmental factors, such as humidity, temperature, pH, and food abundance, which can induce changes in their community composition [100,101]. Since ciliates need water to be active, the daily fluctuations of temperature and humidity can induce cycles of encystment and excystment so that, due to the different tolerance to abiotic conditions and food preferences, at any moment the habitat is occupied by active and inactive individuals [96].
The ciliate trophic groups in deadwood are very similar to those in soil with the prevalence of bacterivores and predators in the most decomposed deadwood [102]. Through the predation and secretion of metabolites, ciliates regulate the size and composition of bacterial communities and influence C and N cycling [19]. The nutrients temporarily immobilized in the bacterial biomass are released by the ciliate predatory action. Overall, deadwood ciliates affect the rates of nutrients released into the soil and atmosphere and significantly contribute to improving plant growth [100]. Part of the ingested C is used for new ciliated biomass production while the rest is returned to the atmosphere as CO2. Organic molecules not used by any species of fungi, bacteria, or ciliates become a non-recycled end-product and accumulate in the soil, contributing to humus formation [96]. Nitrogen excesses are excreted as NH4+, readily available to other organisms, improving the total N content of deadwood and soil fertility [100]. Consistently, the abundance of ciliates in deadwood was found to be positively correlated with CO2 production, N content, and bacterial abundance [98].
The ciliate species identified in deadwood generally belong to soil-inhabiting genera within the Colpodea and Spirotrichea classes [98]. Colpodea are abundant in soils, especially in polluted soil [103,104], and are typically bacterivorous [95]. Interestingly, Jia et al. [19] showed a vesicular Colpodea strongly related to catalase and polyphenol oxidase, both enzymes involved in the degradation of refractory C sources, such as lignin. This finding suggested a potential role of Colpodea in deadwood degradation. Spirotrichea are common in soil, freshwater, and marine environments. Bartošová and Tirjaková [102] identified Colpodea, Spirotrichea, and Lithostomes as dominant systematic groups in decaying bark and wood.
3. Factors Affecting Decomposition
The wood–soil contact enhances the ability of the necrobiome to colonize deadwood by facilitating the microbial species to move from soil to wood residues and by increasing the deadwood’s moisture [105]. Jaroszewicz et al. [106] found that the composition of fungal assemblage strongly depends on the degree of deadwood debris’ contact with soil (on the ground, underground, or aboveground). Several studies on Mediterranean forests have shown that humidity has a higher influence on the activity of enzymes such as endoglucanase, cellulose, and peroxidase than temperature. The activity of these enzymes is strongly correlated to cold/hot or humid/dry alternations, typical characteristics of the Mediterranean climate [107].
Baldrian et al. [108] found high variability in deadwood chemistry with the general trend of increasing N content and decreasing pH as decomposition progresses. The N content appears to be a factor that significantly affects the composition of the fungal community. In the early stages of decomposition, the low wood N content represents a more important and limiting factor for fungal growth than the recalcitrance of structural biopolymers such as lignin.
Many saproxylic fungi are host-specific, e.g., Fomitopsis rosea and Daedalea quercina exhibited preferences for spruce and oak deadwood, respectively. Furthermore, several taxa have a distinct predilection for deadwood from gymnosperms or angiosperms [106]. Alongside decomposition, deadwood undergoes changes mainly related to the reduction in density and the accumulation of nutrients and lignin compounds [29,57]. The rate of density reduction depends on the wood’s initial density and varies according to the tree species [107], as different species contain different concentrations of structural compounds such as lignin. Lignin represents a barrier to wood degradation because it protects cellulose and hemicellulose from microbial attack [109]. By degrading lignin, the specialized ligninolytic microorganisms (Basidiomycota and some Ascomycota) increase the accessibility of the non-ligninolytic microorganisms to the other structural carbohydrates [109]. In conifers, the wood density decreases more slowly than—for example—in birch, which decomposes completely in a rather short period of about 25–40 years [110]. In natural boreal forests, Picea abies wood density is considered an indicator of the rate of decomposition [108]. The composition of the woody debris of broad-leaved species differs considerably from that of conifers [32]. Deciduous trees generally have high-quality organic components, with a higher nutrient content, the presence of more labile compounds, and a lower concentration of lignin and polyphenolic compounds than conifers [32]. Therefore, the woody structure of broadleaf trees is better suited to promote the faster decomposition of wood residues [107]. Fomitopsis rosea and Xeromphalina campanella have been proposed as saproxylic indicator species for spruce and general conifer deadwood, respectively [106]. Daedalea quercina, Peniophora quercina, Panellus stipticus, Xylobolus frustulatus, Jackrogersella multiformis, and Hymenochaete rubiginosa have been hypothesized as saproxylic indicator species for the deciduous oak and hornbeam [106]. Lee et al. [28] suggested that fungal taxa may be more specialized to a specific wood substrate than bacterial taxa. The substrate specificity expressed by the bacterial and fungal species is more evident in the early stages of decomposition than in the late ones, suggesting the progressive homogenization of woody substrates as the decay proceeds [28].
Furthermore, concerning the relation between C concentration and decomposition, C content can vary within individual trees and between species [111]. Deadwood has a high initial C/N ratio which tends to decrease during decomposition because of N inputs due to fungal N translocation [65], N2-fixation by bacteria [112], and N secretion by protozoa [100,101]. Carbon concentration increases during the decay process with the loss of polysaccharides and the related accumulation of lignin during the decay process [113]. However, in some cases, the percentage of C may not differ significantly between the decay classes. Potassium is released faster than other nutrients by leaching, but it is also subject to immobilization by microorganisms, while the concentration of phosphorous increases with a species-specific pattern [114].
4. Interspecific Interactions within the Necrobiome
Within the necrobiome, various microbial members can coexist, interact, and influence each other. Numerous intra- and inter-specific interactions occur within the forest necrobiome. The efficient cellulose-degrading Trichoderma species may benefit from the delignification process carried out by white rot fungi [42]. On the other hand, by covering naked wood, fungi belonging to Sistonema and Resiniium spp. may slow down the development of other fungal species and hinder the xylophagous invertebrate deadwood colonization [106].
During the deadwood decomposition process, both mutualistic and antagonistic interactions have been observed. These interactions are the driving forces of the functioning of nutrient recycling [115,116]. By degrading the structural biopolymers of deadwood, fungi weaken the lignin barriers and release readily degradable oligomers, providing opportunities for bacterial cell access and growth [117]. At the same time, bacteria can also make wood more accessible to fungi [118], particularly in the early stages of decomposition. Fungi could also facilitate colonization by soil bacterial cells adhering to their exploratory hyphae [73]. On the other hand, the decomposition processes operated by fungi lower the pH and generate reactive oxygen species, creating an adverse and selective environment for bacterial colonization [54]. It has been found that the white rot fungi H. fasciculare and R. bicolor can produce toxic compounds, sesquiterpenoid antibiotics, with bactericidal and/or bacteriostatic effects that counteract the deadwood colonization by bacteria or reduce the number of bacteria that have already colonized it [73]. Bacteria can also have negative effects on the fungal community by competing for low-molecular-weight nutrients. On the other hand, bacteria provide fungi with limiting nutrients, such as N (through N2 fixation), and growth factors, such as vitamins [54]. Furthermore, bacteria can detoxify the deadwood environment by eliminating fungal growth inhibitory compounds [116]. Fungi may meet their N needs for vegetative and generative growth through association with N2-fixing bacteria. In the deadwood logs of Fagus sylvatica and P. abies, Hoppe et al. [84] found positive correlations between fungal sporocarps and the abundance of nifH genes (coding for dinitrogen reductase, a key enzyme in the N2-fixing process). Along the same lines, Gómez-Brandón et al. [42] observed that in P. abies coarse woody debris at different stages of natural decay, the abundance of fungi was strongly correlated with the abundance of nifH gene copies.
All the organisms inhabiting deadwood contribute in some way to transforming the debris of autotrophic origin and introducing the released nutrients and organic matter into the trophic network of the forest ecosystem. The term “microbial loop”, or microbial chain, proposed by Azam et al. [119], describes the process that acts as a link between necromass and the classical trophic network, ensuring the recycling of nutrients and energy contained in the organic matter [100]. Bacteria and fungi are primary saprotrophs. By degrading the structural biopolymers of wood, they contribute to the release of nutrients [96]. A certain number of primary saprotrophic taxa is necessary to provide a diversity of enzymes degrading the various wood substrates and to occupy the different microhabitats characterized by a variety of environmental conditions. Fungal and bacterial number and activity increase as decomposition progresses. In the late decay classes, most of the deadwood biomass is immobilized within their biomass, in particular as cell wall molecules (chitin and murein, respectively) [96]. Primary saprotrophs are preyed upon by secondary saprotrophs, including specialized ciliates [102]. Fungivores and bacterivores digest the primary saprotrophs and transform these cells and their walls into new biomass. Thus, biomass is transferred to a subsequent trophic level. Therefore, the increase in bacteria and fungi is followed by an increase in ciliate predators [98]. Different species of ciliates have specific food preferences, thus influencing the dynamics of bacterial and fungal communities and, consequently, the expression of genes encoding for enzymes associated with decomposition [19].
The microbial chain that develops in deadwood promotes the mineralization of organic matter, increases the availability of nutrients (C, N, P, and S), and supports plant growth [96,100]. The transfer of nutrients to plants through the predation of microbial biomass by ciliates has been demonstrated in numerous studies. In the plant rhizosphere, ciliates can mobilize about one-third of the consumed N as NH4+ by consuming rapidly growing bacterial biomass [120].
5. Role of the Necrobiome in Climate Change
Deadwood represents a significant fraction of the forest C stock, accounting for 10–20% of the total C pool in mature forests [121]. However, this C pool is transient because, through the transformations operated by saproxylic microorganisms, it is released into the atmosphere mainly as CO2, while the rest is transferred to litter, leached in soil water, or sequestered in the soil [65]. Therefore, microorganisms play a crucial role in maintaining the balance between the decomposition of organic matter and C sequestration in forests [27]. The double role of deadwood as a sink and source of C has a high impact on biogeochemical cycling and climate change [122]. Deadwood decomposition, together with forest fires, is one of the main processes that release CO2 from the earth’s surface with strong implications for temperature increases. In situ measurements of CO2 emissions from deadwood highlighted an increase in deadwood respiration correlated with the progression of decomposition and bacterial abundance [35] (Figure 8).

Figure 8.
Portable infrared gas analyzer operating in closed-path mode for the in situ measurement of CO2 production from deadwood [35].
Climate change is expected to alter forest species distribution, particularly at higher latitudes and altitudes, affecting groups of decomposers that colonize specific forest species [123]. Where temperature is a limiting factor, climate warming can increase forest productivity, leading to the increased storage of C [124]. However, microbial activity, the faster decomposition of organic matter, and net CO2 release are expected to increase as temperatures rise. Microbial activity increases under high CO2 concentrations, favoring a more rapid decomposition of the organic substance [125].
Climate changes are also believed to affect N dynamics in forests, in particular in boreal ecosystems. A slight increase in N availability could improve deadwood decomposition by influencing the response of fungi forming mycelial cords [126]. However, other studies have shown that an N excess can reduce the activity of fungal ligninolytic enzymes [127]. Higher temperatures may also cause an acceleration of the nitrification and denitrification processes [27]. Denitrification is a process that leads to N2O emissions operated by bacteria widely distributed in the environment and showing high taxonomic diversity [85].
Deadwood can also be a significant source of CH4 emissions [34,36,128]. The production of CH4 is mainly the result of deadwood decomposition mediated by members belonging to the archaea domain (Figure 6). Recently, brown rot fungi have been also identified to produce CH4 under anaerobic conditions [129].
However, there is currently little understanding of the relationship between microbial diversity and deadwood decomposition. All the information obtained on the microbial species or functional groups involved in the decomposition processes is of fundamental importance for the prediction of the responses of forest ecosystems to climate change. New data are needed to help in clarifying the processes that regulate global warming and the flows of greenhouse gases from deadwood [130].
6. Conclusions
New knowledge on the deadwood necrobiome is of critical importance to improve forestry strategies for deadwood management during silvicultural operations. In fact, during the mechanical operations of harvesting deadwood, only the trunks in the first stages of decomposition are generally taken away, because the removal of the most decomposed trunks is difficult. By adopting the principles of naturalistic forest management, an amount of 20–50 m3 ha−1 of deadwood should be preserved, preferably concentrating in networks of senescence islands (1–2 ha each) with an equal distribution by decay classes. Forest managers should consider this aspect to maintain and improve biodiversity in managed and semi-natural forests.
Author Contributions
Conceptualization, R.P., I.D.M. and A.L.; writing—original draft preparation and reviewing, R.P., I.D.M. and A.L. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financially supported by the LIFE program, in the context of FoResMit (LIFE14/CCM/IT/905) “Recovery of Degraded Coniferous Forests for Environmental Sustainability Restoration and Climate Change Mitigation” and SelPiBio (LIFE13 BIO/IT/000282) “Innovative silvicultural treatments to enhance soil biodiversity in artificial black pine stands, i.e., for demonstration of innovative silvicultural treatments in artificial black pine stands” projects.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data for the review are from the original publications, which can be found in the reference list.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fichtner, A.; Härdtle, W. Forest Ecosystems: A Functional and Biodiversity Perspective. In Perspectives for Biodiversity and Ecosystems, Environmental Challenges and Solutions, 1st ed.; Hobohm, C., Ed.; Springer: Cham, Switzerland, 2021; Volume 16, pp. 383–405. [Google Scholar]
- Lassauce, A.; Paillet, Y.; Jactel, H.; Bouget, C. Deadwood as a surrogate for forest biodiversity: Meta-analysis of correlations between deadwood volume and species richness of saproxylic organisms. Ecol. Indic. 2011, 11, 1027–1039. [Google Scholar] [CrossRef]
- Penman, J.; Gytarsky, M.; Hiraishi, T.; Kruger, D.; Pipatti, R.; Buendia, L.; Miwa, K.; Ngara, T.; Tanabe, K.; Wagner, F. (Eds.) IPCC–Good Practice Guidance for Land Use, Land-Use Change and Forestry; Intergovernmental Panel on Climate Change (IPCC), IPCC/IGES: Hayama, Japan, 2003. [Google Scholar]
- FAO 2004. Global Forest Resources Assessment Update 2005: Terms and definitions (Working Papers 83/E); Forest Resources Assessment Programme: Rome, Italy, 2004. [Google Scholar]
- Hagemann, U.; Moroni, M.T.; Makeschin, F. Deadwood abundance in Labrador high-boreal black spruce forests. Can. J. For. Res. 2009, 39, 131–142. [Google Scholar] [CrossRef]
- La Fauci, A.; Bagnato, S.; Gugliotta, O.I.; Mercurio, R. Osservazioni preliminari sulla necromassa in popolamenti di pino laricio nel Parco Nazionale dell’Aspromonte. Forest@ 2006, 3, 54–62. [Google Scholar] [CrossRef]
- Camia, A.; Barbosa, P.; Amatulli, G.; San-Miguel-Ayanz, J. Fire danger rating in the European Forest Fire Information System (EFFIS): Current developments. For. Ecol. Manag. 2006, 234, S20. [Google Scholar] [CrossRef]
- Harmon, M.E.; Franklin, J.F.; Swanson, F.J.; Sollins, P.; Gregory, S.V.; Lattin, J.D.; Anderson, N.H.; Cline, S.P.; Aumen, N.G.; Sedell, J.R.; et al. Ecology of coarse woody debris in temperate ecosystems. Adv. Ecol. Res. 1986, 15, 133–302. [Google Scholar]
- Deng, Y.; Bai, Y.; Cao, R.; Jiang, Y.; Wang, Z.; Li, F.; Gong, H.; Yang, W. Key drivers of soil arthropod community shift across a subalpine forest series vary greatly with litter and topsoil layers. Eur. J. Soil Biol. 2022, 111, 103421. [Google Scholar] [CrossRef]
- Herrero, C.; Krankina, O.; Monleon, V.J.; Bravo, F. Amount and distribution of coarse woody debris in pine ecosystem of north-western Spain, Russia and the United States. IForest 2014, 7, 53–60. [Google Scholar] [CrossRef]
- Krankina, O.N.; Harmon, M.E. The impact of intensive forest management on carbon stores in forest ecosystems. World Res. Rev. 1994, 6, 161–177. [Google Scholar]
- Bragg, D.C.; Kershner, J.L. Coarse woody debris in riparian zones. J. For. 1999, 4, 30–35. [Google Scholar]
- Ravindranath, N.H.; Ostwald, M. Carbon Inventory Methods: Handbook for Greenhouse Gas Inventory, Carbon Mitigation and Roundwood Production Projects; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Piaszczyk, W.; Lasota, J.; Błońska, E. Effect of organic matter released from deadwood at different decomposition stages on physical properties of forest soil. Forests 2019, 11, 24. [Google Scholar] [CrossRef]
- Dunker, R.; Bullock, D.; Bollero, G.; Armstrong, K. A system to evaluate prime farmland proclamation success based on spatial soil properties. In Proceedings of the American Society for Surface Mining and Reclamation, Tupelo, MS, USA, 8–15 June 2012; pp. 103–132. [Google Scholar]
- Fridman, J.; Walheim, M. Amount, structure, and dynamics of dead wood on managed forestland in Sweden. For. Ecol. Manag. 2000, 131, 23–36. [Google Scholar] [CrossRef]
- Paletto, A.; Ferretti, F.; De Meo, I.; Cantiani, P.; Focacci, M. Ecological and environmental role of deadwood in managed and unmanaged forests. In Sustainable Forest Management–Current Research; García, J.M., Diez Casero, J.J., Eds.; InTech: Reijeka, Croatia, 2012; pp. 219–238. [Google Scholar]
- Foissner, W.; Berger, H.; Xu, K.; Zechmeister-Boltenstern, S. A huge, undescribed soil ciliate (Protozoa: Ciliophora) diversity in natural forest stands of Central Europe. Biodivers. Conserv. 2005, 14, 617–701. [Google Scholar] [CrossRef]
- Jia, T.; Liang, X.; Guo, T.; Chai, B. Impact of nutrients on protozoa community diversity and structure in litter of two natural grass species in a copper tailings dam, China. Microorganisms 2021, 9, 2250. [Google Scholar] [CrossRef] [PubMed]
- Wolynski, A. Close-to-Nature Forestry in the Trentino/North Italy; Forst und Holz: Berlin, Germany, 2001. [Google Scholar]
- Benbow, M.E.; Lewis, A.J.; Tomberlin, J.K.; Pechal, J.L. Seasonal necrophagous insect community assembly during vertebrate carrion decomposition. J. Med. Entomol. 2013, 50, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Benbow, M.E.; Barton, P.S.; Ulyshen, M.D.; Beasley, J.C.; DeVault, T.L.; Strickland, M.S.; Tomberlin, J.K.; Jordan, H.R.; Pechal, J.L. Necrobiome framework for bridging decomposition ecology of autotrophically and heterotrophically derived organic matter. Ecol. Monogr. 2019, 89, e01331. [Google Scholar] [CrossRef]
- Mali, T.; Mäki, M.; Hellen, H.; Heinonsalo, J.; Bäck, J.; Lundell, T. Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns. FEMS Microb. Ecol. 2019, 95, fiz135. [Google Scholar] [CrossRef]
- Parisi, F.; Pioli, S.; Lombardi, F.; Fravolini, G.; Marchetti, M.; Tognetti, R. Linking deadwood traits with saproxylic invertebrates and fungi in European forests-a review. iForest 2018, 11, 423. [Google Scholar] [CrossRef]
- Domonell, A.; Brabender, M.; Nitsche, F.; Bonkowski, M.; Arndt, H. Community structure of cultivable protists in different grassland and forest soils of Thuringia. Pedobiologia 2013, 56, 1–7. [Google Scholar] [CrossRef]
- Ho, A.; Angel, R.; Veraart, A.J.; Daebeler, A.; Jia, Z.; Kim, S.Y.; Kerckhof, F.M.; Boon, N.; Bodelier, P.L. Biotic interactions in microbial communities as modulators of biogeochemical processes: Methanotrophy as a model system. Front. Microbiol. 2016, 7, 1285. [Google Scholar] [CrossRef]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Forest soil bacteria: Diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef]
- Lee, M.R.; Oberle, B.; Olivas, W.; Young, D.F.; Zanne, A.E. Wood construction more strongly shapes deadwood microbial communities than spatial location over 5 years of decay. Environ. Microbiol. 2020, 22, 4702–4717. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, M.; Cherubini, P.; Sartori, G.; Abiven, S.; Ascher, J.; Bertoldi, D.; Camin, F.; Barbero, A.; Larcher, R.; Egli, M. Decomposition of Norway spruce and European larch coarse woody debris (CWD) in relation to different elevation and exposure in an Alpine setting. iForest-Biogeosci. For. 2015, 9, 154. [Google Scholar] [CrossRef]
- Zhou, L.; Dai, L.M.; Gu, H.Y.; Zhong, L. Review on the decomposition and influence factors of coarse woody debris in forest ecosystem. J. For. Res. 2007, 18, 48–54. [Google Scholar] [CrossRef]
- Rajala, T.; Tuomivirta, T.; Pennanen, T.; Mäkipää, R. Habitat models of wood-inhabiting fungi along a decay gradient of Norway spruce logs. Fungal Ecol. 2015, 18, 48–55. [Google Scholar] [CrossRef]
- Magnússon, R.Í.; Tietema, A.; Cornelissen, J.H.C.; Hefting, M.M.; Kalbitz, K. Tamm Review: Sequestration of carbon from coarse woody debris in forest soils. For. Ecol. Manag. 2016, 377, 1–15. [Google Scholar] [CrossRef]
- Purahong, W.; Wubet, T.; Lentendu, G.; Hoppe, B.; Jariyavidyanont, K.; Arnstadt, T.; Baber, K.; Otto, P.; Kellner, H.; Hofrichter, M.; et al. Determinants of deadwood-inhabiting fungal communities in temperate forests: Molecular evidence from a large scale deadwood decomposition experiment. Front. Microbiol. 2018, 9, 2120. [Google Scholar] [CrossRef]
- Pastorelli, R.; Agnelli, A.E.; De Meo, I.; Graziani, A.; Paletto, A.; Lagomarsino, A. Analysis of microbial diversity and greenhouse gas production of decaying pine logs. Forests 2017, 8, 224. [Google Scholar] [CrossRef]
- Pastorelli, R.; Paletto, A.; Agnelli, A.E.; Lagomarsino, A.; De Meo, I. Microbial communities associated with decomposing deadwood of downy birch in a natural forest in Khibiny Mountains (Kola Peninsula, Russian Federation). For. Ecol. Manag. 2020, 455, 117643. [Google Scholar] [CrossRef]
- Pastorelli, R.; Paletto, A.; Agnelli, A.E.; Lagomarsino, A.; De Meo, I. Microbial diversity and ecosystem functioning in deadwood of black pine of a temperate forest. Forests 2021, 12, 1418. [Google Scholar] [CrossRef]
- Strukelj, M.; Brais, S.; Quideau, S.A.; Angers, V.A.; Kebli, H.; Drapeau, P.; Oh, S.W. Chemical transformations in downed logs and snags of mixed boreal species during decomposition. Can. J. For. Res. 2013, 43, 785–798. [Google Scholar] [CrossRef]
- Moll, J.; Kellner, H.; Leonhardt, S.; Stengel, E.; Dahl, A.; Bässler, C.; Buscot, F.; Hofrichter, M.; Hoppe, B. Bacteria inhabiting deadwood of 13 tree species are heterogeneously distributed between sapwood and heartwood. Environ. Microb. 2018, 20, 3744–3756. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.; Badeck, F.W.; Harmon, M.E. Estimating decomposition rate constants for European tree species from literature sources. Eur. J. For. Res. 2008, 127, 301–313. [Google Scholar] [CrossRef]
- Wei, Y.; Dai, Y. Ecological function of wood-inhabiting fungi in forest ecosystem. J. Appl. Ecol. 2004, 15, 1935–1938. [Google Scholar]
- Longa, C.M.O.; Francioli, D.; Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Pietramellara, G.; Insam, H. Culturable fungi associated with wood decay of Picea abies in subalpine forest soils: A field-mesocosm case study. iForest 2018, 11, 781. [Google Scholar] [CrossRef]
- Gómez-Brandón, M.; Probst, M.; Siles, J.A.; Peintner, U.; Bardelli, T.; Egli, M.; Insam, H.; Ascher-Jenull, J. Fungal communities and their association with nitrogen-fixing bacteria affect early decomposition of Norway spruce deadwood. Sci. Rep. 2020, 10, 8025. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Osono, T.; Takeda, H. Dynamics of physicochemical properties and occurrence of fungal fruit bodies during decomposition of coarse woody debris of Fagus crenata. J. For. Res. 2009, 14, 20–29. [Google Scholar] [CrossRef]
- Voříšková, J.; Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013, 7, 477–486. [Google Scholar] [CrossRef]
- Fukami, T.; Dickie, I.A.; Paula Wilkie, J.; Paulus, B.C.; Park, D.; Roberts, A.; Buchanan, P.K.; Allen, R.B. Assembly history dictates ecosystem functioning: Evidence from wood decomposer communities. Ecol. Lett. 2010, 13, 675–684. [Google Scholar] [CrossRef]
- Yang, S.; Limpens, J.; Sterck, F.J.; Sass-Klaassen, U.; Cornelissen, J.H.; Hefting, M.; van Logtestijn, R.S.P.; Goudzwaard, L.; Dam, N.; Dam, M.; et al. Dead wood diversity promotes fungal diversity. Oikos 2021, 130, 2202–2216. [Google Scholar] [CrossRef]
- Paliwal, R.; Giri, K.; Rai, J.P.N. Microbial ligninolysis: Avenue for natural ecosystem management. In Biotechnology: Concepts, Methodologies, Tools, and Applications; IGI Global: Hershey, PA, USA, 2019; pp. 1399–1423. [Google Scholar]
- Berg, B.; McClaugherty, C. Decomposer organisms. In Plant Litter; Springer: Cham, Switzerland, 2020; pp. 45–65. [Google Scholar]
- Savory, J.G. Breakdown of timber by ascomycetes and fungi imperfecti. Ann. Appl. Biol. 1954, 41, 336–347. [Google Scholar] [CrossRef]
- Daniel, G.; Nilsson, T. Developments in the study of soft rot and bacterial decay. In Forest Products Biotechnology, 1st ed.; Bruce, A., Palfreyman, J.W., Eds.; Taylor and Francis: London, UK, 1997; pp. 47–72. [Google Scholar]
- Ward, G.; Hadar, Y.; Dosoretz, C.G. The biodegradation of lignocellulose by white rot fungi. Mycol. Ser. 2004, 21, 393–408. [Google Scholar]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. In Industrial applications. The Mycota, 1st ed.; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 10, pp. 319–340. [Google Scholar]
- Allen, R.B.; Buchanan, P.K.; Clinton, P.W.; Cone, A.J. Composition and diversity of fungi on decaying logs in a New Zealand temperate beech (Nothofagus) forest. Can. J. For. Res. 2000, 30, 10251033. [Google Scholar] [CrossRef]
- Kielak, A.M.; Scheublin, T.R.; Mendes, L.W.; Van Veen, J.A.; Kuramae, E.E. Bacterial community succession in pine-wood decomposition. Front. Microbiol. 2016, 7, 231. [Google Scholar] [CrossRef]
- Schwarze, F.W.M.R.; Engels, J.; Mattheck, C. Fungal Strategies of Wood Decay in Trees; Springer: Berlin/Hedelberg, Germany; New York, NY, USA,, 2000. [Google Scholar]
- Blanchette, R.A. A review of microbial deterioration found in archaeological wood from different environments. Int. Biodeterior. Biodegrad. 2000, 46, 189–204. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Osono, T.; Takeda, H. Wood decomposing abilities of diverse lignicolous fungi on nondecayed and decayed beech wood. Mycologia 2011, 3, 474–482. [Google Scholar] [CrossRef]
- Blaser, S.; Prati, D.; Senn-Irlet, B.; Fischer, M. Effects of forest management on the diversity of deadwood-inhabiting fungi in Central European forests. For. Ecol. Manag. 2013, 304, 42–48. [Google Scholar] [CrossRef]
- Parfitt, D.; Hunt, J.; Dockrell, D.; Rogers, H.J.; Boddy, L. Do all trees carry the seeds of their own destruction? PCR reveals numerous wood decay fungi latently present in sapwood of a wide range of angiosperm trees. Fungal Ecol. 2010, 3, 338–346. [Google Scholar] [CrossRef]
- Prewitt, L.; Kang, Y.; Kakumanu, M.L.; Williams, M. Fungal and bacterial community succession differs for three wood types during decay in a forest soil. Microb. Ecol. 2014, 68, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Kubartová, A.; Ottosson, E.; Dahlberg, A.; Stenlid, J. Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Mol. Ecol. 2012, 21, 4514–4532. [Google Scholar]
- Ovaskainen, O.; Schigel, D.; Ali-Kovero, H.; Auvinen, P.; Paulin, L.; Nordén, B.; Nordén, J. Combining high-throughput sequencing with fruit body surveys reveals contrasting life-history strategies in fungi. ISME J. 2013, 7, 1696–1709. [Google Scholar] [CrossRef]
- Hoppe, B.; Purahong, W.; Wubet, T.; Kahl, T.; Bauhus, J.; Arnstadt, T.; Hofrichter, M.; Buscot, F.; Krüger, D. Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers. 2016, 77, 367–379. [Google Scholar] [CrossRef]
- Purahong, W.; Pietsch, K.A.; Lentendu, G.; Schöps, R.; Bruelheide, H.; Wirth, C.; Buscot, F.; Wubet, T. Characterization of unexplored deadwood mycobiome in highly diverse subtropical forests using culture-independent molecular technique. Front. Microbiol. 2017, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Tláskal, V.; Brabcová, V.; Větrovský, T.; Jomura, M.; López-Mondéjar, R.; Oliveira Monteiro, L.M.; Saraiva, J.P.; Human, Z.R.; Cajthaml, T.; da Rocha, U.N.; et al. Complementary roles of wood-inhabiting fungi and bacteria facilitate deadwood decomposition. mSystems 2021, 6, e01078-20. [Google Scholar] [CrossRef]
- Probst, M.; Gómez-Brandón, M.; Bardelli, T.; Egli, M.; Insam, H.; Ascher-Jenull, J. Bacterial communities of decaying Norway spruce follow distinct slope exposure and time-dependent trajectories. Environ. Microbiol. 2018, 20, 3657–3670. [Google Scholar] [CrossRef]
- Díaz-García, L.; Bugg, T.D.; Jiménez, D.J. Exploring the lignin catabolism potential of soil-derived lignocellulolytic microbial consortia by a gene-centric metagenomic approach. Microb. Ecol. 2020, 80, 885–896. [Google Scholar] [CrossRef]
- Greaves, H. The bacterial factor in wood decay. Wood Sci. Technol. 1971, 5, 6–16. [Google Scholar] [CrossRef]
- Clausen, C.A. Bacterial associations with decaying wood: A review. Int. Biodeterior. Biodegrad. 1996, 37, 101–107. [Google Scholar] [CrossRef]
- Singh, A.P.; Kim, Y.S.; Singh, T. Chapter 9-Bacterial Degradation of Wood. In Secondary Xylem Biology-Origins, Functions, and Applications; Kim, Y.S., Funada, R., Singh, A.P., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 169–190. [Google Scholar]
- Hagge, J.; Bässler, C.; Gruppe, A.; Hoppe, B.; Kellner, H.; Krah, F.S.; Müller, J.; Seibold, S.; Stengel, E.; Thorn, S. Bark coverage shifts assembly processes of microbial decomposer communities in deadwood. Proc. Royal Soc. B 2019, 286, 20191744. [Google Scholar] [CrossRef]
- De Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Folman, L.B.; Klein Gunnewiek, P.J.; Boddy, L.; De Boer, W. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol. Ecol. 2008, 63, 181–191. [Google Scholar] [CrossRef]
- Sun, H.; Terhonen, E.; Kasanen, R.; Asiegbu, F.O. Diversity and community structure of primary wood-inhabiting bacteria in boreal forest. Geomicrobiol. J. 2014, 31, 315–324. [Google Scholar] [CrossRef]
- Větrovský, T.; Steffen, K.T.; Baldrian, P. Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS ONE 2014, 9, e89108. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology, Microbiol. Mol. Biol. Rev. 2020, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Yang, M.X.; Tu, R. Unexpectedly high bacterial diversity in decaying wood of a conifer as revealed by a molecular method. Int. Biodeterior. Biodegrad. 2008, 62, 471–474. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Kleinsteuber, S.; Müller, F.D.; Chatzinotas, A.; Wendt-Potthoff, K.; Harms, H. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. FEMS Microbiol. Ecol. 2008, 63, 107–117. [Google Scholar] [CrossRef][Green Version]
- Radajewski, S.; Webster, G.; Reay, D.S.; Morris, S.A.; Ineson, P.; Nedwell, D.B.; Prosser, J.I.; Murrell, J.C. Identification of active methylotroph populations in an acidic forest soil by stable isotope probing. Microbiology 2002, 148, 2331–2342. [Google Scholar] [CrossRef]
- Vorob’ev, A.V.; de Boer, W.; Folman, L.B.; Bodelier, P.L.; Doronina, N.V.; Suzina, N.E.; Trotsenko, Y.A.; Dedysh, S.N. Methylovirgula ligni gen. nov., sp. nov., an obligately acidophilic, facultatively methylotrophic bacterium with a highly divergent mxaF gene. Int. J. System Evol. Microbiol. 2009, 59, 2538–2545. [Google Scholar] [CrossRef]
- Mäkipää, R.; Leppänen, S.M.; Munoz, S.S.; Smolander, A.; Tiirola, M.; Tuomivirta, T.; Fritze, H. Methanotrophs are core members of the diazotroph community in decaying Norway spruce logs. Soil Biol. Biochem. 2018, 120, 230–232. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Semrau, J.D.; Liesack, W.; Tiedje, J.M. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 2002, 52, 251–261. [Google Scholar] [CrossRef]
- Hoppe, B.; Kahl, T.; Karasch, P.; Wubet, T.; Bauhus, J.; Buscot, F.; Krüger, D. Network analysis reveals ecological links between N-fixing bacteria and wood-decaying fungi. PLoS ONE 2014, 9, E88141. [Google Scholar] [CrossRef]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [PubMed]
- Shaw, L.J.; Nicol, G.W.; Smith, Z.; Fear, J.; Prosser, J.I.; Baggs, E.M. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 2006, 8, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Wainø, M.; Ingvorsen, K. Production of β-xylanase and β-xylosidase by the extremely halophilic archaeon Halorhabdus utahensis. Extremophiles 2003, 7, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Rinta-Kanto, J.M.; Sinkko, H.; Rajala, T.; Al-Soud, W.A.; Sørensen, S.J.; Tamminen, M.V.; Timonen, S. Natural decay process affects the abundance and community structure of Bacteria and Archaea in Picea abies logs. FEMS Microbiol. Ecol. 2016, 92, 087. [Google Scholar] [CrossRef]
- Topp, E.; Pattey, E. Soils as sources and sinks for atmospheric methane. Can. J. Soil Sci. 1997, 77, 167–177. [Google Scholar] [CrossRef]
- Zeikus, J.G.; Henning, D.L. Methanobacterium arbophilicum sp. nov. An obligate anaerobe isolated from wetwood if living trees. Anton Leeuw. 1975, 41, 543–552. [Google Scholar] [CrossRef]
- Covey, K.R.; de Mesquita, C.P.; Oberle, B.; Maynard, D.S.; Bettigole, C.; Crowther, T.W.; Duguid, M.C.; Steven, B.; Zanne, A.E.; Lapin, M.; et al. Greenhouse trace gases in deadwood. Biogeochemistry 2016, 130, 215–226. [Google Scholar] [CrossRef]
- Mukhin, V.A.; Voronin, P.Y. A new source of methane in boreal forests. Appl. Biochem. Microbiol. 2008, 44, 297–299. [Google Scholar] [CrossRef]
- Kudo, T. Termite-microbe symbiotic system and its efficient degradation of lignocellulose. Biosci. Biotechnol. Biochem. 2009, 73, 2561–2567. [Google Scholar] [CrossRef]
- Hongoh, Y.; Ohkuma, M. Termite gut flagellates and their methanogenic and eubacterial symbionts. In Endosymbiotic Methanogenic Archaea; Hackstein, J.H.P., Ed.; Springer: Heidelberg, Germany, 2010; Volume 19, pp. 55–79. [Google Scholar]
- Foissner, W. An updated compilation of world soil ciliates (Protozoa, Ciliophora), with ecological notes, new records, and descriptions of new species. Eur. J. Protistol. 1998, 34, 195–235. [Google Scholar] [CrossRef]
- Adl, M.S.; Gupta, V.S. Protists in soil ecology and forest nutrient cycling. CA J. For. Res. 2006, 36, 1805–1817. [Google Scholar] [CrossRef]
- Rivera, F.; Lugo, A.; Ramirez, E.; Bonilla, P.; Calderon, A.; Rodriguez, S.; Ortiz, R.; Gallegos, E.; Labastida, A.; Chavez, M.P. Seasonal distribution of air-borne protozoa in Mexico City and its suburbs. Water Air Soil Poll. 1992, 61, 17–36. [Google Scholar] [CrossRef]
- Pastorelli, R.; Cucu, M.A.; Lagomarsino, A.; Paletto, A.; De Meo, I. Analysis of ciliate community diversity in decaying Pinus nigra logs. Forests 2022, 13, 642. [Google Scholar] [CrossRef]
- Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef]
- Bonkowski, M. Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 2004, 162, 617–631. [Google Scholar] [CrossRef]
- Adl, S.M.; Coleman, D.C. Dynamics of soil protozoa using a direct count method. Biol. Fertil. Soils 2005, 42, 168–171. [Google Scholar] [CrossRef]
- Bartošová, P.; Tirjaková, E. Diversity and ecology of ciliates (Alveolata: Ciliophora) living in the bark and decaying wood mass in Slovakia. Acta Protozool. 2008, 47, 173–187. [Google Scholar]
- Lara, E.; Berney, C.; Harms, H.; Chatzinotas, A. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol. Ecol. 2007, 62, 365–373. [Google Scholar] [CrossRef]
- Jousset, A.; Lara, E.; Nikolausz, M.; Harms, H.; Chatzinotas, A. Application of the denaturing gradient gel electrophoresis (DGGE) technique as an efficient diagnostic tool for ciliate communities in soil. Sci. Total Environ. 2010, 408, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Gora, E.M.; Lucas, J.M.; Yanoviak, S.P. Microbial composition and wood decomposition rates vary with microclimate from the ground to the canopy in a tropical forest. Ecosystems 2019, 22, 1206–1219. [Google Scholar] [CrossRef]
- Jaroszewicz, B.; Cholewińska, O.; Chećko, E.; Wrzosek, M. Predictors of diversity of deadwood-dwelling macrofungi in a European natural forest. For. Ecol. Manag. 2021, 490, 119123. [Google Scholar] [CrossRef]
- Bani, A.; Pioli, S.; Ventura, M.; Panzacchi, P.; Borruso, L.; Tognetti, R.; Tonon, G.; Brusetti, L. The role of microbial community in the decomposition of leaf litter and deadwood. Appl. Soil Ecol. 2018, 126, 75–84. [Google Scholar] [CrossRef]
- Baldrian, P.; Zrůstová, P.; Tláskal, V.; Davidová, A.; Merhautová, V.; Vrška, T. Fungi associated with decomposing deadwood in a natural beech-dominated forest. Fungal Ecol. 2016, 23, 109–122. [Google Scholar] [CrossRef]
- Arnstadt, T.; Hoppe, B.; Kahl, T.; Kellner, H.; Krüger, D.; Bauhus, J.; Hofrichter, M. Dynamics of fungal community composition, decomposition and resulting deadwood properties in logs of Fagus sylvatica, Picea abies and Pinus sylvestris. For. Ecol. Manag. 2016, 382, 129–142. [Google Scholar] [CrossRef]
- Mäkinen, H.; Hynynen, J.; Siitonen, J.; Sievänen, R. Predicting the decomposition of Scots pine, Norway spruce, and birch stems in Finland. Ecol. Appl. 2006, 16, 1865–1879. [Google Scholar] [CrossRef]
- Russell, M.B.; Fraver, S.; Aakala, T.; Gove, J.H.; Woodall, C.W.; D’amato, A.W.; Ducey, M.J. Quantifying carbon stores and decomposition in deadwood: A review. For. Ecol. Manag. 2015, 350, 107–128. [Google Scholar] [CrossRef]
- Rinne, K.T.; Rajala, T.; Peltoniemi, K.; Chen, J.; Smolander, A.; Mäkipää, R. Accumulation rates and sources of external nitrogen in decaying wood in a Norway spruce dominated forest. Funct. Ecol. 2017, 31, 530–541. [Google Scholar] [CrossRef]
- Palviainen, M.; Finér, L.; Laiho, R.; Shorohova, E.; Kapitsa, E.; Vanha-Majamaa, I. Carbon and nitrogen release from decomposing Scots pine, Norway spruce and silver birch stumps. For. Ecol. Manag. 2010, 259, 390–398. [Google Scholar] [CrossRef]
- Laiho, R.; Prescott, C.E. Decay and nutrient dynamics of coarse woody debris in northern coniferous forests: A synthesis. Can. J. For. Res. 2004, 34, 763–777. [Google Scholar] [CrossRef]
- Birkemoe, T.; Jacobsen, R.M.; Sverdrup-Thygeson, A.; Biedermann, P.H. Insect-fungus interactions in dead wood systems. In Saproxylic Insects; Springer: Cham, Switzerland, 2018; pp. 377–427. [Google Scholar]
- Johnston, S.R.; Boddy, L.; Weightman, A.J. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol. Ecol. 2016, 92, 179. [Google Scholar] [CrossRef] [PubMed]
- Valášková, V.; de Boer, W.; Klein Gunnewiek, P.J.A.; Pospíšek, M.; Baldrian, P. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 2009, 3, 1218–1221. [Google Scholar] [CrossRef] [PubMed]
- Frey-Klett, P.; Burlinson, P.; Deveau, A.; Barret, M.; Tarkka, M.; Sarniguet, A. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 2011, 75, 583–609. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Fenchel, T.; Field, J.G.; Gray, J.S.; Meyer-Reil, L.A.; Thingstad, F. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 1983, 10, 257–263. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.A.; Adl, S.; Bonkowski, M.; Dunthorn, M.; Ekelund, F.; Fernández, L.D.; Jousset, A.; Krashevska, V.; Singer, D.; et al. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 2018, 42, 293–323. [Google Scholar] [CrossRef] [PubMed]
- Weedon, J.T.; Cornwell, W.K.; Cornelissen, J.H.; Zanne, A.E.; Wirth, C.; Coomes, D. Global meta-analysis of wood decomposition rates: A role for trait variation among tree species? Ecol. Lett. 2009, 12, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Gough, C.M.; Vogel, C.S.; Kazanski, C.; Nagel, L.; Flower, C.E.; Curtis, P.S. Coarse woody debris and the carbon balance of a north temperate forest. For. Ecol. Manag. 2007, 244, 60–67. [Google Scholar] [CrossRef]
- Treseder, K.K.; Bent, E.; Borneman, J.; McGuire, K.L. Shifts in fungal communities during decomposition of boreal forest litter. Fungal Ecol. 2014, 10, 58–69. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Prime time for microbes. Nat. Clim. Chang. 2011, 1, 295–297. [Google Scholar] [CrossRef]
- Baldrian, P.; Šnajdr, J.; Merhautová, V.; Dobiášová, P.; Cajthaml, T.; Valášková, V. Responses of the extracellular enzyme activities in hardwood forest to soil temperature and seasonality and the potential effects of climate change. Soil Biol. Biochem. 2013, 56, 60–68. [Google Scholar] [CrossRef]
- Bebber, D.P.; Watkinson, S.C.; Boddy, L.; Darrah, P.R. Simulated nitrogen deposition affects wood decomposition by cord-forming fungi. Oecologia 2011, 167, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Waldrop, M.P.; Zak, D.R.; Sinsabaugh, R.L.; Gallo, M.; Lauber, C. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity. Ecol. Appl. 2004, 14, 1172–1177. [Google Scholar] [CrossRef]
- Covey, K.R.; Wood, S.A.; Warren, R.J.; Lee, X.; Bradford, M.A. Elevated methane concentrations in trees of an upland forest. Geophys. Res. Lett. 2012, 39, L15705. [Google Scholar] [CrossRef]
- Lenhart, K.; Bunge, M.; Ratering, S.; Neu, T.R.; Schüttmann, I.; Greule, M.; Kammann, C.; Schnell, S.; Müller, C.; Zorn, Z.; et al. Evidence for methane production by saprotrophic fungi. Nat. Commun. 2012, 3, 1046. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Bardgett, R.D.; Smith, P.; Reay, D.S. Microorganisms and climate change: Terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 2010, 8, 779–790. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).