Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC)
Abstract
1. Introduction
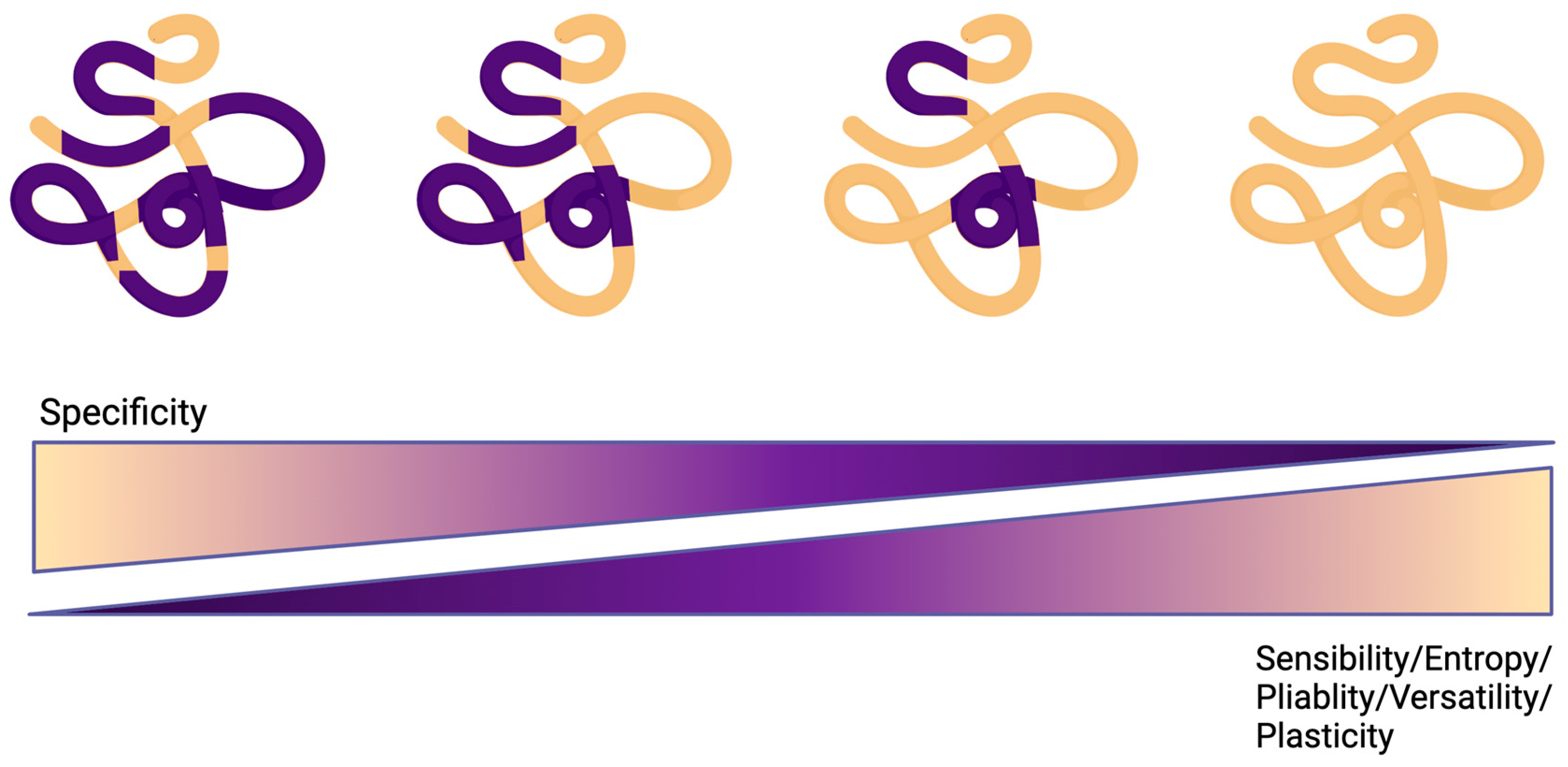
2. Intrinsically Disordered Regions (IDRs)
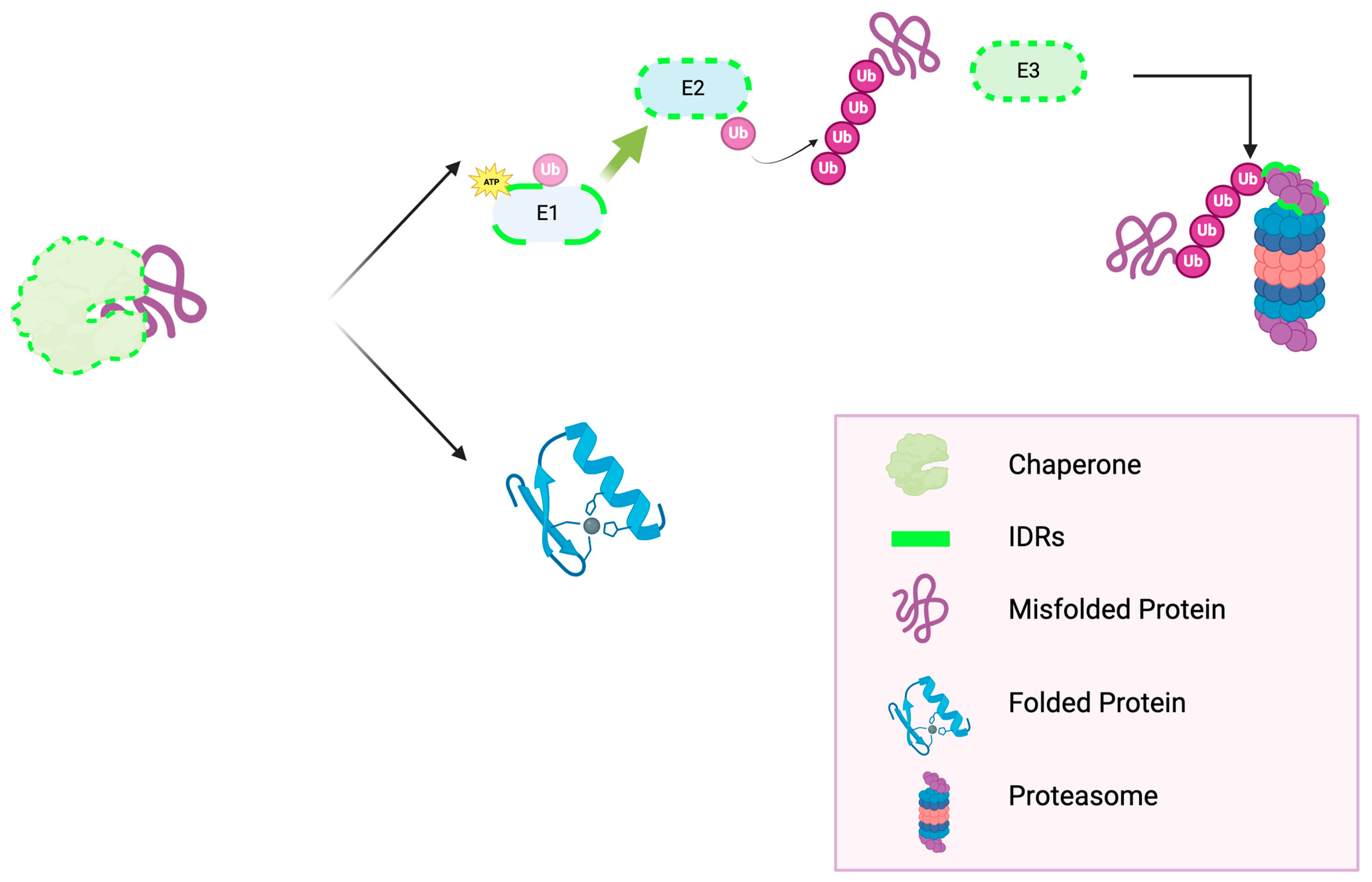
3. The Ubiquitin Proteasome System—UPS
4. The IDRs in UPS- and Chaperone-Mediated PQC
5. Outlook and Outstanding Questions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Morris, R.; Black, K.A.; Stollar, E.J. Uncovering Protein Function: From Classification to Complexes. Essays Biochem. 2022, 66, 255–285. [Google Scholar] [CrossRef] [PubMed]
- Englander, S.W.; Mayne, L. The Nature of Protein Folding Pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 15873–15880. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.N.; Khan, R.H. Protein Misfolding and Related Human Diseases: A Comprehensive Review of Toxicity, Proteins Involved, and Current Therapeutic Strategies. Int. J. Biol. Macromol. 2022, 223, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Retzlaff, M.; Roos, T.; Frydman, J. Cellular Strategies of Protein Quality Control. Cold Spring Harb. Perspect. Biol. 2011, 3, a004374. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, J.B.; Nunez-Castilla, J.; Siltberg-Liberles, J. Evolution of Intrinsic Disorder in Eukaryotic Proteins. Cell. Mol. Life Sci. 2017, 74, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.; Nagarajaram, H.A. Intrinsically Disordered Proteins: An Overview. Int. J. Mol. Sci. 2022, 23, 14050. [Google Scholar] [CrossRef]
- Vladimir, N.; Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically Disordered Proteins: A 10-Year Recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Collins, M.O.; Yu, L.; Campuzano, I.; Grant, S.G.N.; Choudhary, J.S. Phosphoproteomic Analysis of the Mouse Brain Cytosol Reveals a Predominance of Protein Phosphorylation in Regions of Intrinsic Sequence Disorder. Mol. Cell. Proteom. 2008, 7, 1331–1348. [Google Scholar] [CrossRef]
- Iakoucheva, L.M. The Importance of Intrinsic Disorder for Protein Phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Diella, F. Understanding Eukaryotic Linear Motifs and Their Role in Cell Signaling and Regulation. Front. Biosci. 2008, 13, 6580. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Van Roey, K.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of Short Linear Motifs. Mol. BioSyst. 2012, 8, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Daughdrill, G.W.; Narayanaswami, P.; Gilmore, S.H.; Belczyk, A.; Brown, C.J. Dynamic Behavior of an Intrinsically Unstructured Linker Domain Is Conserved in the Face of Negligible Amino Acid Sequence Conservation. J. Mol. Evol. 2007, 65, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.C.; Leclerc, N.; Flanagan, L.A.; Lu, M.; Janmey, P.A.; Kosik, K.S. Microtubule-Associated Protein 2c Reorganizes Both Microtubules and Microfilaments into Distinct Cytological Structures in an Actin-Binding Protein-280–Deficient Melanoma Cell Line. J. Cell Biol. 1997, 136, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.; Tidow, H.; Rutherford, T.J.; Markwick, P.; Jensen, M.R.; Mylonas, E.; Svergun, D.I.; Blackledge, M.; Fersht, A.R. Structure of Tumor Suppressor P53 and Its Intrinsically Disordered N-Terminal Transactivation Domain. Proc. Natl. Acad. Sci. USA 2008, 105, 5762–5767. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, F.; Qiu, W.; Liu, R. P130Cas Substrate Domain Is Intrinsically Disordered as Characterized by Single-Molecule Force Measurements. Biophys. Chem. 2013, 180–181, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.H.; Stewart, D.B.; Laurents, D.V.; Nelson, W.J.; Weis, W.I. The Cadherin Cytoplasmic Domain Is Unstructured in the Absence of β-Catenin. J. Biol. Chem. 2001, 276, 12301–12309. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Kutateladze, T.G. Characterization of Functional Disordered Regions within Chromatin-Associated Proteins. iScience 2021, 24, 102070. [Google Scholar] [CrossRef]
- Bardwell, J.C.A.; Jakob, U. Conditional Disorder in Chaperone Action. Trends Biochem. Sci. 2012, 37, 517–525. [Google Scholar] [CrossRef]
- Levengood, J.D.; Tolbert, B.S. Idiosyncrasies of hnRNP A1-RNA Recognition: Can Binding Mode Influence Function. Semin. Cell Dev. Biol. 2019, 86, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Sudnitsyna, M.V.; Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. The Role of Intrinsically Disordered Regions in the Structure and Functioning of Small Heat Shock Proteins. Curr. Protein Pept. Sci. 2012, 13, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Matthews, M.; Pang, X.; Zhou, H. The Dock-and-coalesce Mechanism for the Association of a WASP Disordered Region with the Cdc42 GTP Ase. FEBS J. 2017, 284, 3381–3391. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Gadhave, K.; Kumar, P.; Giri, R. Transactivation Domain of Adenovirus Early Region 1A (E1A): Investigating Folding Dynamics and Aggregation. Curr. Res. Struct. Biol. 2022, 4, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Dreier, J.E.; Prestel, A.; Martins, J.M.; Brøndum, S.S.; Nielsen, O.; Garbers, A.E.; Suga, H.; Boomsma, W.; Rogers, J.M.; Hartmann-Petersen, R.; et al. A Context-Dependent and Disordered Ubiquitin-Binding Motif. Cell. Mol. Life Sci. 2022, 79, 484. [Google Scholar] [CrossRef]
- Uversky, A.V.; Xue, B.; Peng, Z.; Kurgan, L.; Uversky, V.N. On the Intrinsic Disorder Status of the Major Players in Programmed Cell Death Pathways. F1000Res 2013, 2, 190. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.J.; Nogales, E. The Structures of Eukaryotic Transcription Pre-Initiation Complexes and Their Functional Implications. In Macromolecular Protein Complexes II: Structure and Function; Harris, J.R., Marles-Wright, J., Eds.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2019; Volume 93, pp. 143–192. ISBN 978-3-030-28150-2. [Google Scholar]
- Peng, Z.; Oldfield, C.J.; Xue, B.; Mizianty, M.J.; Dunker, A.K.; Kurgan, L.; Uversky, V.N. A Creature with a Hundred Waggly Tails: Intrinsically Disordered Proteins in the Ribosome. Cell. Mol. Life Sci. 2014, 71, 1477–1504. [Google Scholar] [CrossRef]
- Choi, K.; Han, M.; Kim, S.J. A Systematic Review of Chromogranin A (CgA) and Its Biomedical Applications, Unveiling Its Structure-Related Functions. J. Korean Phys. Soc. 2021, 78, 427–441. [Google Scholar] [CrossRef]
- Redwan, E.; Xue, B.; Almehdar, H.; Uversky, V. Disorder in Milk Proteins: Caseins, Intrinsically Disordered Colloids. Curr. Protein Pept. Sci. 2015, 16, 228–242. [Google Scholar] [CrossRef]
- Grzybowska, E. Calcium-Binding Proteins with Disordered Structure and Their Role in Secretion, Storage, and Cellular Signaling. Biomolecules 2018, 8, 42. [Google Scholar] [CrossRef]
- Schwartz, A.L.; Ciechanover, A. The Ubiquitin-Proteasome Pathway and Pathogenesis of Human Diseases. Annu. Rev. Med. 1999, 50, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Pergolizzi, B.; Bozzaro, S.; Bracco, E. Dictyostelium as Model for Studying Ubiquitination and Deubiquitination. Int. J. Dev. Biol. 2019, 63, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, R.B. The Ubiquitin System: From Cell Signalling to Disease Biology and New Therapeutic Opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Santamarta, M.; Bouvier, C.; Rodriguez, M.S.; Xolalpa, W. Ubiquitin-Chains Dynamics and Its Role Regulating Crucial Cellular Processes. Semin. Cell Dev. Biol. 2022, 132, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, G.G.; Hipp, M.S.; Hartl, F.U. Functional Modules of the Proteostasis Network. Cold Spring Harb. Perspect. Biol. 2020, 12, a033951. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Schulman, B.A. An Expanded Lexicon for the Ubiquitin Code. Nat. Rev. Mol. Cell Biol. 2023, 24, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Kliza, K.; Husnjak, K. Resolving the Complexity of Ubiquitin Networks. Front. Mol. Biosci. 2020, 7, 21. [Google Scholar] [CrossRef]
- Hrdinka, M.; Gyrd-Hansen, M. The Met1-Linked Ubiquitin Machinery: Emerging Themes of (De)Regulation. Mol. Cell 2017, 68, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 Ubiquitin Ligases: Styles, Structures and Functions. Mol. Biomed. 2021, 2, 23. [Google Scholar] [CrossRef]
- Iconomou, M.; Saunders, D.N. Systematic Approaches to Identify E3 Ligase Substrates. Biochem. J. 2016, 473, 4083–4101. [Google Scholar] [CrossRef]
- Cowan, A.D.; Ciulli, A. Driving E3 Ligase Substrate Specificity for Targeted Protein Degradation: Lessons from Nature and the Laboratory. Annu. Rev. Biochem. 2022, 91, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Fushman, D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004, 8, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Urbé, S.; Komander, D. Breaking the Chains: Deubiquitylating Enzyme Specificity Begets Function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Samant, R.S.; Livingston, C.M.; Sontag, E.M.; Frydman, J. Distinct Proteostasis Circuits Cooperate in Nuclear and Cytoplasmic Protein Quality Control. Nature 2018, 563, 407–411. [Google Scholar] [CrossRef]
- Yau, R.G.; Doerner, K.; Castellanos, E.R.; Haakonsen, D.L.; Werner, A.; Wang, N.; Yang, X.W.; Martinez-Martin, N.; Matsumoto, M.L.; Dixit, V.M.; et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933.e20. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, J.; Das, K.P. Molecular Chaperone-like Properties of an Unfolded Protein, As-Casein. J. Biol. Chem. 1999, 274, 15505–15509. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.D.; Paik, S.R.; Yang, C.-H. Structural and Functional Implications of C-Terminal Regions of α-Synuclein. Biochemistry 2002, 41, 13782–13790. [Google Scholar] [CrossRef]
- Jaya, N.; Garcia, V.; Vierling, E. Substrate Binding Site Flexibility of the Small Heat Shock Protein Molecular Chaperones. Proc. Natl. Acad. Sci. USA 2009, 106, 15604–15609. [Google Scholar] [CrossRef]
- Haslbeck, M.; Ignatiou, A.; Saibil, H.; Helmich, S.; Frenzl, E.; Stromer, T.; Buchner, J. A Domain in the N-Terminal Part of Hsp26 Is Essential for Chaperone Function and Oligomerization. J. Mol. Biol. 2004, 343, 445–455. [Google Scholar] [CrossRef]
- Stromer, T.; Fischer, E.; Richter, K.; Haslbeck, M.; Buchner, J. Analysis of the Regulation of the Molecular Chaperone Hsp26 by Temperature-Induced Dissociation. J. Biol. Chem. 2004, 279, 11222–11228. [Google Scholar] [CrossRef]
- van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; Sánchez de Groot, N.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.P.; Barbour, K.W.; Berger, F.G. Cooperation between an Intrinsically Disordered Region and a Helical Segment Is Required for Ubiquitin-Independent Degradation by the Proteasome. J. Biol. Chem. 2011, 286, 36559–36567. [Google Scholar] [CrossRef]
- Aufderheide, A.; Unverdorben, P.; Baumeister, W.; Förster, F. Structural Disorder and Its Role in Proteasomal Degradation. FEBS Lett. 2015, 589, 2552–2560. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Sussman, J.L.; Silman, I.; Shaul, Y. Context-dependent Resistance to Proteolysis of Intrinsically Disordered Proteins. Protein Sci. 2011, 20, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, P.; Pancsa, R.; Guharoy, M.; Tompa, P. Functional Diversity and Structural Disorder in the Human Ubiquitination Pathway. PLoS ONE 2013, 8, e65443. [Google Scholar] [CrossRef]
- Gadhave, K.; Kumar, P.; Kapuganti, S.; Uversky, V.; Giri, R. Unstructured Biology of Proteins from Ubiquitin-Proteasome System: Roles in Cancer and Neurodegenerative Diseases. Biomolecules 2020, 10, 796. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.C.; Fredrickson, E.K.; Oeser, M.L.; Garrett-Engele, C.M.; Locke, M.N.; Richardson, L.A.; Nelson, Z.W.; Hetrick, E.D.; Milac, T.I.; Gottschling, D.E.; et al. Disorder Targets Misorder in Nuclear Quality Control Degradation: A Disordered Ubiquitin Ligase Directly Recognizes Its Misfolded Substrates. Mol. Cell 2011, 41, 93–106. [Google Scholar] [CrossRef]
- Blount, J.R.; Johnson, S.L.; Todi, S.V. Unanchored Ubiquitin Chains, Revisited. Front. Cell Dev. Biol. 2020, 8, 582361. [Google Scholar] [CrossRef]
- Husnjak, K.; Dikic, I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Lambrughi, M.; Maiani, E.; Aykac Fas, B.; Shaw, G.S.; Kragelund, B.B.; Lindorff-Larsen, K.; Teilum, K.; Invernizzi, G.; Papaleo, E. Ubiquitin Interacting Motifs: Duality Between Structured and Disordered Motifs. Front. Mol. Biosci. 2021, 8, 676235. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Falquet, L. A Ubiquitin-Interacting Motif Conserved in Components of the Proteasomal and Lysosomal Protein Degradation Systems. Trends Biochem. Sci. 2001, 26, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A. From UBA to UBX: New Words in the Ubiquitin Vocabulary. Trends Cell Biol. 2002, 12, 216–221. [Google Scholar] [CrossRef]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781.e24. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Babu, M.M.; Blackledge, M.; Bridge, A.; Capella-Gutierrez, S.; Dosztanyi, Z.; Drysdale, R.; Edwards, R.J.; Elofsson, A.; Felli, I.C.; et al. An intrinsically disordered proteins community for ELIXIR. F1000Res 2019, 8, ELIXIR-1753. [Google Scholar] [CrossRef]
- Lotthammer, J.M.; Ginell, G.M.; Griffith, D.; Emenecker, R.J.; Holehouse, A.S. Direct prediction of intrinsically disordered protein conformational properties from sequence. Nat. Methods 2024, 21, 465–476. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magnati, S.; Bracco, E. Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC). Biophysica 2024, 4, 158-167. https://doi.org/10.3390/biophysica4020011
Magnati S, Bracco E. Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC). Biophysica. 2024; 4(2):158-167. https://doi.org/10.3390/biophysica4020011
Chicago/Turabian StyleMagnati, Stefano, and Enrico Bracco. 2024. "Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC)" Biophysica 4, no. 2: 158-167. https://doi.org/10.3390/biophysica4020011
APA StyleMagnati, S., & Bracco, E. (2024). Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC). Biophysica, 4(2), 158-167. https://doi.org/10.3390/biophysica4020011





